Bioactive Efficacy of Identified Phytochemicals Solasodine, Lupeol and Quercetin from Solanum xanthocarpum against the RgpB Protein of Porphyromonas gingivalis—A Molecular Docking and Simulation Analysis
Abstract
1. Introduction
2. Materials and Methods
2.1. Collection of S. xanthocarpum Fruits and Extraction
2.2. Preliminary Screening of Activities of Solanum xanthocarpum Extracts
2.2.1. Antioxidant Assay of Crude Extract
2.2.2. Anti-Inflammatory Assay of Crude Extract
2.2.3. Antibacterial Activity of the Crude Extract
2.3. Column Chromatographic Fractionation of the Aqueous Extract
2.3.1. Separation and Identification of Bioactive Compounds
2.3.2. GC-MS Analysis of S. xanthocarpum
2.4. Molecular Docking Analysis
2.5. Molecular Dynamics Simulation of Apo-Protein and Protein–Ligand Complexes
3. Results and Discussion
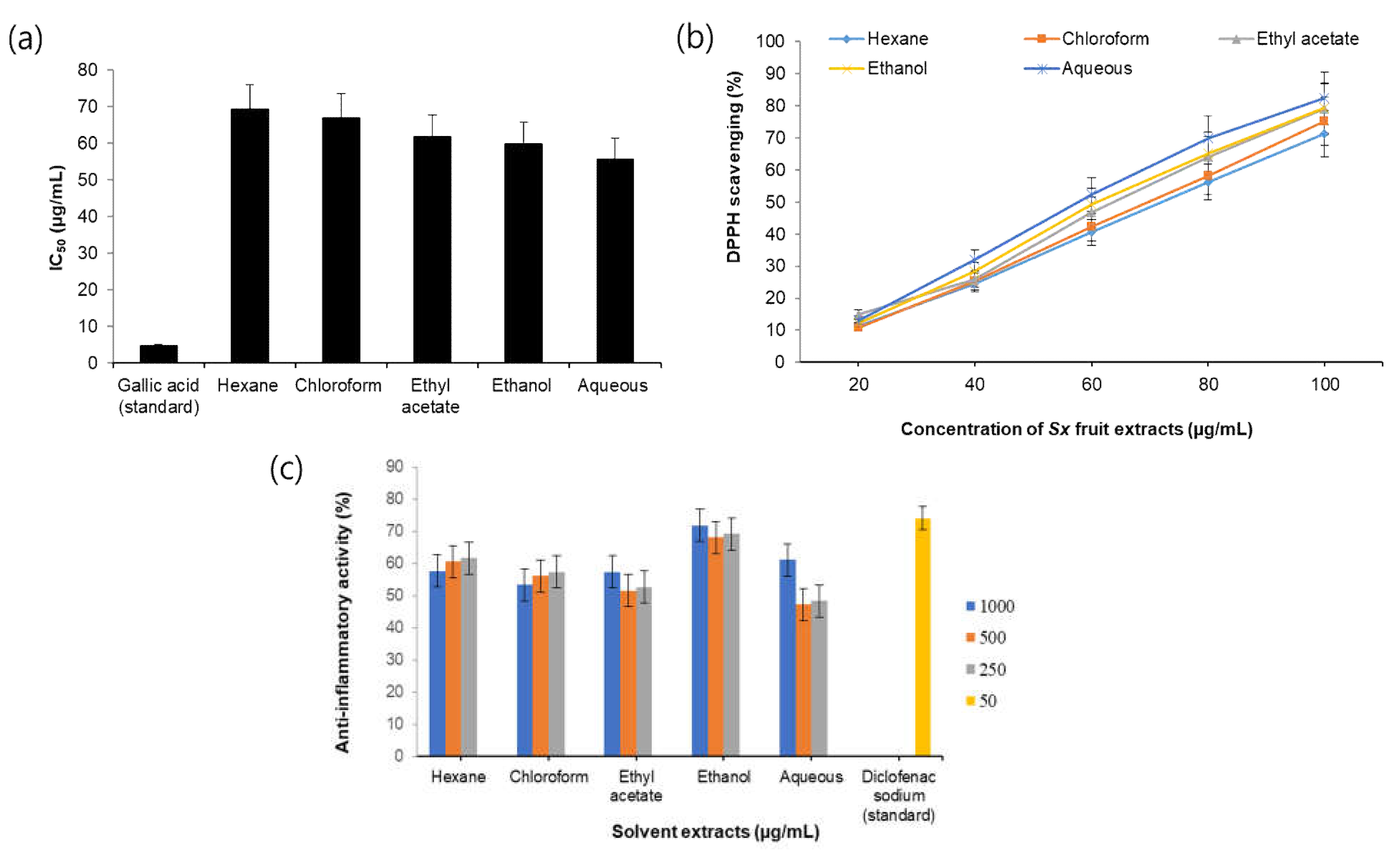
3.1. Antioxidant Activity of S. xanthocarpum Extracts
3.2. Anti-Inflammatory Activity of S. xanthocarpum Extracts
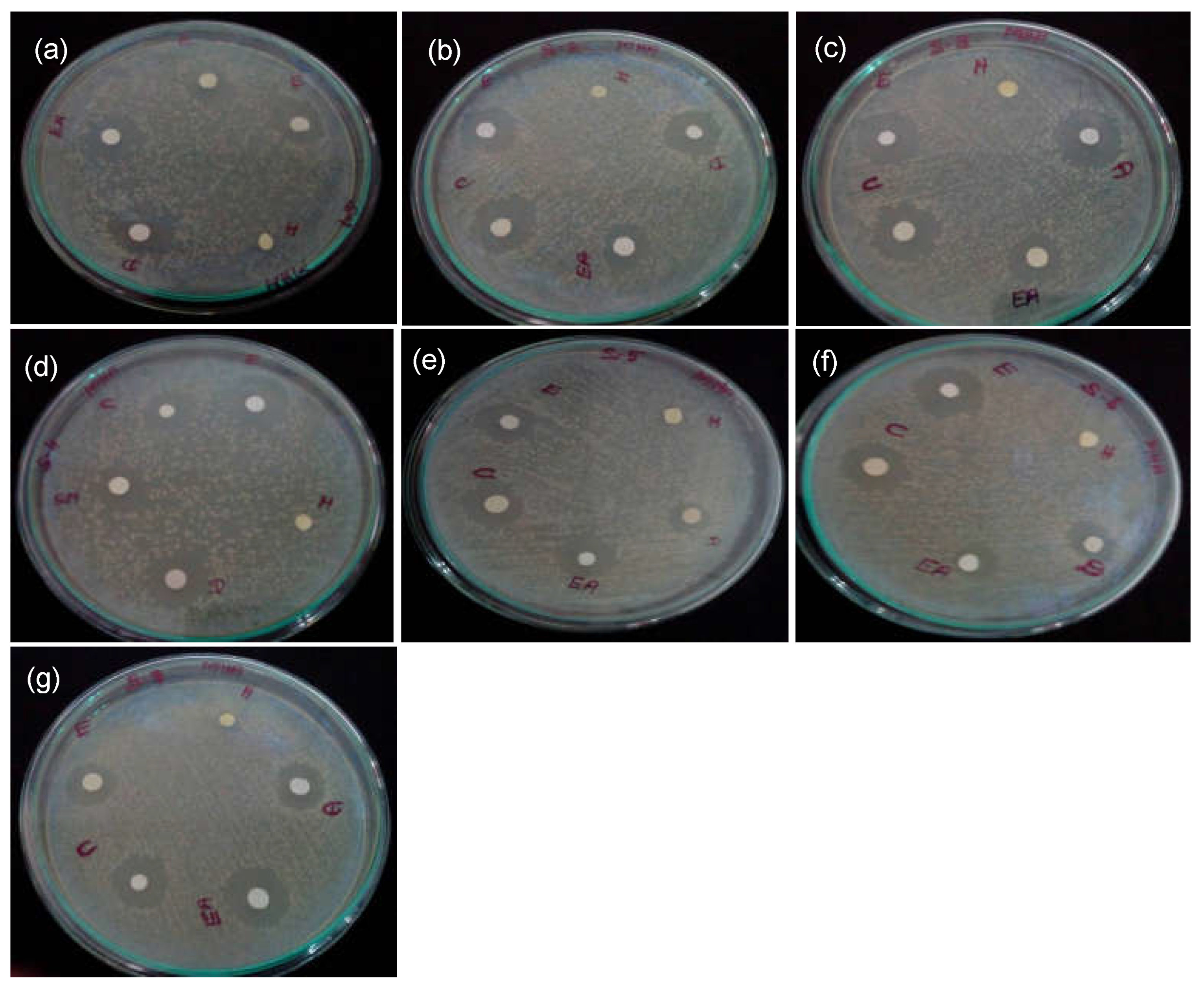
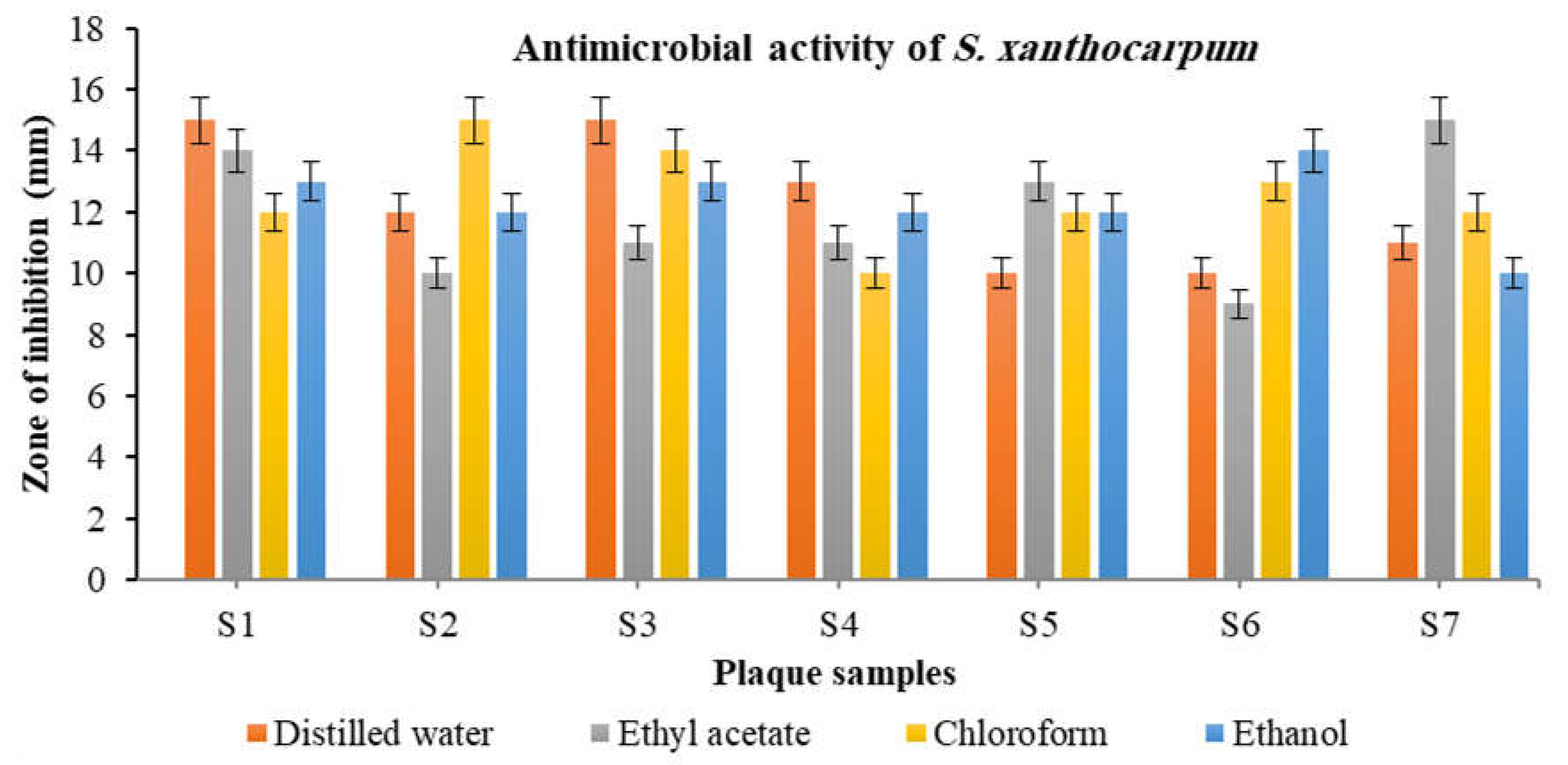
3.3. Antibacterial Activity of S. xanthocarpum Extracts
3.4. Gas Chromatography–Mass Spectrometry (GC-MS) Analysis
3.5. InSilico Analysis
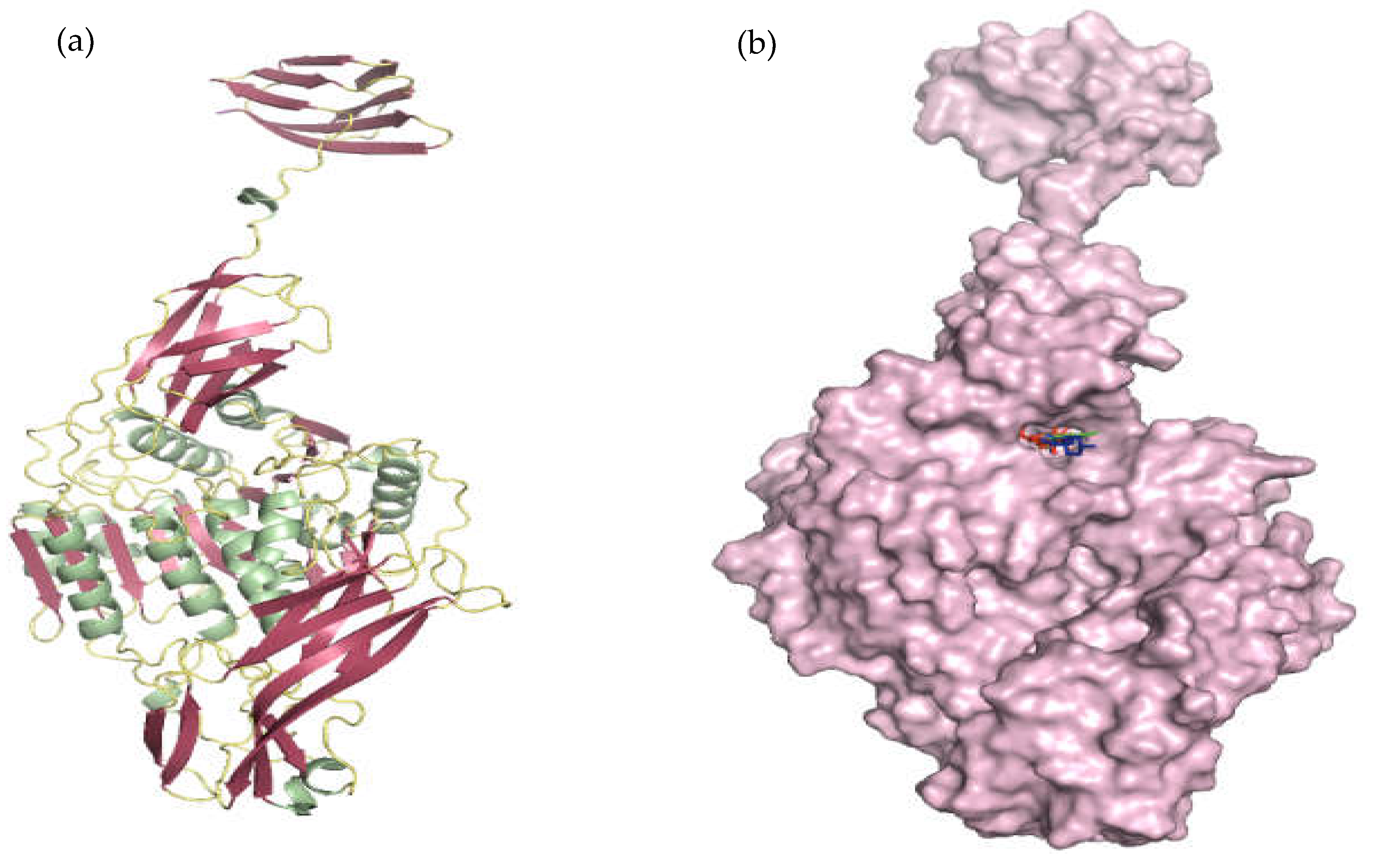
3.5.1. Molecular Docking Using CDOCKER
3.5.2. Molecular Dynamics and Simulation Analysis
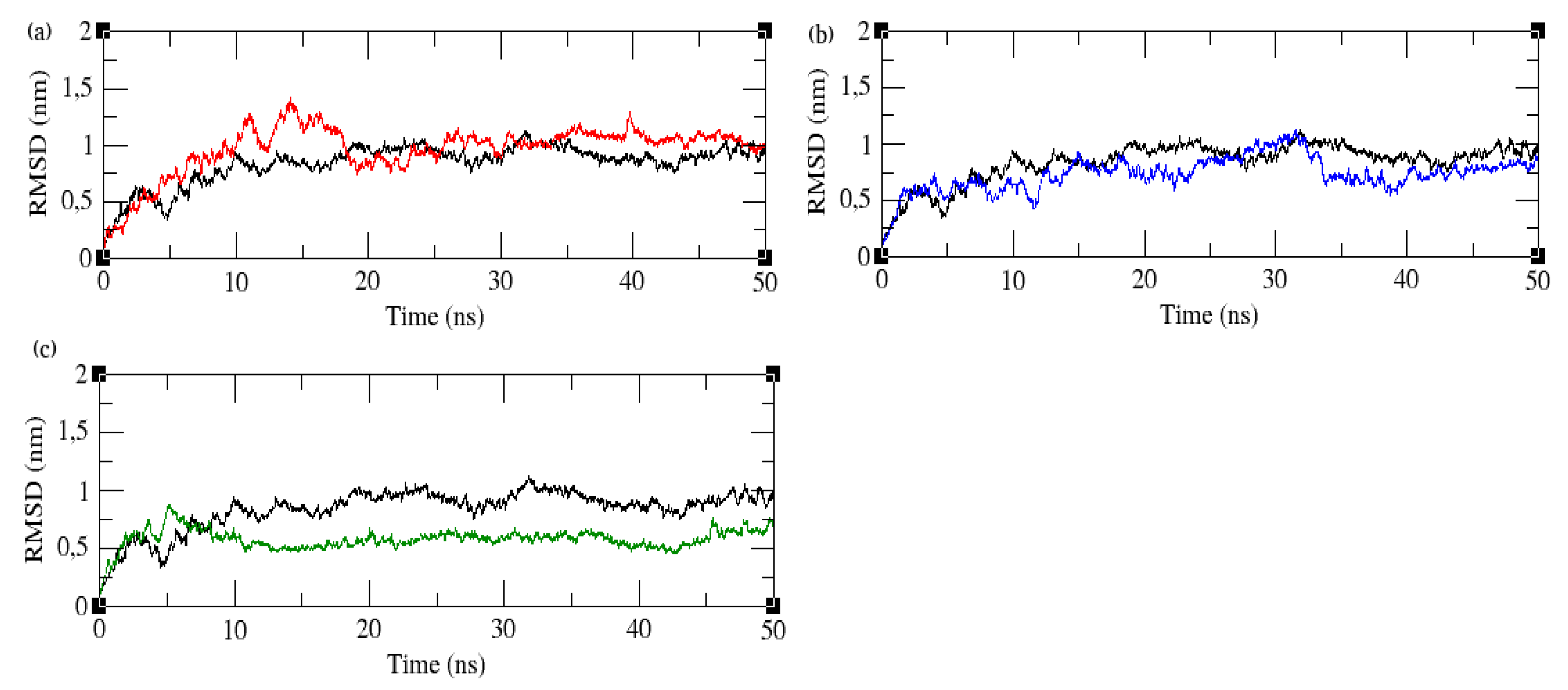
3.6. Root Mean Square Deviation
3.7. Root Mean Square Fluctuation
3.8. Radius of Gyration
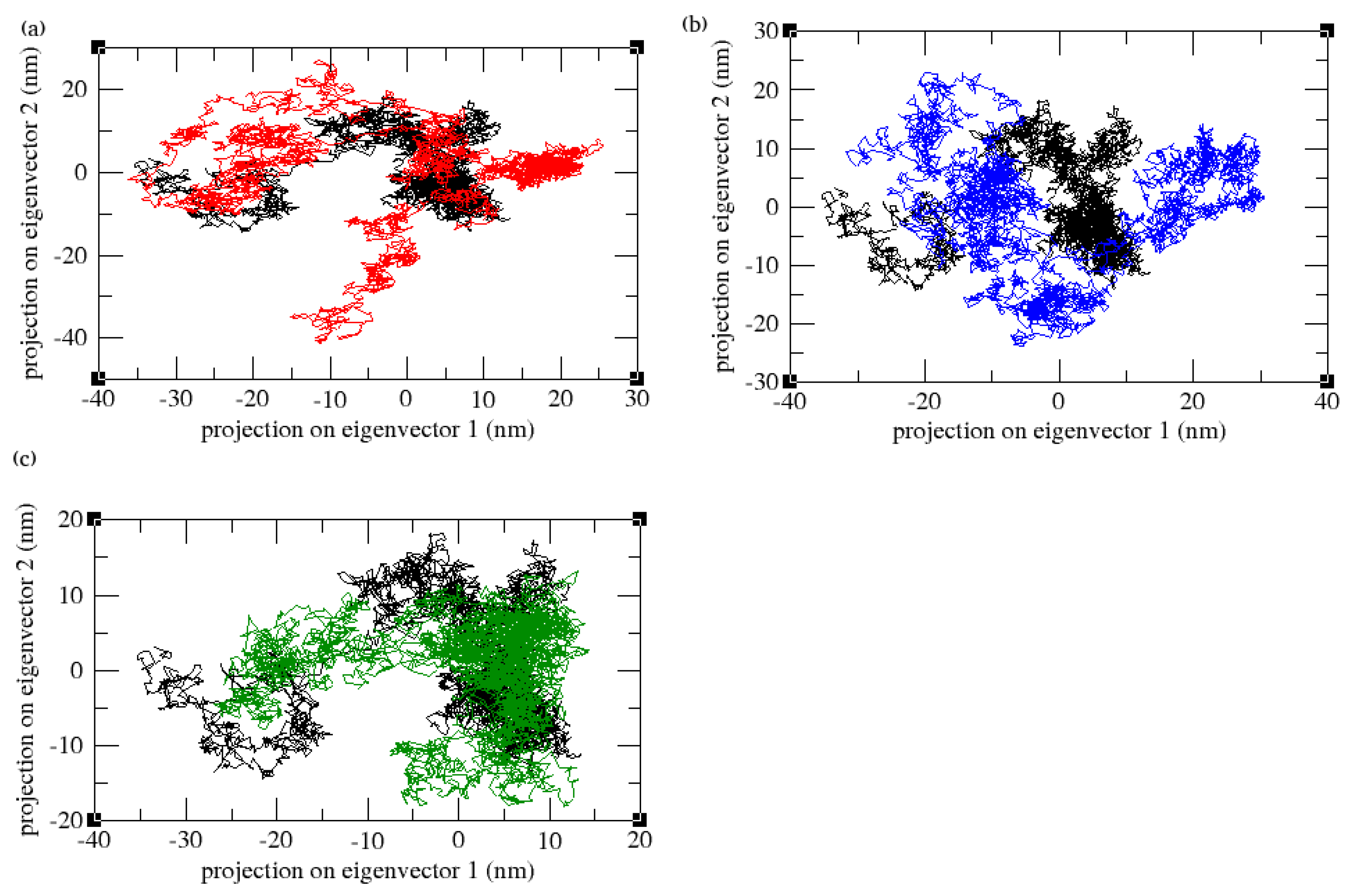
3.9. Principal Component Analysis
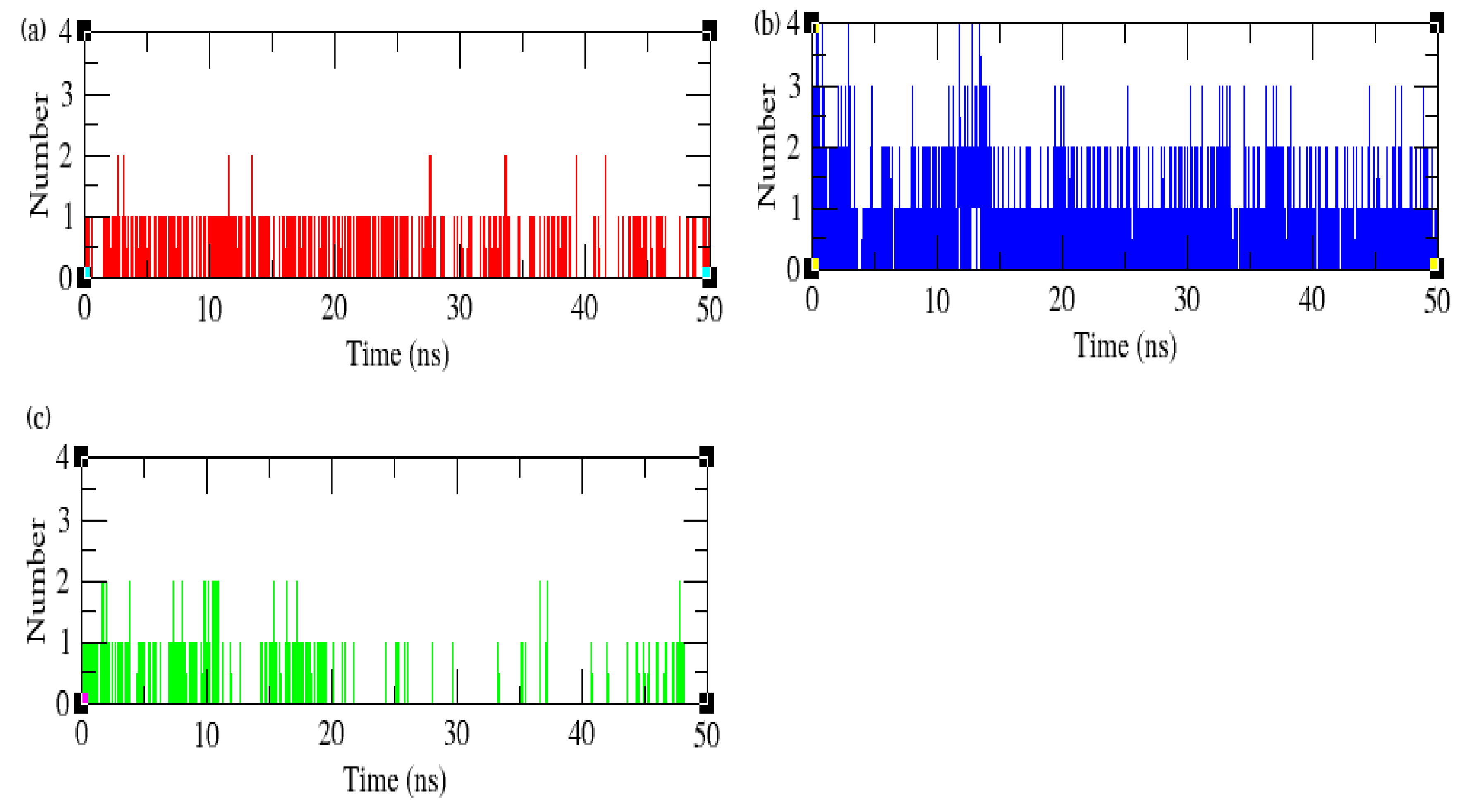
3.10. Hydrogen Bonds and Interaction Energy Analysis
4. Side Effects, Limitations and Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Könönen, E.; Gursoy, M.; Gursoy, U.K. Periodontitis: A Multifaceted Disease of Tooth-Supporting Tissues. J. Clin. Med. 2019, 8, 1135. [Google Scholar] [CrossRef] [PubMed]
- Kinane, D.F.; Stathopoulou, P.G.; Papapanou, P.N. Periodontal diseases. Nat. Rev. Dis. Primers 2017, 3, 17038. [Google Scholar] [CrossRef] [PubMed]
