Post Acid Treatment on Pressurized Liquid Extracts of Sorghum (Sorghum bicolor L. Moench) Grain and Plant Material Improves Quantification and Identification of 3-Deoxyanthocyanidins
Abstract
1. Introduction
2. Materials and Methods
2.1. Raw Material and Chemicals
2.2. Pressurized Liquid Extraction (PLE)
2.2.1. Global Yield of Soluble Extract
2.2.2. Acidification of PLE Extracts
2.3. Characterization of Extracts
2.3.1. Color Analysis
2.3.2. Total Phenolic Content Assay
2.3.3. DPPH Antioxidant Capacity Assay
2.3.4. High-Performance Liquid Chromatography
2.3.5. Thin-Layer Chromatography (TLC)
2.3.6. LC-MS Identification of 5-Methoxy Luteolinidin
2.4. Statistical Analysis
3. Results and Discussion
3.1. Selection of Temperature for Obtaining Phenolic Compounds via PLE
3.2. Acid Hydrolysis of PLE Extracts
3.2.1. Color Analysis
3.2.2. Effect of Acidification on Total Phenolic Content
3.2.3. Effect of Acidification on DPPH Antioxidant Capacity
3.2.4. Phenolic Profile Detected by HPLC
Flavonoids: 3-Deoxyanthocyanidins
Flavonoids: Other Classes
Non-Flavonoids
3.2.5. Thin-Layer Chromatography (TLC)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Disclaimer
Abbreviations
| ASE | Accelerated solvent extraction |
| HCl | Hydrochloric acid |
| HPLC | High-performance liquid chromatography |
| LC-MS | Liquid chromatography–mass spectrometry |
| PLE | Pressurized liquid extraction |
| TLC | Thin-layer chromatography |
| TPC | Total phenolic content |
| 3-DA | 3-deoxyanthocyanidins |
References
- Gleave, G.L.; Roethe, N.J.; Kemp, D.W.; Richter, B.E.; Ezzel, J.L. Automated Accelerated Solvent Extraction Apparatus and Method. U.S. Patent 5,785,856, 28 July 1998. [Google Scholar]
- Cardenas-Toro, F.P.; Forster-Carneiro, T.; Rostagno, M.A.; Petenate, A.J.; Maugeri Filho, F.; Meireles, M.A.A. Integrated supercritical fluid extraction and subcritical water hydrolysis for the recovery of bioactive compounds from pressed palm fiber. J. Supercrit. Fluids 2014, 93, 42–48. [Google Scholar] [CrossRef]
- Lachoz-Perez, D.; Brown, A.B.; Mudhoo, A.; Martinez, J.; Timko, M.T.; Rostagno, M.A.; Forster-Carneiro, T. Applications of subcritical and supercritical water conditions for extraction, hydrolysis, gasification, and carbonization of biomass: A critical review. Biofuel Res. J. 2017, 14, 611–626. [Google Scholar] [CrossRef]
