Bioaugmentation of Aerobic Granular Sludge with Dye-Decolorizing Yeast for Textile Industrial Wastewater
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Materials
2.2. Cultivation Conditions
2.3. AGS Sequencing Batch Reactor (SBR) Set-Up
2.4. Analytical Methods
2.5. Recovery of Y. lipolytica from AGS and Identification
2.6. Microbial Community Analysis of AGS
2.7. EPS Extraction
2.8. Image Analysis
2.9. AGS Ability for Dye Removal in Batch Conditions
3. Results and Discussion
3.1. Reactor Performance—Removal of C and N
3.2. AGS Settling Properties
3.3. Dye Removal
3.4. Ability for Dye Removal in Batch Conditions
3.5. Recovery of the Bioaugmented Yeast along Reactor Operation
3.6. Microbial Community of AGS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dellamatrice, P.M.; Silva-Stenico, M.E.; de Moraes, L.A.B.; Fiore, M.F.; Monteiro, R.T.R. Degradation of textile dyes by cyanobacteria. Braz. J. Microbiol. 2017, 48, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, D.A.; Scholz, M. Textile dye wastewater characteristics and constituents of synthetic effluents: A critical review. Inter. J. Environ. Sci. Technol. 2019, 16, 1193–1226. [Google Scholar] [CrossRef]
- Saratale, R.G.; Saratale, G.D.; Chang, J.S.; Govindwar, S.P. Bacterial decolorization and degradation of azo dyes: A review. J. Taiwan Inst. Chem. Eng. 2011, 42, 138–157. [Google Scholar] [CrossRef]
- Sarvajith, M.; Reddy, G.K.K.; Nancharaiah, Y.V. Textile dye biodecolourization and ammonium removal over nitrite in aerobic granular sludge sequencing batch reactors. J. Hazard. Mater. 2018, 342, 536–543. [Google Scholar] [CrossRef]
- Adav, S.S.; Lee, D.J.; Show, K.Y.; Tay, J.H. Aerobic granular sludge: Recent advances. Biotechnol. Adv. 2008, 26, 411–423. [Google Scholar] [CrossRef] [PubMed]
- Ely, C.; Moreira, I.S.; Bassin, J.P.; Dezotti, M.W.C.; Mesquita, D.P.; Costa, J.; Ferreira, E.C.; Castro, P.M.L. Treatment of saline wastewater amended with endocrine disruptors by aerobic granular sludge: Assessing performance and microbial community dynamics. J. Environ. Chem. Eng. 2022, 10, 107272. [Google Scholar] [CrossRef]
- Amorim, C.L.; Alves, M.; Castro, P.M.L.; Henriques, I. Bacterial community dynamics within an aerobic granular sludge reactor treating wastewater loaded with pharmaceuticals. Ecotoxicol. Environ. Saf. 2018, 147, 905–912. [Google Scholar] [CrossRef]
- Oliveira, A.S.; Amorim, C.L.; Ramos, M.A.; Mesquita, D.P.; Inocêncio, P.; Ferreira, E.C.; Van Loosdrecht, M.; Castro, P.M.L. Variability in the composition of extracellular polymeric substances from a full-scale aerobic granular sludge reactor treating urban wastewater. J. Environ. Chem. Eng. 2020, 8, 104156. [Google Scholar] [CrossRef]
- Bassin, J.P.; Pronk, M.; Muyzer, G.; Kleerebezem, R.; Dezotti, M.; van Loosdrecht, M.C.M. Effect of elevated salt concentrations on the aerobic granular sludge process: Linking microbial activity with microbial community structure. Appl. Environ. Microbiol. 2011, 77, 7942–7953. [Google Scholar] [CrossRef]
- de Kreuk, M.K.; Heijnen, J.J.; Van Loosdrecht, M.C.M. Simultaneous COD, nitrogen, and phosphate removal by aerobic granular sludge. Biotechnol. Bioeng. 2005, 90, 761–769. [Google Scholar] [CrossRef]