- Ferreira, M.C.; Dias-Pereira, A.C.; Branco-de-Almeida, L.S.; Martins, C.C.; Paiva, S.M. Impact of periodontal disease on quality of life: A systematic review. J. Periodontal. Res. 2017, 52, 651–665. [Google Scholar] [CrossRef]
- Lertpimonchai, A.; Rattanasiri, S.; Arj-Ong Vallibhakara, S.; Attia, J.; Thakkinstian, A. The association between oral hygiene and periodontitis: A systematic review and meta-analysis. Int. Dent. J. 2017, 67, 332–343. [Google Scholar] [CrossRef]
- Eivazi, M.; Falahi, N.; Eivazi, N.; Eivazi, M.A.; Raygani, A.V.; Rezaei, F. The Effect of Scaling and Root Planning on Salivary TNF-α and IL-1α Concentrations in Patients with Chronic Periodontitis. Open Dent. J. 2017, 11, 573–580. [Google Scholar] [CrossRef]
- Brookes, Z.L.S.; Bescos, R.; Belfield, L.A.; Ali, K.; Roberts, A. Current uses of chlorhexidine for management of oral disease: A narrative review. J. Dent. 2020, 103, 103497. [Google Scholar] [CrossRef]
- Guerrini, L.; Monaco, A.; Pietropaoli, D.; Ortu, E.; Giannoni, M.; Marci, M.C. Antibiotics in Dentistry: A Narrative Review of Literature and Guidelines Considering Antibiotic Resistance. Open Dent. J. 2019, 13, 383–398. [Google Scholar] [CrossRef]
- Bardají, D.K.R.; Reis, E.B.; Medeiros, T.C.T.; Lucarini, R.; Crotti, A.E.M.; Martins, C.H.G. Antibacterial activity of commercially available plant-derived essential oils against oral pathogenic bacteria. Nat. Prod. Res. 2016, 30, 1178–1181. [Google Scholar] [CrossRef]
- Livada, R.; Shiloah, J.; Tipton, D.A.; Dabbous, M.K. The Potential Role of Curcumin in Periodontal Therapy: A Review of the Literature. J. Int. Acad. Periodontol. 2017, 19, 70–79. [Google Scholar]
- Chandra Shekar, B.R.; Nagarajappa, R.; Suma, S.; Thakur, R. Herbal extracts in oral health care—A review of the current scenario and its future needs. Pharmacogn. Rev. 2015, 9, 87–92. [Google Scholar] [CrossRef]