- Luo, X.; Cui, J.; Zhang, H.; Duan, Y. Subcritical water extraction of polyphenolic compounds from sorghum (Sorghum bicolor L.) bran and their biological activities. Food Chem. 2018, 262, 14–20. [Google Scholar] [CrossRef]
- Dykes, L.; Seitz, L.M.; Rooney, W.L.; Rooney, L.W. Flavonoid composition of red sorghum genotypes. Food Chem. 2009, 116, 313–317. [Google Scholar] [CrossRef]
- Cox, S.; Noronha, L.; Herald, T.; Bean, S.; Lee, S.-H.; Perumal, R.; Wang, W.; Smolensky, D. Evaluation of ethanol-based extraction conditions of sorghum bran bioactive compounds with downstream anti-proliferative properties in human cancer cells. Heliyon 2019, 5, e01589. [Google Scholar] [CrossRef]
- Hou, F.; Su, D.; Xu, J.; Gong, Y.; Zhang, R.; Wei, Z.; Chi, J.; Zhang, M. Enhanced Extraction of Phenolics and Antioxidant Capacity from Sorghum (Sorghum bicolor L. Moench) Shell Using Ultrasonic-Assisted Ethanol–Water Binary Solvent. J. Food Process. Preserv. 2016, 40, 1171–1179. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, Y.; Liu, Z.; Wang, J. Extraction, Identification and Antioxidant Activity of 3-Deoxyanthocyanidins from Sorghum bicolor L. Moench Cultivated in China. Antioxidants 2023, 12, 468. [Google Scholar] [CrossRef] [PubMed]
- Herrman, D.A.; Brantsen, J.F.; Ravisankar, S.; Lee, K.-M.; Awika, J.M. Stability of 3-deoxyanthocyanin pigment structure relative to anthocyanins from grains under microwave assisted extraction. Food Chem. 2020, 333, 127494. [Google Scholar] [CrossRef]
- Barros, F.; Dykes, L.; Awika, J.M.; Rooney, L.W. Accelerated solvent extraction of phenolic compounds from sorghum brans. J. Cereal Sci. 2013, 58, 305–312. [Google Scholar] [CrossRef]
- Hefni, M.E.; Amann, L.S.; Witthöft, C.M. A HPLC-UV Method for the Quantification of Phenolic Acids in Cereals. Food Anal. Methods 2019, 12, 2802–2812. [Google Scholar] [CrossRef]
- İlbay, Z.; Şahin, S.; Büyükkabasakal, K. A novel approach for olive leaf extraction through ultrasound technology: Response surface methodology versus artificial neural networks. Korean J. Chem. Eng. 2014, 31, 1661–1667. [Google Scholar] [CrossRef]