- Beun, J.J.; Hendriks, A.; Van Loosdrecht, M.C.M.; Morgenroth, E.; Wilderer, P.A.; Heijnen, J.J. Aerobic granulation in a sequencing batch reactor. Water Res. 1999, 33, 2283–2290. [Google Scholar] [CrossRef]
- Flemming, H.C. Eps—Then and now. Microorganisms 2016, 4, 41. [Google Scholar] [CrossRef]
- Amorim, C.L.; Moreira, I.S.; Ribeiro, A.R.; Santos, L.H.M.L.M.; Delerue-Matos, C.; Tiritan, M.E.; Castro, P.M.L. Treatment of a simulated wastewater amended with a chiral pharmaceuticals mixture by an aerobic granular sludge sequencing batch reactor. Int. Biodeterior. Biodegrad. 2016, 115, 277–285. [Google Scholar] [CrossRef]
- Duque, A.F.; Bessa, V.S.; Carvalho, M.F.; de Kreuk, M.K.; van Loosdrecht, M.C.M.; Castro, P.M.L. 2-Fluorophenol degradation by aerobic granular sludge in a sequencing batch reactor. Water Res. 2011, 45, 6745–6752. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, A.S.; Amorim, C.L.; Zlopasa, J.; van Loosdrecht, M.; Castro, P.M.L. Recovered granular sludge extracellular polymeric substances as carrier for bioaugmentation of granular sludge reactor. Chemosphere 2021, 275, 130037. [Google Scholar] [CrossRef] [PubMed]
- Quan, X.; Ma, J.; Xiong, W.; Wang, X. Bioaugmentation of half-matured granular sludge with special microbial culture promoted establishment of 2,4-dichlorophenoxyacetic acid degrading aerobic granules. Bioprocess Biosyst. Eng. 2015, 38, 1081–1090. [Google Scholar] [CrossRef]
- Maqbool, Z.; Shahid, M.; Azeem, F.; Shahzad, T.; Mahmood, F.; Rehman, M.; Ahmed, T.; Imran, M.; Hussain, S. Application of a Dye-Decolorizing Pseudomonas aeruginosa Strain ZM130 for Remediation of Textile Wastewaters in Aerobic/Anaerobic Sequential Batch Bioreactor and Soil Columns. Water. Air. Soil Pollut. 2020, 231, 386. [Google Scholar] [CrossRef]
- Franca, R.D.G.; Vieira, A.; Carvalho, G.; Oehmen, A.; Pinheiro, H.M.; Barreto Crespo, M.T.; Lourenço, N.D. Oerskovia paurometabola can efficiently decolorize azo dye Acid Red 14 and remove its recalcitrant metabolite. Ecotoxicol. Environ. Saf. 2020, 191, 110007. [Google Scholar] [CrossRef]
- Lourenço, N.D.; Franca, R.D.G.; Moreira, M.A.; Gil, F.N.; Viegas, C.A.; Pinheiro, H.M. Comparing aerobic granular sludge and flocculent sequencing batch reactor technologies for textile wastewater treatment. Biochem. Eng. J. 2015, 104, 57–63. [Google Scholar] [CrossRef]
- Louhasakul, Y.; Cheirsilp, B.; Treu, L.; Kougias, P.G.; Angelidali, I. Metagenomic insights into bioaugmentation and biovalorization of oily industrial wastes by lipolytic oleaginous yeast Yarrowia lipolytica during successive batch fermentation. Biotech. App. Biochem. 2019, 67, 1020–1029. [Google Scholar] [CrossRef]
- Wen, H.; Xiong, K.; Yang, H.; Zhang, P.; Wang, X. Dynamic mechanism of the microbiota of high-salinity organic wastewater with salt-tolerant yeast and its application. J. Environ. Chem. Eng. 2022, 10, 107377. [Google Scholar] [CrossRef]
