1. Introduction
Statistics from the World Health Organization (WHO) indicate that heart disease is a major threat to humans worldwide [
1]. Heart disease can be caused by many different things, including high blood pressure, obesity, excessive cholesterol, smoking, unhealthy eating habits, diabetes, and abnormal heart rhythms [
2]. Most patients die from heart disease as a result of an inadequate diagnosis at the initial phase. Therefore, it is imperative to use efficient disease classification and prediction algorithms to comprehend disease prediction. In contrast, it is necessary to implement a more accurate model in order to predict heart disease. An assessment of the accuracy of a model to predict heart-related diseases is based on its precision, F1 score, and recall performance. Association rules can also improve the prediction accuracy for heart disease models. The use of association rules on medical datasets produces a number of regulations. Most of these rules do not have any medical relevance. Furthermore, finding them can be time-consuming and impractical. This is due to the fact that the association rules are drawn from the available dataset rather than being based on an independent sample. Hence, to identify early-stage predictions for heart disease, search constraints are applied to actual datasets containing patients with heart disease. Using search constraints, a rule-generation algorithm has been used for the early detection of heart attacks [
3]. Moreover, recent advances in healthcare technology have driven the development of machine learning (ML) systems for the prediction of human health diseases [
4,
5,
6]. There have been many researchers working on the development of improved ML models. The primary objective of the ML technique is to generate computer code that can access and use current data to predict future data [
7]. Additionally, there are some tried-and-true methods for improving the accuracy of the model. These include adding more information to the dataset, treating missing and outlier values, feature selection, algorithm tuning, cross-validation, and ensembling. This paper implements GridsearchCV hyperparameter tuning and five-fold cross-validation to evaluate the model’s performance on both benchmark datasets. It also employs an ensemble voting classifier to improve model accuracy, aiming to enhance ML model accuracy. This article presents the following significant work:
This work examines and implements six major ML algorithms on the Cleveland and IEEE Dataport heart disease datasets, analyzing performance classification metrics.
In the early phase, various ML classifier techniques, including random forest (RF), K-nearest neighbor (KNN), logistic regression (LR), Naive Bayes (NB), gradient boosting (GB), and AdaBoost (AB) were trained.
The GridsearchCV hyperparameter tuning method with five-fold cross-validation and performance assessment using accuracy and negative log loss metrics was employed to achieve the highest level of accuracy.
Finally, all classifiers were combined using a soft voting ensemble method in order to increase the accuracy of the model.
2. Literature Review
Several new research opportunities in healthcare have been enabled by advances in ML and advances in computing capabilities [
8]. Various researchers have proposed ML algorithms to enhance the accuracy of disease prediction [
9,
10,
11]. To refine the precision of the outcomes, much of the research has meticulously evaluated the presence of missing data in the dataset, a crucial aspect in the data preprocessing process. Gupta et al. [
12] used Pearson correlation coefficients and different ML classifiers to replace missing values in the Cleveland dataset. Rani et al. [
13] have investigated multiple imputations by the chained equations (MICE) method to deal with the missing values problem. In this case, missing values are imputed through a series of iterative predictive models. During each iteration, each variable in the dataset is assigned using the other variables. In another work, Jordanov et al. [
14] proposed a KNN imputation method for the prediction of both continuous (average of the nearest neighbors) and categorical variables (most frequent). Another study used an LR model to classify cardiac disease with an accuracy of 87.1% after cleaning the dataset and identifying missing values at the time of preprocessing [
15]. In contrast, some researchers have eliminated missing values. Based on DT, LR, and Gaussian NB algorithms, the features are reduced from 13 to 4 using feature selection method and reported an accuracy of 82.75% [
16]. A hybrid random forest (RF) with the linear model was developed by Mohan et al. [
17] and improved the accuracy of 297 records and 13 characteristics of the Cleveland dataset for heart disease prediction. Kodati et al. [
18] tested several types of classifiers using Orange and Weka data-mining tools to predict heart disease with 297 records and 13 features.
In addition, the feature selection method plays an important role in improving the accuracy of the model. To select features, Shah et al. [
19] utilized probabilistic principal component analysis (PCA). The Cleveland dataset was used by R. Perumal et al. [
20] to develop LR and support vector machine (SVM) models with similar accuracy levels (87% and 85%, respectively). To train the ML classifiers, they used a dataset of 303 data instances and standardized and reduced features using PCA. In another study, a particle swarm optimization (PSO) technique was used to select features [
21]. In contrast, Yekkala et al. [
22] used a rough set-based feature selection method along with the RF algorithm and obtained an accuracy of 84%. Saw et al. [
23] used a random search to find the best parameters to build an accurate prediction model. It was found that this approach uses LR for classification and is 87% accurate at predicting heart attacks. Other works have used both methods and predicted the accuracy using different algorithms. The model presented by Otoom et al. [
24] used NB, SVMs, and available trees to achieve an accuracy of 84.5%. Vembandasamy et al. [
25] proposed an NB classifier for predicting heart disease and achieved an accuracy of 84.4%.