- Safiaghdam, H.; Oveissi, V.; Bahramsoltani, R.; Farzaei, M.H.; Rahimi, R. Medicinal plants for gingivitis: A review of clinical trials. Iran. J. Basic Med. Sci. 2018, 21, 978–991. [Google Scholar] [PubMed]
- Kumar, S.; Pandey, A.K. Medicinal attributes of Solanum xanthocarpum fruit consumed by several tribal communities as food: An in vitro antioxidant, anticancer and anti HIV perspective. BMC Complement. Altern. Med. 2014, 14, 112. [Google Scholar] [CrossRef] [PubMed]
- Mahalakshmi, P.; Rameshkumar, A.; Sudha, G.; Dineshkumar, T.; Vinoth, H.; Malar, A. Evaluation of anti-microbial properties of Solanum xanthocarpum and Pistacia lentiscus extracts on Streptococcus mutans, Lactobacillus species and Actinomyces viscosus: An in vitro study. J. Oral MaxillofacPathol. 2019, 23, 383–388. [Google Scholar]
- Singh, O.M.; Singh, T.P. Phytochemistry of Solanum xanthocarpum: An Amazing Traditional Healer. J. Sci. Ind. Res. 2010, 69, 732–740. [Google Scholar]
- Hajishengallis, G.; Darveau, R.P.; Curtis, M.A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 2012, 10, 717–725. [Google Scholar] [CrossRef]
- Lee, S.E.; Hwang, H.J.; Ha, J.S.; Jeong, H.S.; Kim, J.H. Screening of medicinal plant extracts for antioxidant activity. Life Sci. 2003, 73, 167–179. [Google Scholar] [CrossRef]
- Shinde, U.A.; Phadke, A.S.; Nair, A.M.; Mungantiwar, A.A.; Dikshit, V.J.; Saraf, M.N. Membrane stabilizing activity—A possible mechanism of action for the anti-inflammatory activity of Cedrus deodara wood oil. Fitoterapia 1999, 70, 251–257. [Google Scholar] [CrossRef]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H.; Tonetti, M.S. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S162–S170. [Google Scholar] [CrossRef]
- How, K.Y.; Song, K.P.; Chan, K.G. Porphyromonasgingivalis: An Overview of Periodontopathic Pathogen below the Gum Line. Front. Microbiol. 2016, 7, 53. [Google Scholar] [CrossRef]
- Bostanci, N.; Belibasakis, G.N. Porphyromonasgingivalis: An invasive and evasive opportunistic oral pathogen. FEMS Microbiol. Lett. 2012, 333, 1–9. [Google Scholar] [CrossRef]