- Mendes, M.; Cassoni, A.C.; Alves, S.; Pintado, M.E.; Castro, P.M.; Moreira, P. Screening for a more sustainable solution for decolorization of dyes and textile effluents using Candida and Yarrowia spp. J. Environ. Manag. 2022, 307, 114421. [Google Scholar] [CrossRef] [PubMed]
- Kokabian, B.; Bonakdarpour, B.; Fazel, S. The effect of salt on the performance and characteristics of a combined anaerobic-aerobic biological process for the treatment of synthetic wastewaters containing Reactive Black 5. Chem. Eng. J. 2013, 221, 363–372. [Google Scholar] [CrossRef]
- Mata, A.M.T.; Pinheiro, H.M.; Lourenço, N.D. Effect of sequencing batch cycle strategy on the treatment of a simulated textile wastewater with aerobic granular sludge. Biochem. Eng. J. 2015, 104, 106–114. [Google Scholar] [CrossRef]
- Franca, R.D.G.; Vieira, A.; Mata, A.M.T.; Carvalho, G.S.; Pinheiro, H.M.; Lourenço, N.D. Effect of an azo dye on the performance of an aerobic granular sludge sequencing batch reactor treating a simulated textile wastewater. Water Res. 2015, 85, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Felz, S.; Al-Zuhairy, S.; Aarstad, O.A.; van Loosdrecht, M.C.M.; Lin, Y.M. Extraction of structural extracellular polymeric substances from aerobic granular sludge. J. Vis. Exp. 2016, 115, e54534. [Google Scholar] [CrossRef]
- Lopez-Vazquez, C.M.; Hooijmans, C.M.; Brdjanovic, D.; Gijzen, H.J.; van Loosdrecht, M.C.M. Temperature effects on glycogen accumulating organisms. Water Res. 2009, 43, 2852–2864. [Google Scholar] [CrossRef]
- Oliveira, A.S.; Amorim, C.L.; Mesquita, D.P.; Ferreira, E.C.; van Loosdrecht, M.; Castro, P.M.L. Increased extracellular polymeric substances production contributes for the robustness of aerobic granular sludge during long-term intermittent exposure to 2-fluorophenol in saline wastewater. J. Water Process Eng. 2021, 40, 101977. [Google Scholar] [CrossRef]
- Zhao, Y.; Park, H.D.; Park, J.H.; Zhang, F.; Chen, C.; Li, X.; Zhao, D.; Zhao, F. Effect of different salinity adaptation on the performance and microbial community in a sequencing batch reactor. Bioresour. Technol. 2016, 216, 808–816. [Google Scholar] [CrossRef]
- Li, H.; Tan, L.; Ning, S.; He, M. Reactor performance and microbial community dynamics during aerobic degradation and detoxification of Acid Red B with activated sludge bioaugmented by a yeast Candida tropicalis TL-F1 in MBR. Int. Biodeterior. Biodegrad. 2015, 104, 149–156. [Google Scholar] [CrossRef]
- Song, L.; Shao, Y.; Shi, S.; Tan, L. Continuously Biodegrading High Concentration of Acid Red B under Hypersaline Conditions in a Membrane Bioreactor Bioaugmented by a Halotolerant Yeast Pichia occidentalis G1 and Microbial Community Dynamics. Environ. Eng. Sci. 2019, 36, 1412–1420. [Google Scholar] [CrossRef]
- Assadi, A.; Naderi, M.; Mehrasbi, M.R. Anaerobic–aerobic sequencing batch reactor treating azo dye containing wastewater: Effect of high nitrate ions and salt. J. Water Reuse Desalin. 2018, 8, 251–261. [Google Scholar] [CrossRef]
- Amorim, C.L.; Moreira, I.S.; Duque, A.F.; van Loosdrecht, M.C.M.; Castro, P.M.L. Aerobic Granular Sludge: Treatment of Wastewaters Containing Toxic Compounds. In Technologies for the Treatment and Recovery of Nutrients from Industrial Wastewater; Chapter 9; Mosquera, A., Campos, L., Val, Á., Eds.; IGI-Global: Hershey, PA, USA, 2007; ISBN 9781522510376. [Google Scholar] [CrossRef]