- Chaves, J.O.; Souza, M.C.; Silva, L.C.; Lachoz-Perez, D.; Torres-Mayanga, P.C.; Machado, A.P.F.; Forster-Carneiro, T.; Vazquez-Espinoza, M.; Gonzalez-de-Peredo, A.V.; Barbero, G.F.; et al. Extraction of Flavonoids From Natural Sources Using Modern Techniques. Front. Chem. 2020, 8, 507887. [Google Scholar] [CrossRef] [PubMed]
- Wizi, J.; Wang, L.; Hou, X.; Tao, Y.; Ma, B.; Yang, Y. Ultrasound-microwave assisted extraction of natural colorants from sorghum husk with different solvents. Ind. Crops Prod. 2018, 120, 203–213. [Google Scholar] [CrossRef]
- Paunović, D.Đ.; Mitić, S.S.; Kostić, D.A.; Mitić, M.N.; Stojanović, B.T.; Pavlović, J.L. Kinetics and Thermodynamics of the Solid-Liquid Extraction Process of Total Polyphenols From Barley. Adv. Technol. 2015, 3, 58–63. [Google Scholar] [CrossRef]
- Ju, Z.Y.; Howard, L.R. Effects of Solvent and Temperature on Pressurized Liquid Extraction of Anthocyanins and Total Phenolics from Dried Red Grape Skin. J. Agric. Food Chem. 2003, 51, 5207–5213. [Google Scholar] [CrossRef]
- Thermo Fisher Scientific. Dionex ASE 350; Thermo Fisher Scientific: Waltham, MA, USA, 2011. [Google Scholar]
- Herald, T.J.; Gadgil, P.; Tilley, M. High-throughput micro plate assays for screening flavonoid content and DPPH-scavenging activity in sorghum bran and flour. J. Sci. Food Agric. 2012, 92, 2326–2331. [Google Scholar] [CrossRef]
- Plank, D.W.; Szpylka, J.; Sapirstein, H.; Woolard, D.; Zapf, C.M.; Lee, V.; Chen, C.-Y.O.; Liu, R.H.; Tsao, R.; Dusterloch, A.; et al. Determination of Antioxidant Activity in Foods and Beverages by Reaction with 2,2’-Diphenyl-1-Picrylhydrazyl (DPPH): Collaborative Study First Action 2012.04. J. AOAC Int. 2012, 95, 1562–1569. [Google Scholar] [CrossRef] [PubMed]
- Irakli, M.N.; Samanidou, V.F.; Biliaderis, C.G.; Papadoyannis, I.N. Simultaneous determination of phenolic acids and flavonoids in rice using solid-phase extraction and RP-HPLC with photodiode array detection. J. Sep. Sci. 2012, 35, 1603–1611. [Google Scholar] [CrossRef]
- Lee, H.-S.; Santana, Á.L.; Peterson, J.; Yucel, U.; Perumal, R.; De Leon, J.; Lee, S.-H.; Smolensky, D. Anti-Adipogenic Activity of High-Phenolic Sorghum Brans in Pre-Adipocytes. Nutrients 2022, 14, 1493. [Google Scholar] [CrossRef]
