Isolation and Characterization of Novel Butachlor-Degrading Bacteria from Rice Paddy Soils
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Media and Culture Condition
2.3. Enrichment and Isolation of Butachlor-Degrading Strains
2.4. Identification of Butachlor-Degrading Strain
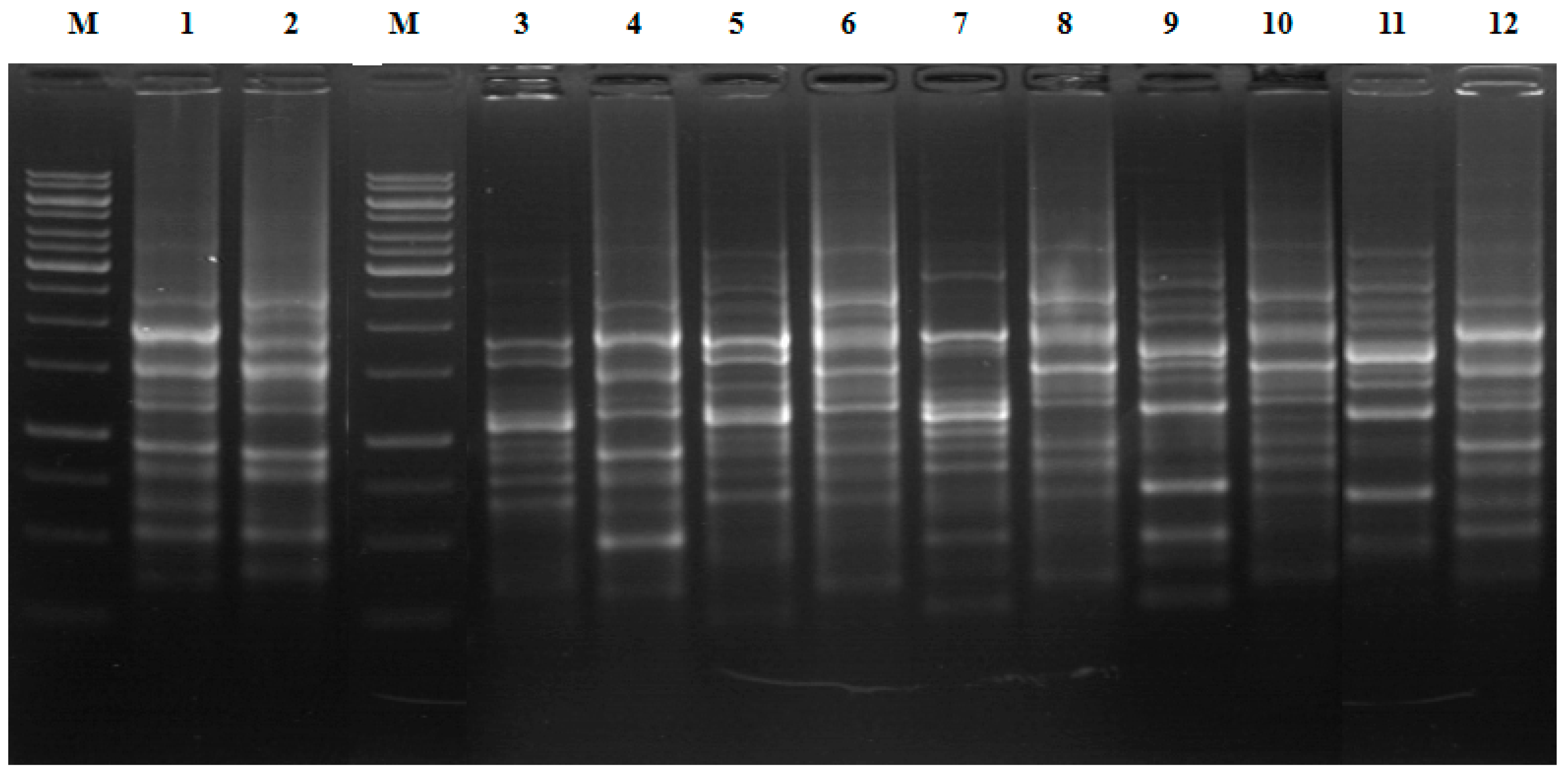
2.5. Colony REP-PCR
2.6. Analysis of Butachlor Degradation
2.7. Identification of Metabolic Intermediates
2.8. Degradation of Butachlor Intermediates
3. Results
3.1. Isolation of Butachlor-Degrading Bacteria
3.2. 16S rRNA Gene Sequence and REP-PCR Analyses
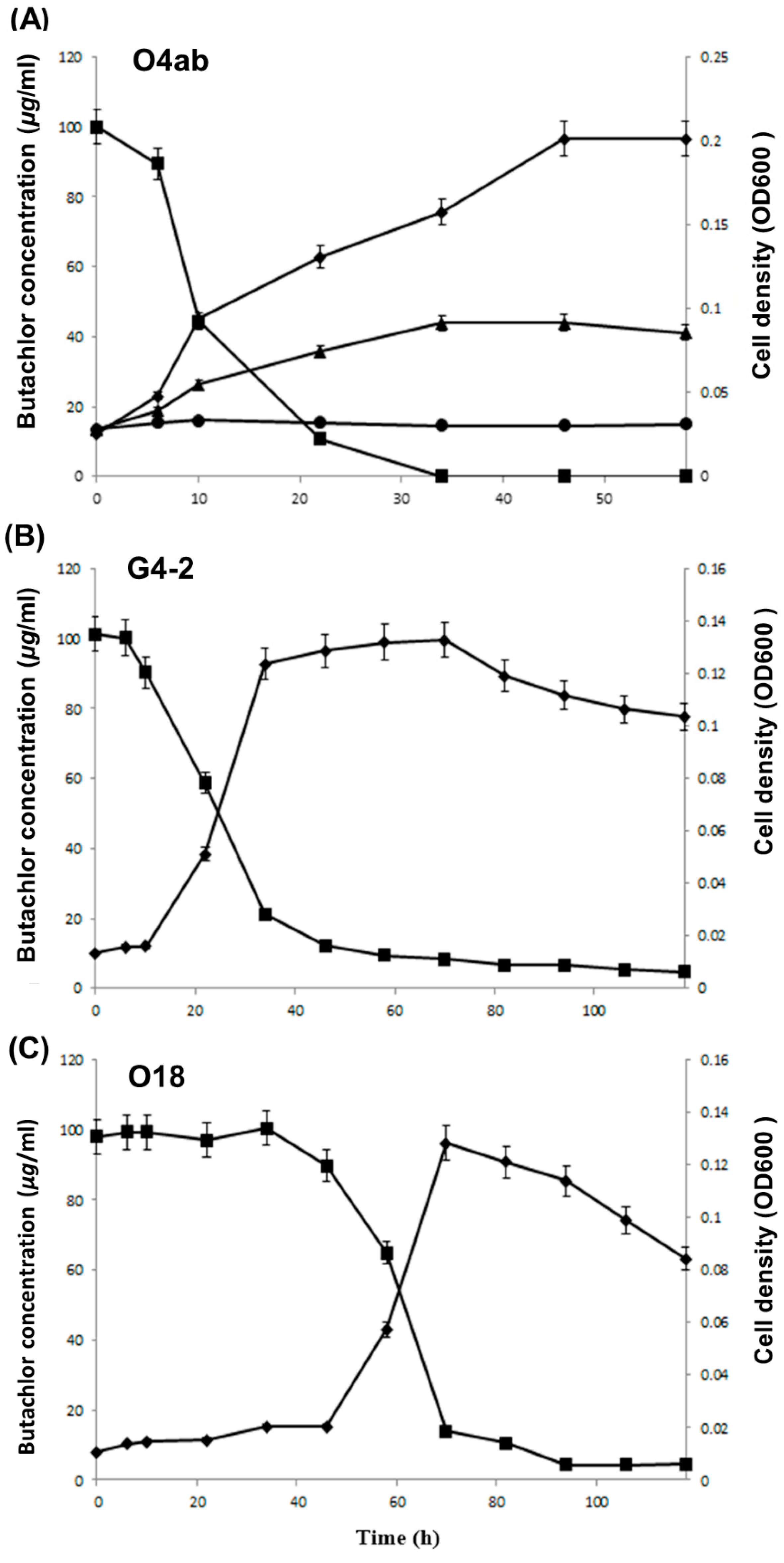
3.3. Growth Characteristics and Degradation Phenotype
3.4. Identification of Metabolites Produced during Butachlor Degradation
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Yu, Y.; Chen, Y.; Luo, Y.; Pan, X.; He, Y.; Wong, M. Rapid degradation of butachlor in wheat rhizosphere soil. Chemosphere 2003, 50, 771–774. [Google Scholar] [CrossRef] [PubMed]
- Dwivedi, S.; Singh, B.R.; Al-Khedhairy, A.A.; Alarifi, S.; Musarrat, J. Isolation and characterization of butachlor-catabolizing bacterial strain Stenotrophomonas acidaminiphila JS-1 from soil and assessment of its biodegradation potential. Lett. Appl. Microbiol. 2010, 51, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Lo, Y.C.; Yang, C.C.; Deng, J.F. Acute alachlor and butachlor herbicide poisoning. Clin. Toxicol. 2008, 46, 716–721. [Google Scholar] [CrossRef]
- Singh, J.; Kadapakkam Nandabalan, Y. Prospecting Ammoniphilus sp. JF isolated from agricultural fields for butachlor degradation. 3 Biotech 2018, 8, 164. [Google Scholar] [CrossRef]
- Kaur, R.; Goyal, D.; Agnihotri, S. Chitosan/PVA silver nanocomposite for butachlor removal: Fabrication, characterization, adsorption mechanism and isotherms. Carbohydra. Polym. 2021, 262, 117906. [Google Scholar] [CrossRef]
- Abigail, M.E.A.; Samuel, S.M.; Ramalingam, C. Addressing the environmental impacts of butachlor and the available remediation strategies: A systematic review. Int. J. Environ. Sci. Technol. 2015, 12, 4025–4036. [Google Scholar] [CrossRef]
- Götz, T.; Böger, P. The very-long-chain fatty acid synthase is inhibited by chloroacetamides. Zeitschrift fur Naturforsch. Sect. C. J. Biosci. 2004, 59, 549–553. [Google Scholar] [CrossRef] [PubMed]