- He, T.; Hua, J.Q.; Chen, R.P.; Yu, L. Adsorption characteristics of methylene blue by a dye-degrading and extracellular polymeric substance -producing strain. J. Environ. Manag. 2021, 288, 112446. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.; Banat, I.M.; McMullan, G.; Nigam, P.; Smyth, F.; Marchant, R. Decolorization of Remazol Black B using a thermotolerant yeast. Kluyveromyces Marx. IMB3 2006, 26, 75–79. [Google Scholar]
- Wang, Y.; Jiang, L.; Shang, H.; Li, Q.; Zhou, W. Treatment of azo dye wastewater by the self-flocculating marine bacterium Aliiglaciecola lipolytica. Environ. Technol. Innov. 2020, 19, 100810. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, B.; Ning, S.; Shi, S.; Tan, L. Magnetically stimulated azo dye biodegradation by a newly isolated osmo-tolerant Candida tropicalis A1 and transcriptomic responses. Ecotoxicol. Environ. Saf. 2021, 209, 111791. [Google Scholar] [CrossRef]
- Charumathi, D.; Das, N. Packed bed column studies for the removal of synthetic dyes from textile wastewater using immobilised dead C. tropicalis. Desalination 2012, 285, 22–30. [Google Scholar] [CrossRef]
- Aksu, Z. Application of biosorption for the removal of organic pollutants: A review. Process Biochem. 2005, 40, 997–1026. [Google Scholar] [CrossRef]
- Aksu, Z.; Dönmez, G. A comparative study on the biosorption characteristics of some yeasts for Remazol Blue reactive dye. Chemosphere 2003, 50, 1075–1083. [Google Scholar] [CrossRef]
- Basri, H.F.; Hakim, M.; Halim, A. Microbial Community Shift and Role of Bacteria in Rapid Granulation By Using Diatomite. Res. Sq. 2022. preprint. [Google Scholar] [CrossRef]
- Wu, L.; Tang, B.; Bin, L.; Chen, G.; Huang, S.; Li, P.; Fu, F. Heterogeneity of the diverse aerobic sludge granules self-cultivated in a membrane bioreactor with enhanced internal circulation. Bioresour. Technol. 2018, 263, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.; Liu, J.; Ma, T.; Zhao, X.; Ma, H.; Li, J. Coupling of sponge fillers and two-zone clarifiers for granular sludge in an integrated oxidation ditch. Environ. Technol. Innov. 2022, 26, 102264. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.; Ahn, D. Effects of carbon to nitrogen ratio on the performance and stability of aerobic granular sludge. Environ. Eng. Res. 2021, 26, 190284. [Google Scholar] [CrossRef]
- Wang, X.T.; Yang, H.; Su, Y.; Liu, X.Y. Characteristics and mechanism of anammox granular sludge with different granule size in high load and low rising velocity sewage treatment. Bioresour. Technol. 2020, 312, 123608. [Google Scholar] [CrossRef] [PubMed]
- Kalavathi, F.D.; Uma, L.; Subramanian, G. Degradation and metabolization of the pigment—Melanoidin in distillery effluent by the marine cyanobacterium Oscillatoria boryana BDU 92181. Enzym. Microb. Technol. 2001, 29, 246–251. [Google Scholar] [CrossRef]
- Sequence Read Archive (SRA), National Center for Biotechnology Information. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 27 September 2022).