- Speranza, S.; Knechtl, R.; Witlaczil, R.; Schönlechner, R. Reversed-Phase HPLC Characterization and Quantification and Antioxidant Capacity of the Phenolic Acids and Flavonoids Extracted From Eight Varieties of Sorghum Grown in Austria. Front. Plant Sci. 2021, 5, 769151. [Google Scholar] [CrossRef] [PubMed]
- Irakli, M.N.; Samanidou, V.F.; Biliaderis, C.G.; Papadoyannis, I.N. Development and validation of an HPLC-method for determination of free and bound phenolic acids in cereals after solid-phase extraction. Food Chem. 2012, 134, 1624–1632. [Google Scholar] [CrossRef]
- Xiong, Y.; Zhang, P.; Warner, R.D.; Shen, S.; Johnson, S. HPLC-DAD-ESI-QTOF-MS/MS qualitative analysis data and HPLC-DAD quantification data of phenolic compounds of grains from five Australian sorghum genotypes. Data Br. 2020, 33, 106584. [Google Scholar] [CrossRef] [PubMed]
- Bae, H.; Jayaprakasha, G.K.; Jifon, J.; Patil, B.S. Extraction efficiency and validation of an HPLC method for flavonoid analysis in peppers. Food Chem. 2012, 130, 751–758. [Google Scholar] [CrossRef]
- Shapla, U.M.; Solayman, M.; Alam, N.; Khalil, M.; Gan, S.H. 5-Hydroxymethylfurfural (HMF) levels in honey and other food products: Effects on bees and human health. Chem. Cent. J. 2018, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Brantsen, J.F.; Hermann, D.; Ravisankar, S.; Awika, J.M. Effect of tannins on microwave-assisted extractability and color properties of sorghum 3-deoxyanthocyanins. Food Res. Int. 2021, 148, 110612. [Google Scholar] [CrossRef] [PubMed]
- Kayodé, A.P.P.; Bara, C.A.; Dalodé-Vieira, G.; Linnemann, A.R.; Mout, M.J.R. Extraction of antioxidant pigments from dye sorghum leaf sheaths. LWT Food Sci. Technol. 2012, 46, 49–55. [Google Scholar] [CrossRef]
- Przybylska-Balcerek, A.; Frankowski, J.; Stuper-Szablewska, K. The influence of weather conditions on bioactive compound content in sorghum grain. Eur. Food Res. Technol. 2020, 246, 13–22. [Google Scholar] [CrossRef]
- Di Meo, F.; Lemaur, V.; Cornil, J.; Lazzaroni, R.; Duroux, J.-L.; Olivier, Y.; Trouillas, P. Free Radical Scavenging by Natural Polyphenols: Atom versus Electron Transfer. J. Phys. Chem. A 2013, 117, 2082–2092. [Google Scholar] [CrossRef]