Further, to determine the optimum combination of heart disease predictors, Gazeloglu et al. [
26] evaluated 18 ML models and three feature selection techniques for the Cleveland dataset of 303 instances and 13 variables. Recently, ten classifiers were trained to identify the most effective prediction models for precise prediction [
27]. The most suitable attributes were identified using three methods of attribute selection, including a feature subset evaluator based on correlation, a chi-squared attribute evaluator, and a relief attribute evaluator. Furthermore, a hybrid feature selection method aimed at enhancing accuracy by incorporating RF, AB, and linear correlation was suggested by Pavithra et al. [
28]. The implementation of this technique led to a 2% increase in the accuracy of the hybrid model, following the selection of 11 features through a combination of filter, wrapper, and embedded methods. To further enhance the accuracy, researchers have used the ensemble technique to combine different algorithms. The ensemble method for detecting heart disease was developed by Latha et al. [
29] by combining NB, RF, multilayer perceptrons (MLP), and Bayesian networks based on majority voting (MV). They achieved an accuracy of 85.48%. It was also employed by an ensemble model with five classifiers, including a memory-based learner (MBI), an SVM, DT induction with information gain (DT-IG), NB, and DT initiation with the Gini index (DT-GI) [
30]. As the datasets in the authors’ study contained only pertinent attributes, there was no feature selection. A pre-processing step has been performed to eliminate outliers and missing values from the data. Tama et al. [
31] developed an ensemble model to diagnose heart disease with an accuracy rate of 85.71%. The ensemble model utilized GB, RF, and extreme GB classifiers. Alqahtani et al. [
32] developed an ensemble of ML and deep learning (DL) models to predict the disease with an accuracy rate of 88.70%. This study employed a total of six classification algorithms. Trigka et al. [
33] developed a stacking ensemble model after applying SVM, NB, and KNN with a 10-fold cross-validation synthetic minority oversampling technique (SMOTE) in order to balance out imbalanced datasets. This study demonstrated that a stacking SMOTE with a 10-fold cross-validation achieved an accuracy of 90.9%. Another study used stochastic gradient descent classifiers, LR, and SVM to develop a model with an accuracy of 93% using multiple datasets [
34]. For further improving accuracy, Cyriac et al. [
35] utilized seven different machine-learning models as well as two ensemble methods (soft voting and hard voting). With this approach, the highest accuracy score was achieved at 94.2%. Another study developed a combined multiple-classifier predictive model approach for better prediction accuracy [
36]. Five classifier models are combined with Cleveland and Hungarian datasets. A total of 590 data-valid instances and 13 attributes were taken into consideration. A baseline accuracy of 93% was achieved using the Weka data-mining tool.
In 2020, Manu Siddhartha created a new dataset by combining five well-known heart disease datasets—Switzerland, Cleveland, Hungary, Statlog, and Long Beach VA. This new dataset includes all the characteristics shared by the five datasets [
37]. In the same dataset, Mert Ozcan et al. [
38] investigated the use of a supervised ML technique known as the Classification and Regression Tree (CART) algorithm to predict the prevalence of heart disease and to extract decision rules that clarify the associations between the input and output variables. The outcomes of the investigation further ranked the heart disease influencing features based on their significance. The model’s reliability was corroborated by an 87% accuracy in the prediction. Other researchers Rüstem Yilmaz et l. [
39] worked to compare the predictive classification performances of ML techniques for coronary heart disease. Three distinct models using RF, LR, and SVM algorithms were developed. Hyper-parameter optimization was performed using a 10-fold repeated cross-validation approach. Model performance was assessed using various metrics. Results showed that the RF model exhibited the highest accuracy of 92.9%, specificity of 92.9%, sensitivity of 92.8%, F1 score of 92.8%, and negative predictive and positive predictive values of 92.9% and 92.8%, respectively.
In the field of predictive modeling, there is a constant pursuit to enhance the accuracy of classification and forecast models. The classification models are deployed to label data points while forecast models are used to predict future values. A suitable combination of models and features can enhance the accuracy of these models. Bhanu Prakash Doppala et.al [
40] proposed a model that was evaluated on diverse datasets to determine its efficacy in improving accuracy. The evaluation involved testing the model on three datasets: the Cleveland dataset, a comprehensive dataset from IEEE Dataport, and a cardiovascular disease dataset from the Mendeley Data Center. The results of the proposed model exhibited high accuracy rates of 96.75%, 93.39%, and 88.24% on the respective datasets.
In contrast to the above work, the ensemble classifier is implemented using six ML models on the Cleveland heart disease dataset [
41] and the IEEE Dataport heart disease datasets (comprehensive) [
42]. This study used six ML algorithms: RF, KNN, LR, NB, GB, and AB. A GridsearchCV hyperparameter method and five-fold cross-validation methods were employed to obtain the best accuracy results before implementing the models. The hyperparameter values provided by GridsearchCV enhance the accuracy of the model. Using these parameters, the accuracy of six different algorithms is verified and the most accurate algorithm is determined. Additionally, the ensemble method was applied to the proposed algorithms in order to enhance their accuracy. This method boosts overall model accuracy from 90.16% (LR) to 93.44%, and from 90% (AB) to 95% using soft-voting ensemble classifiers on Cleveland and IEEE datasets.