- Kantyka, T.; Latendorf, T.; Wiedow, O.; Bartels, J.; Gläser, R.; Dubin, G.; Schröder, J.M.; Potempa, J.; Meyer-Hoffert, U. Elafin is specifically inactivated by RgpB from Porphyromonas gingivalis by distinct proteolytic cleavage. Biol. Chem. 2009, 390, 1313–1320. [Google Scholar] [PubMed]
- Skottrup, P.D.; Leonard, P.; Kaczmarek, J.Z.; Veillard, F.; Enghild, J.J.; O’Kennedy, R.; Sroka, A.; Clausen, R.P.; Potempa, J.; Riise, E. Diagnostic evaluation of a nanobody with picomolar affinity toward the protease RgpB from Porphyromonasgingivalis. Anal. Biochem. 2011, 415, 158–167. [Google Scholar] [CrossRef] [PubMed]
- UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Hassabis, D. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Cryst. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Bolton, E.E. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res. 2021, 49, D1388–D1395. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J.C. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef]
- Van Aalten, D.M.; Bywater, R.; Findlay, J.B.; Hendlich, M.; Hooft, R.W.; Vriend, G. PRODRG, a program for generating molecular topologies and unique molecular descriptors from coordinates of small molecules. J. Comput. Aided Mol. Des. 1996, 10, 255–262. [Google Scholar]
- Kumari, R.; Kumar, R. Open Source Drug Discovery Consortium, Lynn A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model. 2014, 54, 1951–1962. [Google Scholar] [PubMed]
- Muruhan, S.; Selvaraj, S.; Viswanathan, P.K. In vitro antioxidant activities of Solanum surattense leaf extract. Asian Pac. J. Trop. Biomed. 2013, 3, 28–34. [Google Scholar] [CrossRef]
- Narayanan, M.; Jayashree, T.; Kandasamy, S.; Natarajan, D.; Liu, G.; Elesawy, B.H.; Pugazhendhi, A. An in vitro investigation of the antidermatophytic, antioxidant, and nephroprotective activity of Solanum surattense. Process Biochem. 2021, 109, 178–185. [Google Scholar] [CrossRef]