- Zhao, J.; Ou, S.; Ding, S.; Wang, Y.; Wang, Y. Effect of activated charcoal treatment of alkaline hydrolysates from sugarcane bagasse on purification of p-coumaric acid. Chem. Eng. Res. Des. 2011, 89, 2176–2181. [Google Scholar] [CrossRef]
- Nuutila, A.M.; Kammiovirta, K.; Oksman-Caldentey, K.-M. Comparison of methods for the hydrolysis of flavonoids and phenolic acids from onion and spinach for HPLC analysis. Food Chem. 2002, 76, 519–525. [Google Scholar] [CrossRef]
- Wu, G.; Johnson, S.K.; Fang, Z. Sorghum non-extractable polyphenols: Chemistry, extraction and bioactivity. In Non-Extractable Polyphenols and Carotenoids: Importance in Human Nutrition and Health; Saura-Calixto, F., Pérez-Jiménez, J., Eds.; Royal Society of Chemistry: London, UK, 2018; pp. 326–344. [Google Scholar]
- Ofosu, F.K.; Elahi, F.; Daliri, E.B.M.; Yeon, S.-J.; Ham, H.J.; Kim, J.-H.; Han, S.-I.; Oh, D.-W. Flavonoids in Decorticated Sorghum Grains Exert Antioxidant, Antidiabetic and Antiobesity Activities. Molecules 2020, 25, 2854. [Google Scholar] [CrossRef] [PubMed]
- Pontieri, P.; Pepe, G.; Campiglia, P.; Mercial, F.; Basilicata, M.G.; Smolensky, D.; Calcagnile, M.; Troisi, J.; Romano, R.; Del Giudice, F.; et al. Comparison of Content in Phenolic Compounds and Antioxidant Capacity in Grains of White, Red, and Black Sorghum Varieties Grown in the Mediterranean Area. ACS Food Sci. Technol. 2021, 1, 1109–1119. [Google Scholar] [CrossRef]
- Mizuno, H.; Yazawa, T.; Kasuga, S.; Sawada, Y.; Ogata, J.; Ando, Y.; Kanamori, H.; Yonemaru, J.-I.; Wu, J.; Hirai, M.Y.; et al. Expression level of a flavonoid 3’-hydroxylase gene determines pathogen-induced color variation in sorghum. BMC Res. Notes 2014, 7, 761. [Google Scholar] [CrossRef]
- Petti, C.; Kushwaha, R.; Tateno, K.; Harman-Ware, A.E.; Crocker, M.; Awika, J.; DeBolt, S. Mutagenesis Breeding for Increased 3-Deoxyanthocyanidin Accumulation in Leaves of Sorghum bicolor (L.) Moench: A Source of Natural Food Pigment. J. Agric. Food Chem. 2014, 62, 1227–1232. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Zhang, P.; Warner, R.D.; Fang, Z. In vitro and cellular antioxidant activities of 3-deoxyanthocyanidin colourants. Food Biosci. 2021, 42, 101171. [Google Scholar] [CrossRef]