3. Resources and Approaches
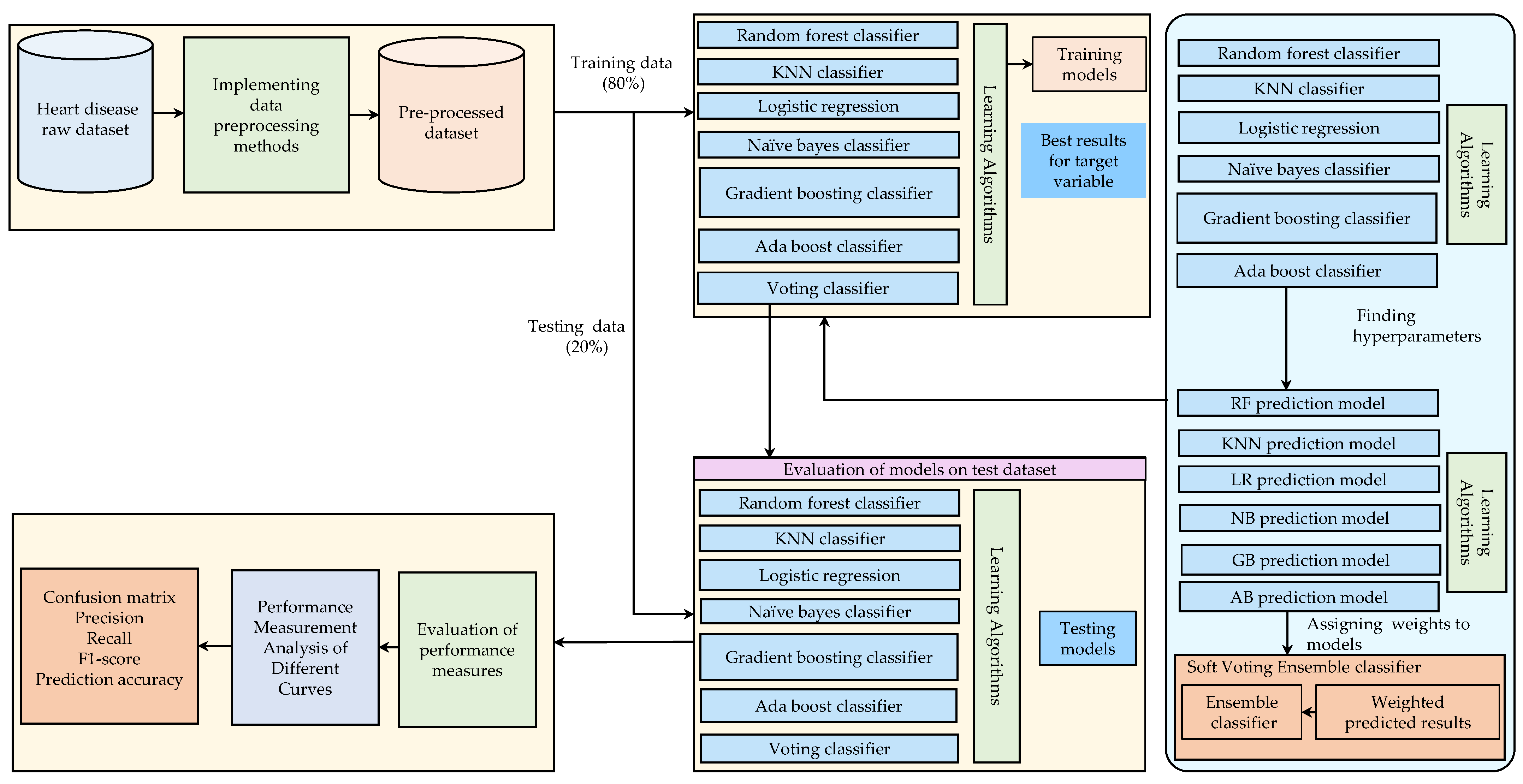
This section describes the methods to predict heart disease using the two benchmark publicly available datasets. This study consists of various phases, from the collection of data to the prediction of heart disease. In the first phase, data can be pre-processed using feature scaling and data transformation methods. The proposed model is built using multiple ML algorithms as the next step. An ensemble approach is used in the next phase of the process to enhance the model’s accuracy.
Figure 1 shows a detailed diagram of the workflow architecture.
3.1. Description of the Datasets
Dataset I: The UC Irvine ML Repository-Cleveland dataset, which contains 303 instances and 14 attributes, is included in this dataset.
Dataset II: The IEEE Dataport heart disease dataset (comprehensive) comprises 12 multivariate attributes and 1190 instances, which are included in this dataset.
The availability of these datasets can accelerate research in the field of heart disease prediction and lead to the development of more accurate and effective diagnostic tools.
3.2. Data Pre-Processing
This study preprocesses Dataset I and Dataset II heart disease datasets before constructing the predictive model with ML. Since these datasets have undergone extensive preprocessing and cleaning, they are easier to use and require less time and effort for data preparation. Additionally, they are well documented and frequently cited in scientific literature.
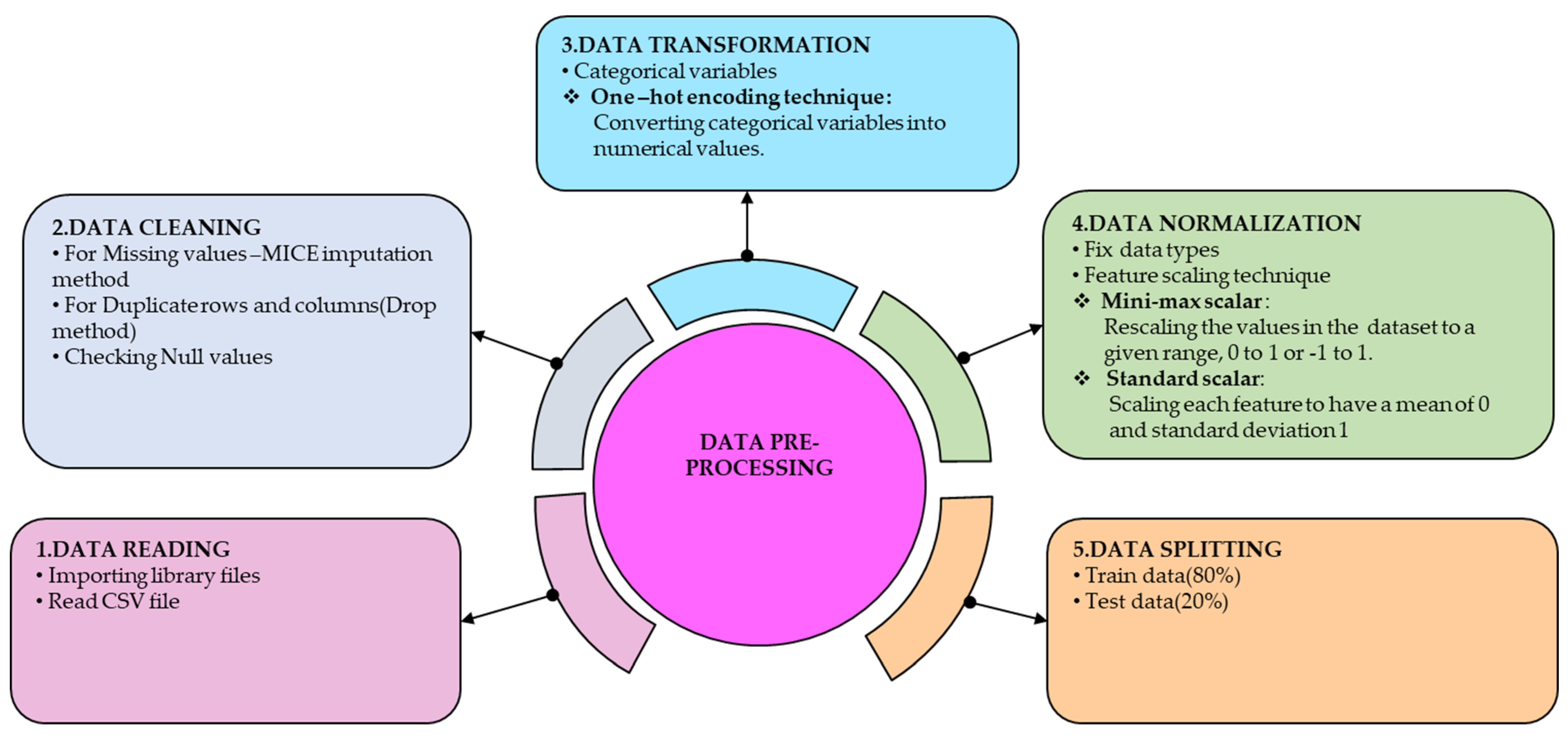
In both datasets, a target attribute integer value indicates the presence of a patient’s heart disease. If the value is 0, there is no heart disease, and if it is 1, there is heart disease. Based on gender, the attribute ‘sex’ consists of two classes: 1 for males and 0 for females. There are four classes of chest pain in the attribute ‘cp’ (chest pain type) two classes of fasting blood sugar in the attribute ‘fbs’ (fasting blood sugar) three classes of resting electrocardiograms in the attribute ‘restecg’ (resting ecg) and two classes of exercise in the attribute ‘exang’ (exercise angina). Additionally, ‘slope’ (ST slope) is composed of three classes. The remaining four attributes, such as ‘trestbps’ (resting bp s), ‘chol’ (cholesterol), ‘age’, and ‘oldpeak’, are considered numerical values. There are different steps involved in data pre-processing, from reading data to splitting data for training and testing. The steps are illustrated in
Figure 2.
The data pre-processing process begins by identifying and addressing missing or duplicate values in the dataset. The presence of missing data in a dataset implies incompleteness, which can affect the statistical significance of the results obtained from the model analysis. When data are missing, the overall accuracy and validity of the analysis may be compromised. Therefore, to maximize the effectiveness of the analysis, it is recommended to fill in missing values either with a user-defined constant or the average value of the dataset, rather than completely suppressing the observations. The detailed explanation of each attribute presented in
Table 1 and
Table 2 describes both datasets. Initially, Dataset I contained 303 instances, out of which one duplicate row was removed. As a result, a dataset of 302 unique instances was obtained, with 164 instances corresponding to patients with heart disease and 138 instances corresponding to patients without heart disease. In Dataset II, we identified no missing values. Additionally, 272 duplicate instances have been identified. Therefore, to complete the dataset, the duplicate instance was removed. Among these, 508 instances correspond to patients with heart disease, and the remaining 410 instances belong to patients without heart disease.