- Prasad, K.N.; Yang, B.; Dong, X.; Jiang, G.; Zhang, H.; Xie, H.; Jiang, Y. Flavonoid contents and antioxidant activities from Cinnamomum species. Innov. Food Sci. Emerg. Technol. 2009, 10, 627–632. [Google Scholar] [CrossRef]
- Gabay, O.; Sanchez, C.; Salvat, C.; Chevy, F.; Breton, M.; Nourissat, G.; Berenbaum, F. Stigmasterol: A phytosterol with potential anti-osteoarthritic properties. Osteoarthr. Cartil. 2010, 18, 106–116. [Google Scholar] [CrossRef]
- Saleem, M. Lupeol, A Novel Anti-inflammatory and Anti-cancer Dietary Triterpene. Cancer Lett. 2009, 285, 109–115. [Google Scholar] [CrossRef]
- Sathasivam, A.; Muthuselvam, M.; Rajasekaran, R. P10 (IN)—Analysis of phyto chemical constituents and anti-microbial activety of some southern Indian medicinal plants. J. Pharm. Res. 2010, 3, 1841–1843. [Google Scholar]
- Abbas, K.; Niaz, U.; Hussain, T.; Saeed, M.A.; Javaid, Z.; Idrees, A.; Rasool, S. Anti-microbial activity of fruits of Solanum nigrum and Solanum xanthocarpum. Acta Pol. Pharm. 2014, 71, 415–421. [Google Scholar]
- Ahmad Khan, M.; Sarwar, A.H.M.G.; Rahat, R.; Ahmed, R.S.; Umar, S. Stigmasterol protects rats from collagen induced arthritis by inhibiting proinflammatory cytokines. Int. Immunopharmacol. 2020, 85, 106642. [Google Scholar] [CrossRef] [PubMed]
- Araldi, E.; Fernández-Fuertes, M.; Canfrán-Duque, A.; Tang, W.; Cline, G.W.; Madrigal-Matute, J.; Suárez, Y. Lanosterol modulates TLR4 mediated innate immune responses in macrophages. Cell Rep. 2017, 19, 2743–2755. [Google Scholar] [CrossRef]
- Jiao, F.; Tan, Z.; Yu, Z.; Zhou, B.; Meng, L.; Shi, X. The phytochemical and pharmacological profile of taraxasterol. Front. Pharmacol. 2022, 13, 927365. [Google Scholar] [CrossRef]
- Santos, C.C.D.M.P.; Salvadori, M.S.; Mota, V.G.; Costa, L.M.; de Almeida, A.A.C.; de Oliveira, G.A.L.; de Almeida, R.N. Antinociceptive and Antioxidant Activities of Phytol In Vivo and In Vitro Models. Neurosci. J. 2013, 2013, 949452. [Google Scholar] [CrossRef]