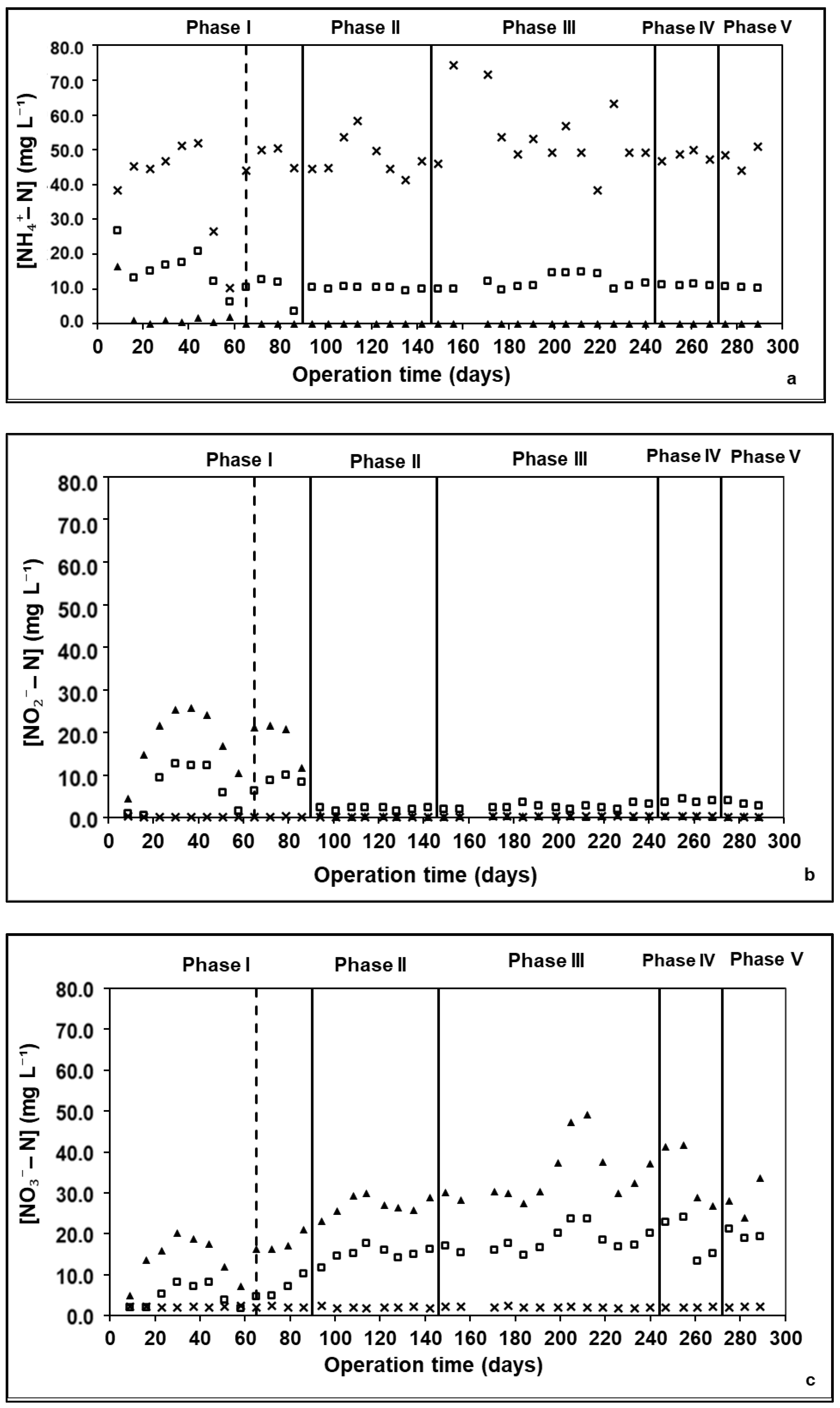


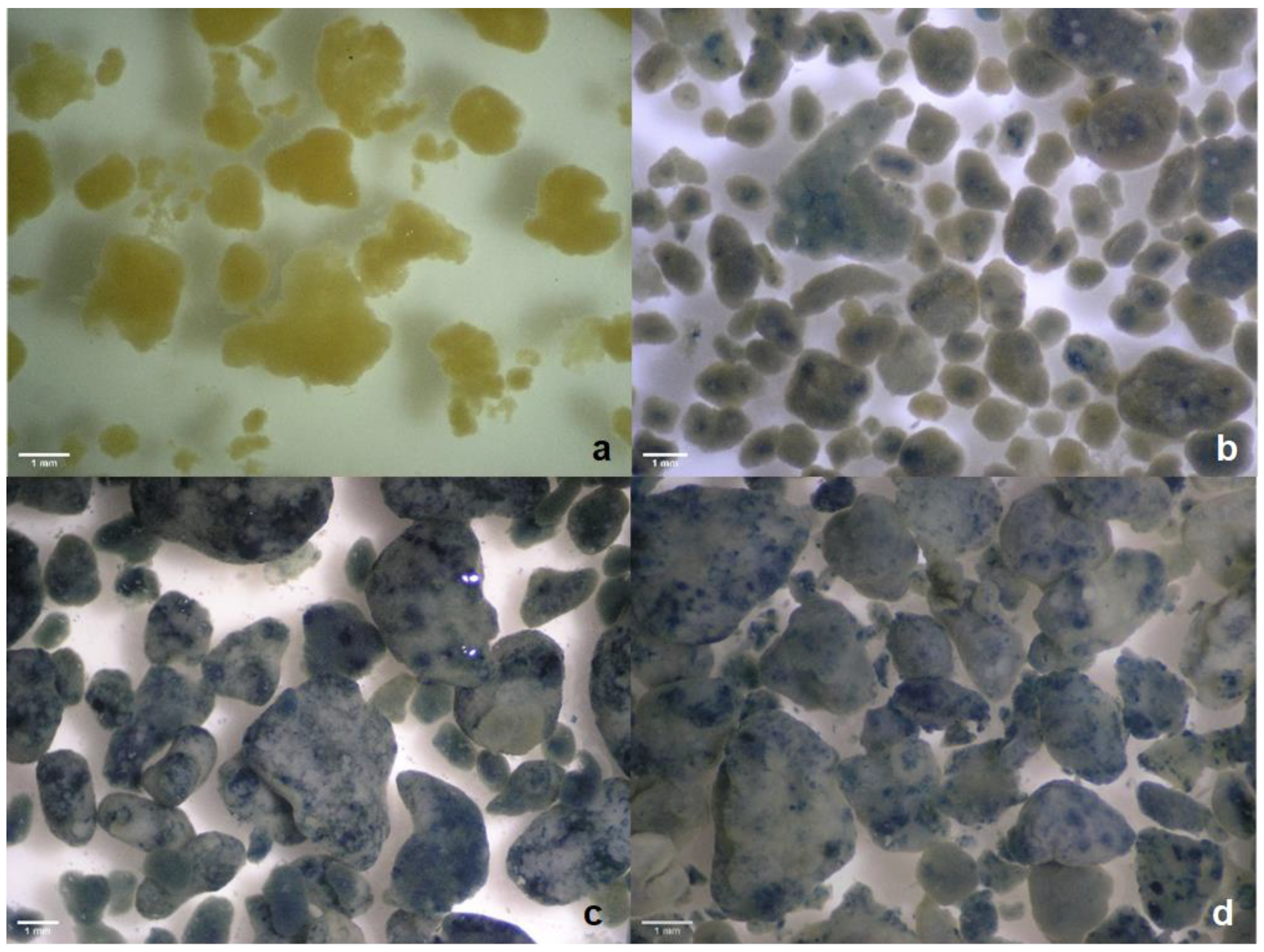
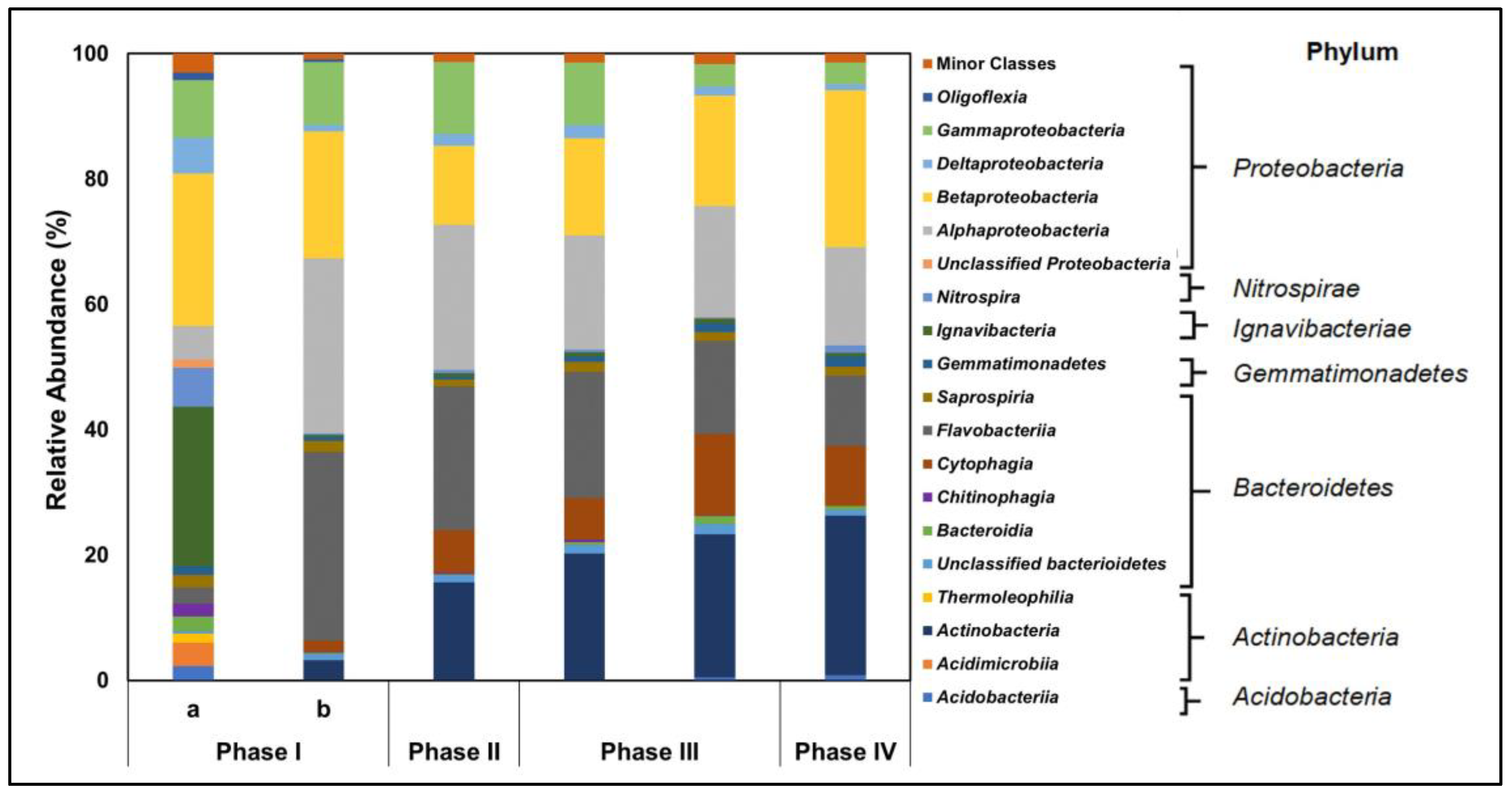
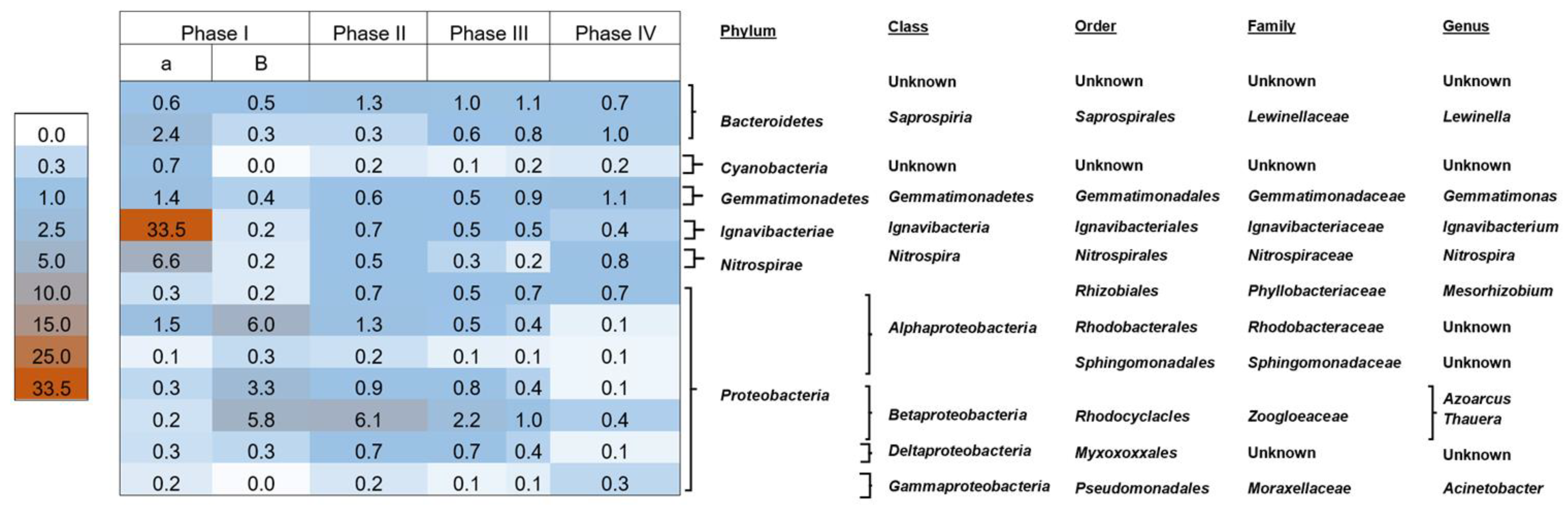
| Phase | Length of Operation (Days) | Days of Operation | Cycle Time (h) | Anaerobic Feeding (min) | Aeration (min) | Inlet Acetate Concentration (mM) | Inlet Dye Concentration (mg L−1) |
|---|---|---|---|---|---|---|---|
| I | 0–89 | 90 | 3 (0–64 d) | 60 | 85–112 | 5.9 | - |
| 6 (65–89 d) | 60 | 292 | |||||
| II a | 90–145 | 55 | 6 | 60 | 292 | 5.9 | 25 |
| III a | 146–243 | 97 | 6 | 60 | 292 | 5.9 | 15 |