However, some attributes in the data have large input values that are incompatible with other attributes, which results in poor learning performance. Therefore, to make it compatible with other attributes, data exploration was performed to visually explore and identify relationships between them. This is accomplished through the use of a one-hot encoding method. One-hot encoding is performed using features such as cp, thal, and slope for the available datasets. Those three features are further subdivided into cp_0 to cp_3,thal_0 to thal_3, and slope_0 to slope_2 features and merged into the original datasets. After exploring the data, the data were scaled for further processing. This is essential when using the dataset for a KNN. In order to make it compatible with all algorithms, a large number of features have been scaled down. As a result, ML models perform better.
Feature scaling involves two essential techniques called standardization and normalization. In standardization, the mean is subtracted from the distribution shifts and divided by the standard deviation. The act of subtracting the average from the data points is referred to as centering while dividing each data point by the standard deviation is called scaling. Standardization helps maintain the presence of outliers, making the resulting algorithm less susceptible to influences compared to one that has not undergone standardization. Standardizing a value can be accomplished using the following equations from (1) to (3).
Here,
x is the participation value,
x’ is the standardized value,
μ the mean, and
σ is the standard deviation. These can be calculated as follows:
When referring to a dataset, N represents the total number of columns in the attribute being scaled. From the available dataset, age, trestbps, chol, and oldpeak features have large dimensional values. Hence, the standard scalar is used to convert these feature values into uniform scaling.
After scaling the large feature values, min-max scaling is applied for normalization. This technique is appropriate for data distributions that do not follow a Gaussian distribution. As a result of normalization, feature values become bounded intervals between the minimum and maximum. For min-max scaling, normalize the data using Equation (4) below.
Here, and are the minimum and maximum values of the respective feature in the dataset. With the use of the above equation, all the features are normalized [0,1]. The last step in pre-processing involved dividing the data into two subsets, known as training and testing data, after normalizing the data. The split was carried out in such a way that 80% of the available data was allocated for training and the remaining 20% for testing. This division enabled the training and evaluation of various ML classifiers by testing their accuracy using the training and testing datasets. An exploratory data analysis (EDA) is also conducted prior to discussing each algorithm used to predict heart disease. A description of the descriptive statistics and the information regarding the correlation matrix cannot be presented here for brevity.
3.3. Performance Measures
This study applied various ML algorithms such as RF, KNN, LR, NB, GB, and AB to predict heart disease. Before using the ML algorithms, a number of matrices such as the confusion matrix, receiver operating characteristics (ROC), the area under curve (AUC), learning curve, and precision-recall curve are briefly described in the following subsection.