- Mooney, E.C.; Holden, S.E.; Xia, X.J.; Li, Y.; Jiang, M.; Banson, C.N.; Sahingur, S.E. Quercetin Preserves Oral Cavity Health by Mitigating Inflammation and Microbial Dysbiosis. Front. Immunol. 2021, 12, 774273. [Google Scholar] [CrossRef] [PubMed]
- Corega, C.; Vaida, L.; Festila, D.G.; Rigoni, G.; Albanese, M.; D’Agostino, A.; Bertossi, D. The benefits of Quercitin for dentistry and maxillofacial surgery: A systematic review. Minerva Stomatol. 2014; ahead of print. [Google Scholar]
- Patel, K.; Singh, R.B.; Patel, D.K. Medicinal significance, pharmacological activities, and analytical aspects of solasodine: A concise report of current scientific literature. J. Acute Dis. 2013, 2, 92–98. [Google Scholar] [CrossRef]
- Revathi, P.; Parimelazhagan, T. Traditional knowledge on medicinal plants used by the Irula tribe of Hasanur hills, Erode District, Tamil Nadu, India. Ethnobot Leaflets. 2010, 14, 136–160. [Google Scholar]
- Narayanan, M.K.R.; Anilkumar, N.; Balakrishnan, V.; Sivadasan, M.; Alfarhan, H.A.; Alatar, A.A. Wild edible plants used by the Kattunaikka, Paniya and Kuruma tribes of Wayanad District, Kerala, India. J. Med. Plants Res. 2011, 5, 3520–3529. [Google Scholar]




| S. No. | RT | Name of the Compound | Molecular Formula | MW | Peak Area % |
|---|---|---|---|---|---|
| 1 | 5.329 | Pyridine | C5H5N | 79.0999 | 3.70 |
| 2 | 6.032 | 3-Hexen-2-one | C7H12O | 112.1696 | 0.67 |
| 3 | 6.7544 | 2-Pentanone,4-Hydroxy-4-methyl- | C6H12O2 | 116.1583 | 62.11 |
| 4 | 17.709 | Diethyl phthalate | C12H14O4 | 222.2372 | 1.54 |
| 5 | 20.457 | Bicyclo[3.1.1]heptane,2,6,6-trimethyl- | C10H18 | 138.2499 | 2.07 |
| 6 | 21.307 | Hexadecanoic acid, methyl ester | C17H34O2 | 270.4507 | 3.22 |
| 7 | 23.021 | 9,12-Octadecadienoic acid (Z,Z)-, methyl ester | C19H34O2 | 294.4721 | 2.97 |
| 8 | 23.080 | 9,12,15-Octadecatrienoic acid, methyl ester | C19H32O2 | 294.4562 | 3.67 |
| 9 | 23.183 | Phytol | C20H40O | 296.5310 | 1.90 |
| 10 | 23.306 | Octadecanoicacid, methylester | C19H34O2 | 294.4721 | 1.00 |