- Oboh, G.; Adewuni, T.M.; Ademosun, A.O.; Olasehinde, T.A. Sorghum stem extract modulates Na+/K+-ATPase, ecto-5′-nucleotidase, and acetylcholinesterase activities. Comp. Clin. Path. 2016, 25, 749–756. [Google Scholar] [CrossRef]
- Parkes, R.; McGee, D.; McDonnell, A.; Gillespie, E.; Touzet, N. Rapid screening of phenolic compounds in extracts of photosynthetic organisms separated using a C18 monolithic column based HPLC-UV method. J. Chromatogr. B 2022, 1213, 123521. [Google Scholar] [CrossRef] [PubMed]


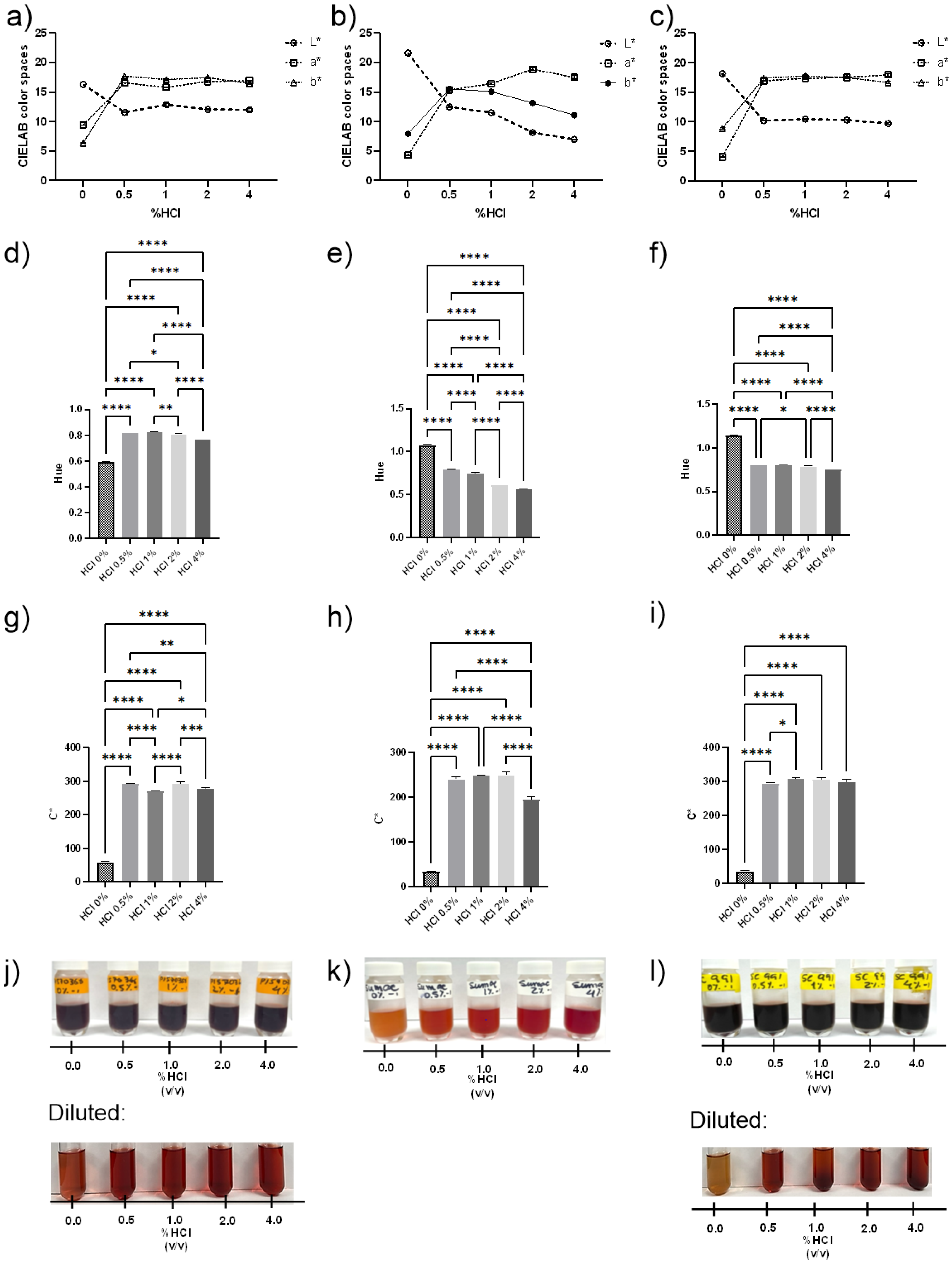



| (A) | |||||||
| Subclass | Compound | λ nm | % HCl | ||||
| 0% | 0.50% | 1% | 2% | 4% | |||
| 3-deoxyanthocyanidin | Luteolinidin | 510 | 60.63 ± 7.88 b | 101.56 ± 14.09 ab | 107.89 ± 20.65 ab | 102.71 ± 30.73 ab | 132.30 ± 21.94 a |
| Apigenidin | 7.62 ± 0.30 b | 14.20 ± 1.77 a | 14.56 ± 0.22 a | 15.19 ± 2.05 a | 17.14 ± 1.85 a | ||
| Benzoic acid | Protocatechuic acid | 280 | 15.96 ± 0.53 b | 26.36 ± 8.71 a | 17.27 ± 3.93 b | 21.19 ± 2.70 ab | 15.63 ± 1.97 b |
| Flavanol | Catechin | 280 | 67.48 ± 2.32 a | 7.50 ± 2.53 b | 4.04 ± 0.91 b | 12.16 ± 4.07 b | 3.88 ± 2.79 b |
| Flavanonol | Taxifolin | 280 | 261.32 ± 11.06 a | 61.22 ± 15.93 b | 76.39 ± 88.72 b | 54.61 ± 20.57 b | 88.14 ± 101.36 b |
| Flavanone | Eriodictyol | 280 | 10.71 ± 0.47 a | 20.92 ± 13.21 a | 58.89 ± 49.82 a | 78.46 ± 35.40 a | 86.02 ± 99.28 a |
| Naringenin | 0 #b | 12.20 ± 5.18 a | 20.12 ± 13.88 a | 24.12 ± 6.17 a | 27.67 ± 27.26 a | ||
| Flavonol | Quercetin | 280 | 26.09 ± 0.56 a | 13.72 ± 2.28 a | 12.35 ± 4.63 a | 11.46 ± 5.19 a | 11.45 ± 14.01 a |
| Cinnamic acid | Chlorogenic acid | 320 | 14.09 ± 0.24 a | 0 #b | 0 #b | 0 #b | 0 #b |
| Caffeic acid | 9.25 ± 0.13 a | 6.16 ± 0.37 b | 6.09 ± 1.31 b | 5.38 ± 0.09 b | 0 #c | ||
| p-Coumaric acid | 2.53 ± 0.49 a | 2.06 ± 1.59 a | 4.23 ± 3.25 a | 3.53 ± 1.13 a | 0.78 ± 0.84 a | ||
| Ferulic acid | 6.28 ± 0.35 a | 6.59 ± 0.52 a | 6.94 ± 3.59 a | 5.87 ± 0.81 a | 5.10 ± 0.09 a | ||
| Flavone | Apigenin | 320 | 5.18 ± 0.21 a | 5.41 ± 0.10 a | 5.42 ± 0.18 a | 5.55 ± 0.20 a | 7.74 ± 4.19 a |
| Luteolin | 27.87 ± 1.26 b | 43.24 ± 3.00 ab | 40.12 ± 6.25 ab | 41.03 ± 4.73 ab | 46.83 ± 10.19 a | ||
| Anthocyanidin | Cyanidin | 510 | 0 #b | 35.95 ± 0.36 ab | 43.51 ± 13.25 a | 41.32 ± 4.79 ab | 60.95 ± 30.81 a |
| (B) | |||||||