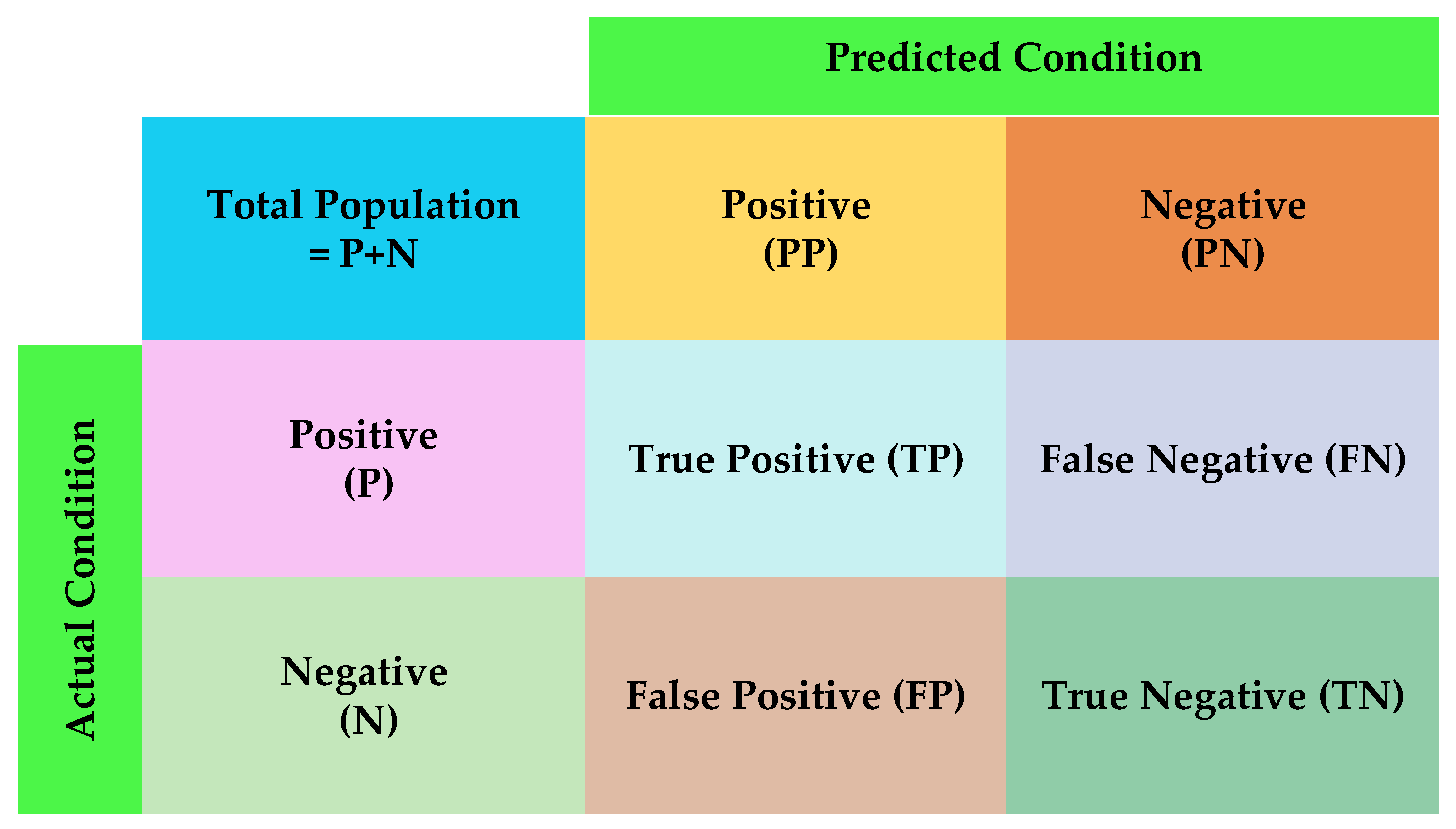
The confusion matrix provides a visual representation of the algorithm’s performance. The confusion matrix table makes it easy to visually inspect the prediction errors. The confusion matrix depicted in
Figure 3 comprises four components: true negatives (TNs), false positives (FPs), false negatives (FNs), and true positives (TPs). The matrix showcases actual class instances as rows and predicted class instances as columns (or vice versa) [
43]. The confusion matrix serves not only as a visual representation of errors, but can also include various metrics such as precision, recall, and F1. Each metric holds its significance and is applied in specific situations.
It is calculated based on the total number of predictions made by the model. The percentage of correct predictions is then divided by the total number of predictions [
44]. This can be defined as the ratio of the TP to the total prediction (TP + FP) made by the model. It can be expressed as an equation in (5).
A second significant metric is recall, which is also known as sensitivity or the true positive rate [
45]. This can be determined by determining the proportion of positive observations that were accurately predicted in relation to the overall number of positive observations. Thus, recall indicates the range of positive classes. As an equation, it can be written as (6).
A good classifier should have precision and recall of one, which corresponds to a FP and FN equal to zero. It is better to consider both precision and recall if the cost of the FP and FN is very different. Consequently, precision and recall need to be considered when there is an uneven distribution of classes. Therefore, the F1 score can be regarded as a measure of both precision and recall [
46].
The F1 score is obtained by taking the average of precision and recall. This metric has generally been considered to be a reliable method for comparing the performance of different classifiers, particularly when the data are unbalanced. F1 scores are calculated by considering both the number of prediction errors and the type of errors the model makes. As an equation, it can be written as (7).
- 2.
ROC curve and AUC
ROC curves are utilized as a means of evaluating the performance of classification algorithms. The curve plots the true positive rate (TPR), also referred to as recall, against the false positive rate (FPR) at various threshold values [
47]. The TPR is calculated using Equation (6), while the FPR is determined through Equation (8). This representation helps to distinguish between the actual positive results and false results (noise).
The TPR is plotted on the
Y-axis, while FPR is plotted on the
X-axis. Thus, it is necessary to utilize a method referred to as AUC in order to calculate the values at any threshold level efficiently [
48]. AUC measures the performance of a classifier across different thresholds as indicated by the ROC curve. In general, the AUC value ranges from 0 to 1, which suggests a good model will have an AUC close to 1, which indicates a high degree of separation. The ROC curve represents how well a classification model performs across all classification thresholds. On this curve, two parameters are plotted. The ROC space is divided by the diagonal. Points above the diagonal indicate successful classification; points below the line indicate unsuccessful classification. The valuation of the AUC curve is explained in
Table 3.