| 11 | 23.705 | Quercetin | C15H10O7 | 302.2360 | 2.76 |
| 12 | 33.203 | Stigmasterol | C29H48O | 412.6908 | 1.72 |
| 13 | 34.003 | Solasodine | C27H43NO2 | 413.6331 | 8.63 |
| 14 | 35.221 | Lanosterol | C30H50O | 426.7174 | 2.08 |
| 15 | 35.437 | Taraxasterol | C30H50O | 426.7174 | 3.10 |
| S. No. | RT | Nameofthe Compound | Molecular Formula | MW | Peak Area % |
|---|---|---|---|---|---|
| 1 | 11.965 | 1-Dodecene | C12H24 | 168.31 | 4.45 |
| 2 | 21.347 | Hexadecanoic acid, methyl ester | C17H34O2 | 270.4507 | 5.75 |
| 3 | 21.762 | n-Hexadecanoicacid | C16H32O2 | 256.4241 | 2.78 |
| 4 | 23.072 | 9,12,15-Octadecatrienoic acid, methyl ester | C19H32O2 | 294.4562 | 4.87 |
| 5 | 33.996 | Solasodine | C27H43NO2 | 413.6331 | 4.41 |
| S. No. | RT | Name of the Compound | Molecular Formula | MW | Peak Area % |
|---|---|---|---|---|---|
| 1 | 17.747 | Diethyl phthalate | C12H14O4 | 222.2372 | 9.36 |
| 2 | 18.967 | Heptadecane | C17H36 | 240.4677 | 5.02 |
| 3 | 20.774 | 1-Nonadecene | C19H38 | 266.5050 | 3.68 |
| 4 | 21.209 | Hexadecanoic acid, methyl ester | C17H34O2 | 270.4507 | 4.25 |
| 5 | 21.337 | n–Hexadecanoicacid | C16H32O2 | 256.4241 | 18.49 |
| 6 | 23.045 | 9,12-Octadecadienoic acid (Z,Z)-, methyl ester | C19H34O2 | 294.4721 | 26.16 |
| 7 | 26.623 | Lupeol | C30H50O | 426.7174 | 6.13 |
| Region | Percentage of Residues (%) |
|---|---|
| Residues in the most favored region | 89.2 |
| Residues in additionally allowed region | 9.8 |
| Residues in the generously allowed region | 0.6 |
| Residues in the disallowed region | 0.3 |
| Compounds | CDOCKER Energy (kcal/mol) |
|---|---|
| Lupeol | −263.879 |
| Solasodine | −102.457 |
| Quercetin | 33.6821 |
| Complex | Binding Energy kJ/mol | Van der Waals Energy kJ/mol | Electrostatic Energy kJ/mol | Polar Solvation Energy kJ/mol | SASA Energy kJ/mol |
|---|---|---|---|---|---|
| Lupeol | −130.003 ± 12.336 | −170.912 ± 7.731 | −2.223 ± 2.030 | 61.079 ± 14.458 | −17.948 ± 0.931 |