- Panneerselvam, N.; Sinha, S.; Shanmugam, G. Butachlor is cytotoxic and genotoxic and induces apoptosis in mammalian cells. Environ. Mol. Mutagen. 1998, 31, 257–262. [Google Scholar]
- Min, H.; Ye, Y.F.; Chen, Z.Y.; Wu, W.X.; Yufeng, D. Effects of butachlor on microbial populations and enzyme activities in paddy soil. J. Environ. Sci. Health 2001, 36, 581–595. [Google Scholar] [CrossRef]
- Muthukaruppan, G.; Janardhanan, S.; Vijayalakshmi, G. Sublethal toxicity of the herbicide butachlor on the earthworm Perionyx sansibaricus and its histological changes. J. Soils. Sediments 2004, 5, 82–86. [Google Scholar] [CrossRef]
- Tilak, K.S.; Veeraiah, K.; Thathaji, P.B.; Butchiram, M.S. Toxicity studies of butachlor to the freshwater fish Channa punctata (Bloch). J. Environ. Biol. 2007, 28, 485–487. [Google Scholar] [PubMed]
- Coleman, S.; Linderman, R.; Hodgson, E.; Rose, R.L. Comparative metabolism of chloroacetamide herbicides and metabolites in human and rat liver microsomes. Environ. Health Perspect. 2000, 108, 1151–1157. [Google Scholar]
- Liu, H.; Cao, L.; Lu, P.; Ni, H.; Li, Y.X.; Yan, X.; Hong, Q.; Li, S.P. Biodegradation of butachlor by Rhodococcus sp. strain B1 and purification of its hydrolase (ChlH) responsible for N-dealkylation of chloroacetamide herbicides. J. Agric. Food. Chem. 2012, 6, 12238–12244. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Lu, Y.T.; Chen, Y.Y. Comparative proteome analysis of butachlor-degrading bacteria. Environ. Geol. 2008, 53, 1339–1344. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, C.H.; Deng, S.K.; Wu, Y.D.; Li, Y.; Yao, L.; Jiang, J.D.; Yan, X.; He, J.; Li, S.P. Novel three-component Rieske non-heme iron oxygenase system catalyzing the N-dealkylation of chloroacetanilide herbicides in Sphingomonads DC-6 and DC-2. Appl. Environ. Microbiol. 2014, 80, 5078–5085. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Chen, Q.; Wang, G.X.; Ni, H.Y.; He, J.; Yan, X.; Gu, J.G.; Li, S.P. Sphingomonas chloroacetimidivorans sp. nov., a chloroacetamide herbicide-degrading bacterium isolated from activated sludge. Antonie Van Leeuwenhoek 2015, 108, 703–710. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zheng, J.W.; Liang, B.; Wang, C.H.; Cai, S.; Ni, Y.Y.; He, J.; Li, S.P. Biodegradation of chloroacetamide herbicides by Paracoccus sp. FLY-8 in vitro. J. Agric. Food Chem. 2011, 59, 4614–4621. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Li, R.; Zhu, J.; Zhang, J.; He, J.; Li, S.; Jiang, J. Degradation of the chloroacetamide herbicide butachlor by Catellibacterium caeni sp. nov DCA-1T. Int. Biodeterior. Biodegrad. 2012, 73, 16–22. [Google Scholar] [CrossRef]
- Baker, G.C.; Smith, J.J.; Cowan, D.A. Review and re-analysis of domain-specific 16S primers. J. Microbiol. Methods 2003, 55, 541–555. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.J. 16S/23S rRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Lee, S.; Malone, C.; Kemp, P. Use of multiple 16S rRNA targeted fluorescent probes to increase signal strength and measure cellular RNA from natural planktonic bacteria. Mar. Ecol. Prog. Ser. 1993, 101, 193–201. [Google Scholar] [CrossRef]
- Kim, M.; Chun, J. 16S rRNA gene-based identification of bacteria and archaea using the EzTaxon server. In Methods in Microbiology; Academic Press: Cambridge, MA, USA, 2014; Volume 41, pp. 61–74. [Google Scholar]
- Bruijin, F.J.D. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl. Environ. Microbiol. 1992, 58, 2180–2187. [Google Scholar] [CrossRef]
- Torsvik, V.; Sorheim, R.; Goksoyr, J. Total bacterial diversity in soil and sediment communities—A review. J. Ind. Microbiol. 1996, 17, 170–178. [Google Scholar] [CrossRef]
- Xiong, M.; Zang, H.; Pan, J.; Xu, C.; Li, C. Isolation of a novel chlorimuron-ethyl-degrading bacterium Rhodococcus sp. D310-1. In Proceedings of the iCBBE, Wuhan, China, 10–12 May 2011; pp. 1–4. [Google Scholar]
- Geminia, V.; Gallegoa, A.; Oliveirab, V.D.; Gomezc, C.; Manfiob, G.; Korola, S. Biodegradation and detoxification of p-nitrophenol by Rhodococcus wratislaviensis. Int. Biodeterior. Biodegrad. 2005, 55, 103–108. [Google Scholar] [CrossRef]
- Osano, O.; Admiraal, W.; Klamer, H.J.C.; Pastor, D.; Bleeker, E.A.J. Comparative toxic and genotoxic effects of chloroacetanilides, formamidines and their degradation products on Vibrio fischeri and Chironomus riparius. Environ. Pollut. 2002, 119, 195–202. [Google Scholar] [CrossRef]
- Feng, P.C.C.; Wratten, S.J. In vitro oxidation of 2, 6-diethylaniline by rat liver microsomal enzymes. J. Agric. Food Chem. 1987, 35, 491–496. [Google Scholar] [CrossRef]