- 3.
ROC curve and AUC
Using a learning curve, we can determine how much more training data will benefit our model. It illustrates the relationship between training and test scores for a ML model with a variable number of training samples. The cross-validation procedure is carried out behind the scenes when we call the learning curve.
- 4.
ROC curve and AUC
Plotting recall on the
x-axis and precision on the
y-axis obtains the precision-recall curve. This curve depicts the false positive to false negative ratio. The precision-recall curve is not constructed using the number of true negative results [
49].
3.4. Accuracy and Loss of Each Fold Measurement
In ML classifiers, the accuracy and loss of each fold have a significant impact on the model’s overall performance. The accuracy of each fold determines how well the model has learned from the training data and how accurately it can predict new data. If the accuracy of a fold is high, it indicates that the model has successfully learned the underlying patterns in the data and can make accurate predictions. However, if the accuracy of a fold is low, it implies that the model needs further improvement and fine-tuning to achieve better results.
Similarly, the loss function of each fold plays a crucial role in determining the model’s performance. The loss function measures how well the model can approximate the actual values of the target variable. A low loss value indicates that the model is fitting the training data well and has the potential to perform well on new data. On the other hand, a high loss value suggests that the model is not fitting the training data well, and more refinement is necessary to improve its performance. Finally, both accuracy and loss of each fold are essential metrics that impact the performance of ML classifiers.
Log loss, also known as cross-entropy loss, is a measure of the performance of a classification model. It measures the difference between the predicted probabilities of the model and the actual outcomes. In binary classification problems, the log loss formula can be expressed as in Equation (9).
where y is the true label (either 0 or 1), p is the predicted probability of the positive class, and the log is the natural logarithm. The log loss ranges from 0 to infinity, with a perfect model having a log loss of 0. A model that always predicts the same probability for all samples would have a log loss of approximately 0.693. Log loss penalizes highly confident but wrong predictions more than it penalizes predictions that are only slightly wrong. As a result, it is a popular loss function for classification problems where the focus is on predicting probabilities rather than hard class labels.
4. ML Classification Algorithms and Experimental Data Analysis
4.1. Hyperparameter Tuning and Experimental Results
Optimizing an ML model’s performance is essential before its implementation to ensure it achieves the highest possible precision. This optimization process entails the careful adjustment of specific variables called hyperparameters, which govern the model’s learning behavior. Fine-tuning a model typically involves fitting it to a training dataset multiple times with various hyperparameter combinations, ultimately determining the ideal configuration for improved performance.
One efficient method for exploring the optimal hyperparameter values is with GridSearchCV, a technique that involves creating a comprehensive grid of potential hyperparameter values.
Table 4 and
Table 5 provide a list of hypermeter tuning values for six ML classifiers.
4.2. Random Forest Classifier
The RF classifier makes predictions by averaging the predictions of their real trees. RF is an ensemble-learning-based method for supervised ML [
50]. It utilizes bagging to combine multiple decision trees, thereby improving the accuracy of predictions. Bagging training is provided on an individual basis to each individual. As part of the training process, each decision tree is evaluated using different samples of data that were generated randomly using replacements from the original dataset. When constructing trees, a random selection of features is also made. A majority vote is used to combine the predictions of multiple trees [
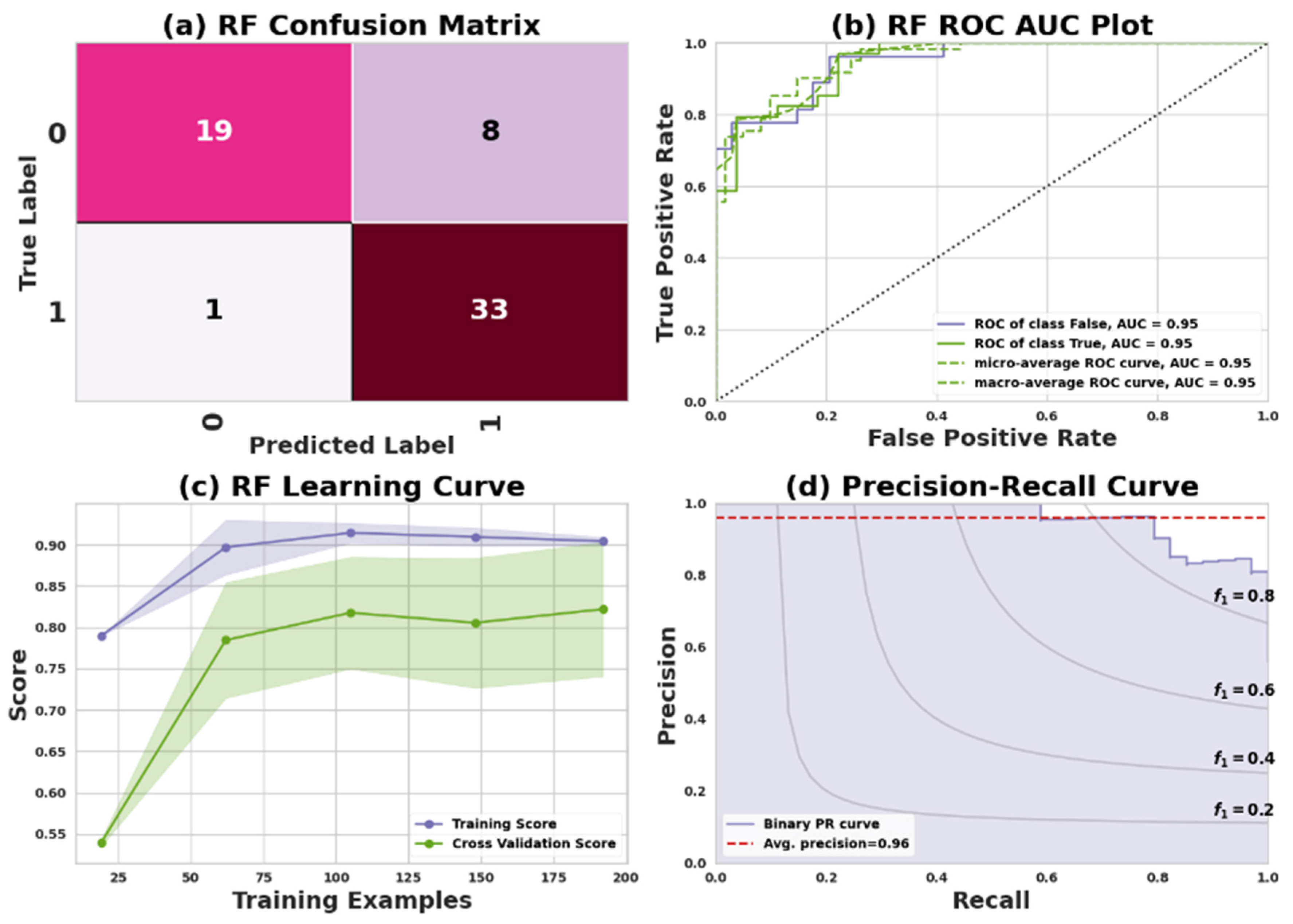
51]. For Dataset I, the model’s confusion matrix revealed that it successfully predicted 19 positive cases and 33 negative cases. However, there were nine incorrect predictions, consisting of eight false negatives and one false positive. In the case of Dataset II, the confusion matrix showed that the model accurately predicted 71 positive cases and 92 negative cases, but it also made 21 incorrect predictions, which included 17 false negatives and 4 false positives.