| Quercetin | −103.109 ± 12.604 | −169.565 ± 12.181 | −6.578 ± 3.867 | 90.583 ± 19.978 | −17.549 ± 0.827 |
| Solasodine | −62.814 ± 18.379 | −70.180 ± 9.281 | −1.051 ± 5.078 | 16.112 ± 19.498 | −7.694 ± 1.561 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arumuganainar, D.; Subramanian, G.; Basavarajappa, S.; Hashem, M.I.; Arun, K.V.; Balaji, S.K.; Yadalam, P.K.; Venkidasamy, B.; Vinayagam, R. Bioactive Efficacy of Identified Phytochemicals Solasodine, Lupeol and Quercetin from Solanum xanthocarpum against the RgpB Protein of Porphyromonas gingivalis—A Molecular Docking and Simulation Analysis. Processes 2023, 11, 1887. https://doi.org/10.3390/pr11071887
Arumuganainar D, Subramanian G, Basavarajappa S, Hashem MI, Arun KV, Balaji SK, Yadalam PK, Venkidasamy B, Vinayagam R. Bioactive Efficacy of Identified Phytochemicals Solasodine, Lupeol and Quercetin from Solanum xanthocarpum against the RgpB Protein of Porphyromonas gingivalis—A Molecular Docking and Simulation Analysis. Processes. 2023; 11(7):1887. https://doi.org/10.3390/pr11071887
Chicago/Turabian StyleArumuganainar, Deepavalli, Gopinath Subramanian, Santhosh Basavarajappa, Mohamed Ibrahim Hashem, Kurumathur Vasudevan Arun, Subbusamy Kanakasabapathy Balaji, Pradeep Kumar Yadalam, Baskar Venkidasamy, and Ramachandran Vinayagam. 2023. "Bioactive Efficacy of Identified Phytochemicals Solasodine, Lupeol and Quercetin from Solanum xanthocarpum against the RgpB Protein of Porphyromonas gingivalis—A Molecular Docking and Simulation Analysis" Processes 11, no. 7: 1887. https://doi.org/10.3390/pr11071887
APA StyleArumuganainar, D., Subramanian, G., Basavarajappa, S., Hashem, M. I., Arun, K. V., Balaji, S. K., Yadalam, P. K., Venkidasamy, B., & Vinayagam, R. (2023). Bioactive Efficacy of Identified Phytochemicals Solasodine, Lupeol and Quercetin from Solanum xanthocarpum against the RgpB Protein of Porphyromonas gingivalis—A Molecular Docking and Simulation Analysis. Processes, 11(7), 1887. https://doi.org/10.3390/pr11071887