| IV a | 244–271 | 27 | 6 | 60 | 292 | 5.9 | 7.5 |
| V | 272–289 | 17 | 6 | 60 | 292 | 5.9 | - |
| Phase | MLSS (g L−1) | MLVSS (g L−1) | MLVSS/ MLSS | SVI5 (mL g−1sst) | SRT (Days) |
|---|---|---|---|---|---|
| I | 9.7 | 8.82 | 0.91 | 20.5 | 117.4 |
| II | 9.0 | 8.26 | 0.92 | 16.7 | 90.2 |
| III | 10.9 | 9.9 | 0.91 | 17.5 | 97.4 |
| IV | 12.3 | 11.2 | 0.90 | 19.3 | 142.2 |
| V | 20.7 | 18.9 | 0.91 | 14.5 | 231.3 |
| [NE] Mass Balance (mg) | ||||
|---|---|---|---|---|
| Phase | [NE] in the Inlet | [NE] in the Effluent | % of Removal | Adsorption (mg g−1 VSS) |
| I | - | - | - | - |
| II | 464.4 | 226.6 | 51,6 ± 8.9 | 3.9 ± 1.1 |
| III | 284.9 | 131.7 | 54.1 ± 7.0 | 1.86 ± 0.25 |
| IV | 53.5 | 21.1 | 60.6 ± 4.1 | 0.98 ± 0.08 |
| V | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mendes, M.; Moreira, I.S.; Moreira, P.; Pintado, M.; Castro, P.M.L. Bioaugmentation of Aerobic Granular Sludge with Dye-Decolorizing Yeast for Textile Industrial Wastewater. Processes 2023, 11, 1654. https://doi.org/10.3390/pr11061654
Mendes M, Moreira IS, Moreira P, Pintado M, Castro PML. Bioaugmentation of Aerobic Granular Sludge with Dye-Decolorizing Yeast for Textile Industrial Wastewater. Processes. 2023; 11(6):1654. https://doi.org/10.3390/pr11061654
Chicago/Turabian StyleMendes, Marta, Irina S. Moreira, Patrícia Moreira, Manuela Pintado, and Paula M. L. Castro. 2023. "Bioaugmentation of Aerobic Granular Sludge with Dye-Decolorizing Yeast for Textile Industrial Wastewater" Processes 11, no. 6: 1654. https://doi.org/10.3390/pr11061654
APA StyleMendes, M., Moreira, I. S., Moreira, P., Pintado, M., & Castro, P. M. L. (2023). Bioaugmentation of Aerobic Granular Sludge with Dye-Decolorizing Yeast for Textile Industrial Wastewater. Processes, 11(6), 1654. https://doi.org/10.3390/pr11061654