| Subclass | Compound | λ nm | % HCl | ||||
| 0% | 0.50% | 1% | 2% | 4% | |||
| 3-deoxyanthocyanidin | Luteolinidin | 510 | 613.87 ± 83.52 b | 1067.40 ± 117.92 a | 1113.65 ± 96.63 a | 1197.85 ± 82.93 a | 1156.78 ± 221.78 a |
| Apigenidin | 85.77 ± 11.17 b | 161.81 ± 33.35 ab | 169.41 ± 22.10 a | 142.17 ± 46.67 ab | 167.34 ± 20.75 a | ||
| 7-methoxyapigenidin | 129.79 ± 4.70 b | 672.37 ± 66.07 a | 744.85 ± 30.73 a | 835.84 ± 110.99 a | 726.24 ± 75.45 a | ||
| Benzoic acid | Protocatechuic acid | 280 | 7.16 ± 0.38 b | 18.84 ± 1.40 a | 18.69 ± 1.05 a | 18.22 ± 4.58 a | 16.39 ± 5.25 a |
| 4-hydroxybenzoic acid | 13.05 ± 3.16 ab | 21.14 ± 3.69 a | 18.59 ± 2.36 a | 17.04 ± 12.36 a | 0 #b | ||
| Flavanol | Catechin | 280 | 41.62 ± 5.97 b | 129.34 ± 21.54 ab | 81.94 ± 23.07 b | 121.70 ± 51.67 b | 308.40 ± 141.11 a |
| Flavanone | Eriodictyol | 280 | 12.11 ± 1.42 b | 75.40 ± 16.50 a | 41.43 ± 15.80 ab | 58.82 ± 27.78 ab | 81.11 ± 10.45 a |
| Naringenin | 0 #c | 21.30 ± 4.64 ab | 17.23 ± 3.40 b | 24.79 ± 7.47 ab | 32.07 ± 2.91 a | ||
| Flavone | Luteolin | 280 | 32.22 ± 6.50 b | 53.17 ± 6.29 ab | 49.19 ± 3.91 ab | 56.70 ± 10 a | 48.16 ± 4.28 ab |
| Apigenin | 5.08 ± 0.23 a | 5.87 ± 0.67 a | 5.70 ± 0.07 a | 5.92 ± 0.18 a | 5.82 ± 0.64 a | ||
| Cinnamic acid | Chlorogenic acid | 320 | 30.05 ± 5.26 a | 11.42 ± 1.08 b | 11.15 ± 0.95 b | 12.13 ± 0.37 b | 12.61 ± 1.28 b |
| p-Coumaric acid | 2.19 ± 1.35 ab | 2.93 ± 0.81 a | 0 #b | 0.94 ± 1.92 ab | 0 #b | ||
| Ferulic acid | 7.19 ± 0.99 a | 12.23 ± 1.93 a | 8.48 ± 1.34 a | 8.60 ± 8.89 a | 7.26 ± 1.09 a | ||
| Anthocyanidin | Cyanidin | 510 | 0 #b | 67.10 ± 17.20 ab | 92.53 ± 11.72 ab | 139.79 ± 67.41 ab | 165.35 ± 108.58 a |
| (C) | |||||||
| Subclass | Compound | λ nm | % HCl | ||||
| 0% | 0.50% | 1% | 2% | 4% | |||
| 3-deoxyanthocyanidin | Luteolinidin | 510 | 295.43 ± 20.10 a | 1499.37 ± 508.47 a | 1486.01 ± 396.13 a | 1514.00 ± 710.02 a | 1595.43 ± 692.57 a |
| Apigenidin | 40.29 ± 7.70 a | NQ | NQ | NQ | NQ | ||
| 7-methoxyapigenidin | 123.13 ± 17.81 b | 1170.42 ± 313.44 a | 1515.29 ± 499.65 a | 1517.42 ± 358.27 a | 1640.87 ± 284.82 a | ||
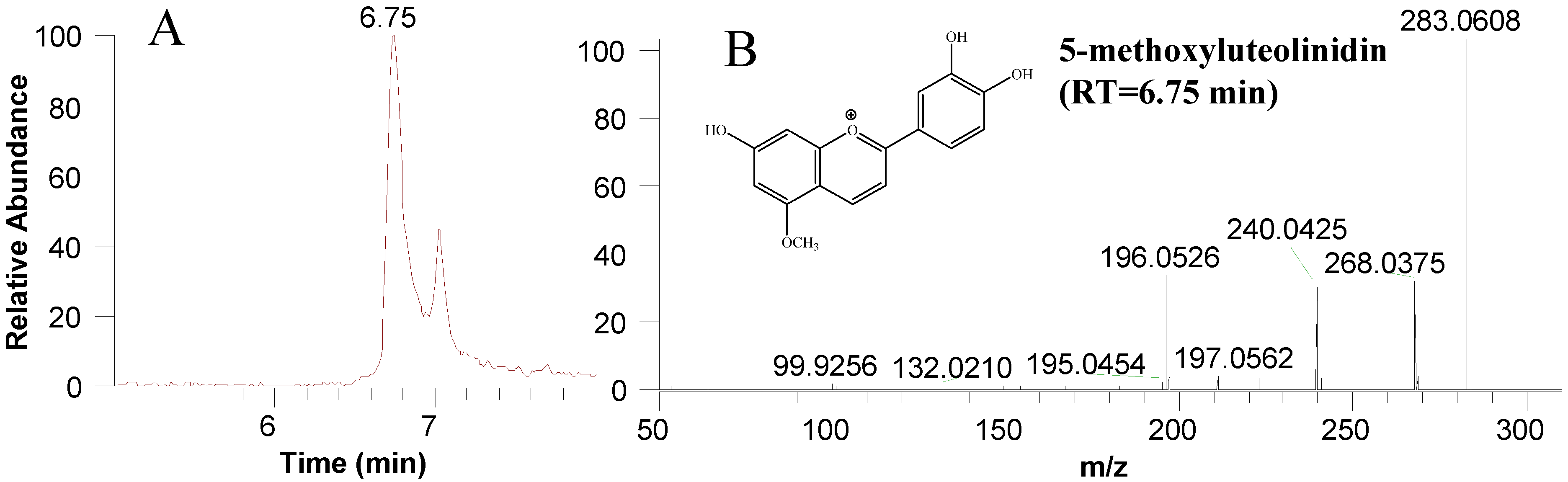
| 5-methoxyluteolinidin | 0 #b | 1733.41 ± 110.50 a | 1861.12 ± 95.90 a | 1881.75 ± 355.67 a | 1765.45 ± 52.60 a | ||