Table 6 showcases the performance of the RF in predicting heart disease for two datasets: Dataset I (Cleveland) and Dataset II (IEEE Dataport). The metrics used to evaluate the model include precision, recall, and F1 score, for both classes, 0 (no heart disease) and 1 (having heart disease).
For Dataset I, Class 0 has a precision of 95%, recall of 70%, F1 score of 81%, and 27 instances. Class 1 has a precision of 80%, recall of 97%, F1 score of 88%, and 34 instances. The overall accuracy, macro average, and weighted average are 85%, 88%, and 87%, respectively, for the 61-instance dataset. For Dataset II, Class 0 has a precision of 94%, recall of 82%, F1 score of 87%, and 88 instances. Class 1 has a precision of 85%, recall of 95%, F1 score of 90%, and 96 instances. The overall accuracy, macro average, and weighted average are 89% for the 184-instance dataset.
Figure 4 and
Figure 5 represent the RF model’s performance measuring plots on Dataset I and Dataset II.
4.3. K-Nearest Neighbor Classifier
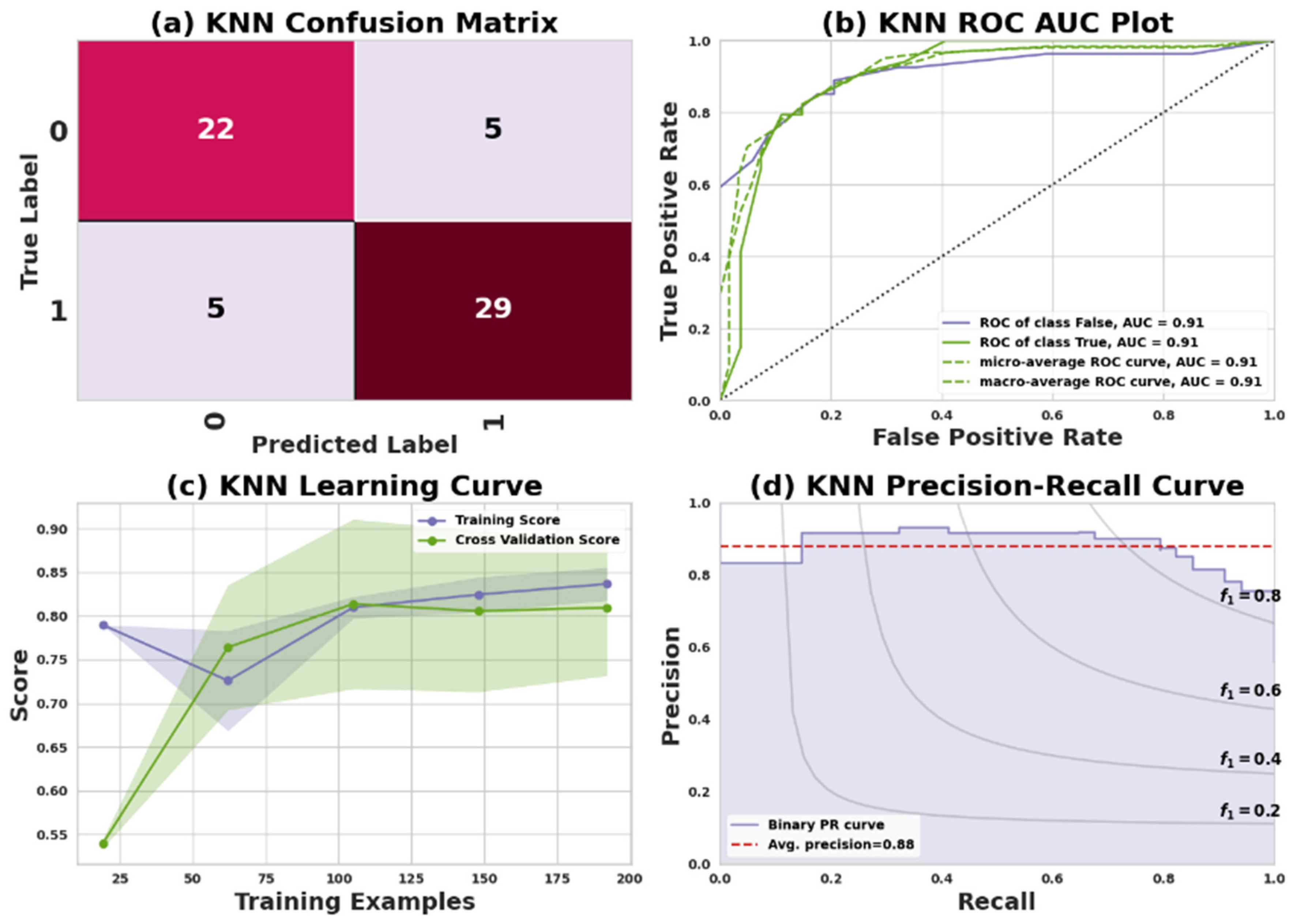
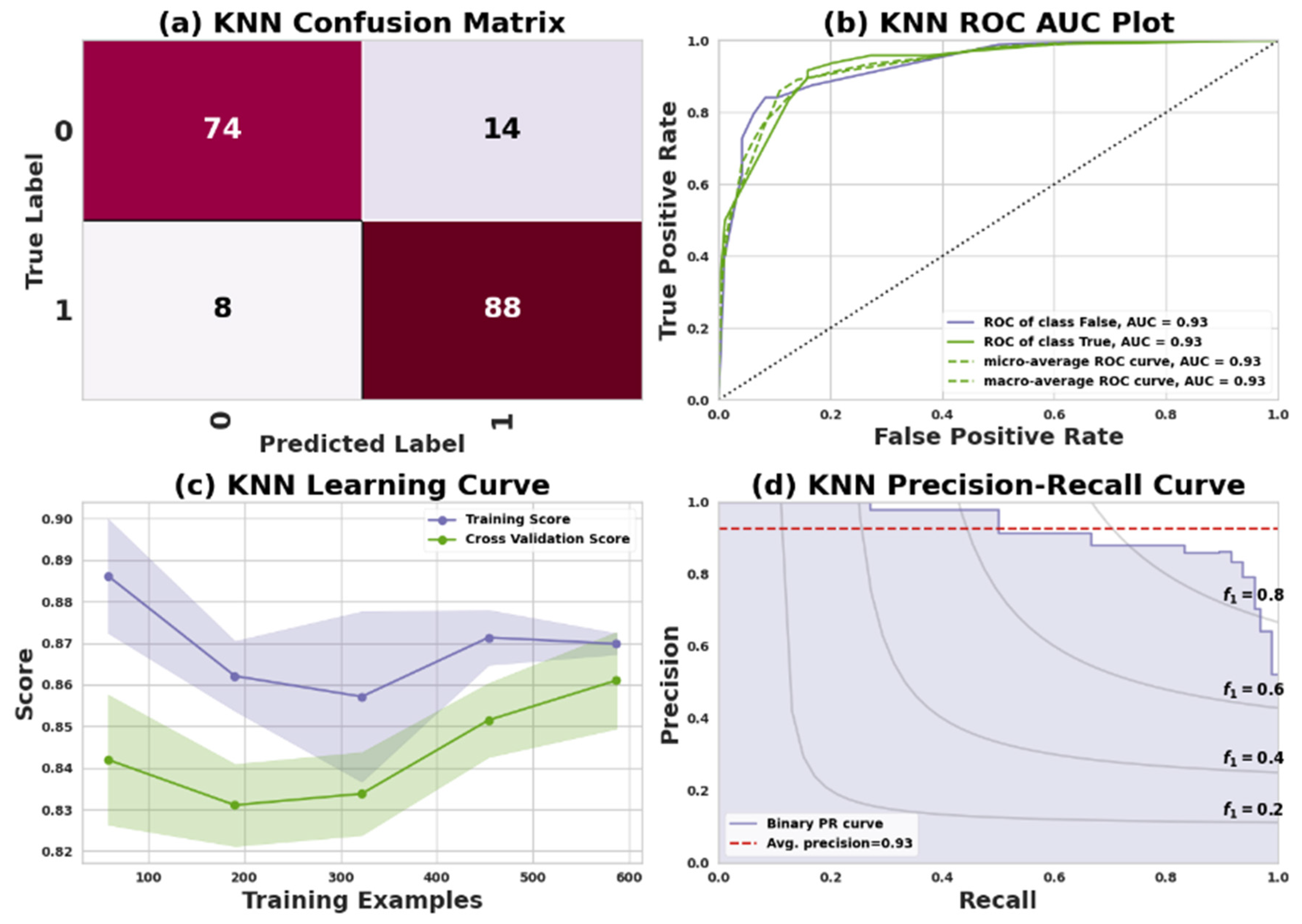
KNN is an instance-based or lazy learning technique. The term lazy learning refers to the process of building a model without the requirement of training data. KNN neighbors are selected from a set of objects with known properties or classes [