| Isolate | Genebank Accession Number | Sampling Site | Nearest Type Strain | Similarity (%) |
|---|---|---|---|---|
| G4-2 | JX101328 | Yeongju | Sphingobium chunbukense DJ77T | 98.4 |
| G3 | JX979142 | Yeongju | Sphingobium chunbukense DJ77T | 98.2 |
| O18 | JX979134 | Wonju | Sphingobium chlorophenilicum ATCC 33790T | 98.4 |
| K8 | JX979135 | Ansan | Sphingobium chunbukense DJ77T | 98.4 |
| O4a | JX979150 | Jangsung | Rhodococcus qingshengii djl-6T | 100.0 |
| O4b | JX979146 | Jangsung | Sphingobium chungbukense DJ77T | 98.6 |
| L9a | JX979140 | Ansan | Rhodococcus qingshengii djl-6T | 99.9 |
| L9b | JX979136 | Ansan | Sphingobium chungbukense DJ77T | 98.8 |
| J12a | JX979132 | Pyeongtaek | Rhodococcus ruber DSM 43338T | 99.7 |
| J12b | JX979141 | Pyeongtaek | Sphingobium chungbukense DJ77T | 98.8 |
| J11a | JX979145 | Pyeongtaek | Rhodococcus ruber DSM 43338T | 100.0 |
| J11b | JX979138 | Pyeongtaek | Sphingobium chungbukense DJ77T | 98.7 |
| H4a | JX979148 | Chungju | Rhodococcus ruber DSM 43338T | 100.0 |
| H4b | JX979144 | Chungju | Sphingobium chungbukense DJ77T | 98.4 |
| Characteristics | G4-2 | O18 | O4a | O4b |
|---|---|---|---|---|
| Utilization of | ||||
| Butachlor | + | + | + | − |
| Alachlor | − | − | − | − |
| CDEPA | + | + | − | + |
| DEA | + | + | + | + |
| Aniline | − | − | − | − |
| Catechol | + | + | + | + |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, H.; Kim, N.H.; Kim, D.-U. Isolation and Characterization of Novel Butachlor-Degrading Bacteria from Rice Paddy Soils. Processes 2023, 11, 1222. https://doi.org/10.3390/pr11041222
Lee H, Kim NH, Kim D-U. Isolation and Characterization of Novel Butachlor-Degrading Bacteria from Rice Paddy Soils. Processes. 2023; 11(4):1222. https://doi.org/10.3390/pr11041222
Chicago/Turabian StyleLee, Hyosun, Nam Hyun Kim, and Dong-Uk Kim. 2023. "Isolation and Characterization of Novel Butachlor-Degrading Bacteria from Rice Paddy Soils" Processes 11, no. 4: 1222. https://doi.org/10.3390/pr11041222
APA StyleLee, H., Kim, N. H., & Kim, D.-U. (2023). Isolation and Characterization of Novel Butachlor-Degrading Bacteria from Rice Paddy Soils. Processes, 11(4), 1222. https://doi.org/10.3390/pr11041222