| Benzoic acid | Protocatechuic acid | 280 | 15.75 ± 0.09 a | 15.85 ± 0.79 a | 16.52 ± 1.18 a | 17.37 ± 2.25 a | 18.03 ± 2.33 a |
| 4-hydroxybenzoic acid | 56.77 ± 0.93 | 84.81 ± 57.03 | 90.72 ± 47.72 | 35.72 ± 47.18 | 12.45 ± 6.57 | ||
| Flavanone | Naringenin | 280 | 9.74 ± 0.40 a | 0 #b | 0 #b | 0 #b | 0 #b |
| Flavone | Luteolin | 320 | 235.44 ± 8.64 a | 362.26 ± 22.19 a | 301.59 ± 71.29 a | 265.14 ± 101.79 a | 260.38 ± 96.03 a |
| Apigenin | 53.49 ± 2.32 a | 41.92 ± 3.33 ab | 34.18 ± 4.73 b | 31.05 ± 7.51 b | 28.94 ± 6.60 b | ||
| Cinnamic acid | Chlorogenic acid | 320 | 43.71 ± 0.37 a | 13.71 ± 2.69 b | 0 #c | 0 #c | 0 #c |
| p-Coumaric acid | 138.67 ± 4.19 ab | 173.01 ± 13.66 a | 90.74 ± 48.62 bc | 33.79 ± 17.46 cd | 21.65 ± 14.56 d | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Santana, Á.L.; Peterson, J.; Perumal, R.; Hu, C.; Sang, S.; Siliveru, K.; Smolensky, D. Post Acid Treatment on Pressurized Liquid Extracts of Sorghum (Sorghum bicolor L. Moench) Grain and Plant Material Improves Quantification and Identification of 3-Deoxyanthocyanidins. Processes 2023, 11, 2079. https://doi.org/10.3390/pr11072079
Santana ÁL, Peterson J, Perumal R, Hu C, Sang S, Siliveru K, Smolensky D. Post Acid Treatment on Pressurized Liquid Extracts of Sorghum (Sorghum bicolor L. Moench) Grain and Plant Material Improves Quantification and Identification of 3-Deoxyanthocyanidins. Processes. 2023; 11(7):2079. https://doi.org/10.3390/pr11072079
Chicago/Turabian StyleSantana, Ádina L., Jaymi Peterson, Ramasamy Perumal, Changling Hu, Shengmin Sang, Kaliramesh Siliveru, and Dmitriy Smolensky. 2023. "Post Acid Treatment on Pressurized Liquid Extracts of Sorghum (Sorghum bicolor L. Moench) Grain and Plant Material Improves Quantification and Identification of 3-Deoxyanthocyanidins" Processes 11, no. 7: 2079. https://doi.org/10.3390/pr11072079
APA StyleSantana, Á. L., Peterson, J., Perumal, R., Hu, C., Sang, S., Siliveru, K., & Smolensky, D. (2023). Post Acid Treatment on Pressurized Liquid Extracts of Sorghum (Sorghum bicolor L. Moench) Grain and Plant Material Improves Quantification and Identification of 3-Deoxyanthocyanidins. Processes, 11(7), 2079. https://doi.org/10.3390/pr11072079