52]. The confusion matrix reveals that for Dataset I, 22 positive records and 29 negative records were accurately predicted, while 10 predictions were inaccurate, specifically consisting of 5 false negatives and 5 false positives. Similarly, in the confusion matrix for Dataset II, 74 positive records and 88 negative records were correctly predicted, but 22 predictions were inaccurate, including 14 false negatives and 8 false positives.
Table 7 presents the performance of the KNN classifier in predicting heart disease for two datasets: Dataset I (Cleveland) and Dataset II (IEEE Dataport). Evaluation metrics include precision, recall, F1 score, and support for both classes: 0 (no heart disease) and 1 (having heart disease).
In Dataset I, Class 0 shows a precision of 81%, recall of 81%, F1 score of 81%, and 27 instances. Class 1 displays a precision of 85%, recall of 85%, F1 score of 85%, and 34 instances. The dataset, containing 61 instances, has an overall accuracy, macro average, and weighted average of 84%, 83%, and 84%, respectively. For Dataset II, Class 0 has a precision of 90%, recall of 84%, F1 score of 87%, and 88 instances. Class 1 demonstrates a precision of 86%, recall of 92%, F1 score of 89%, and 96 instances. The overall accuracy, macro average, and weighted average for the 184-instance dataset are 88%, 88%, and 89%, respectively.
Figure 6 and
Figure 7 represent the KNN model’s performance measuring plots on Dataset I and Dataset II.
4.4. Logistic Regression Classifier
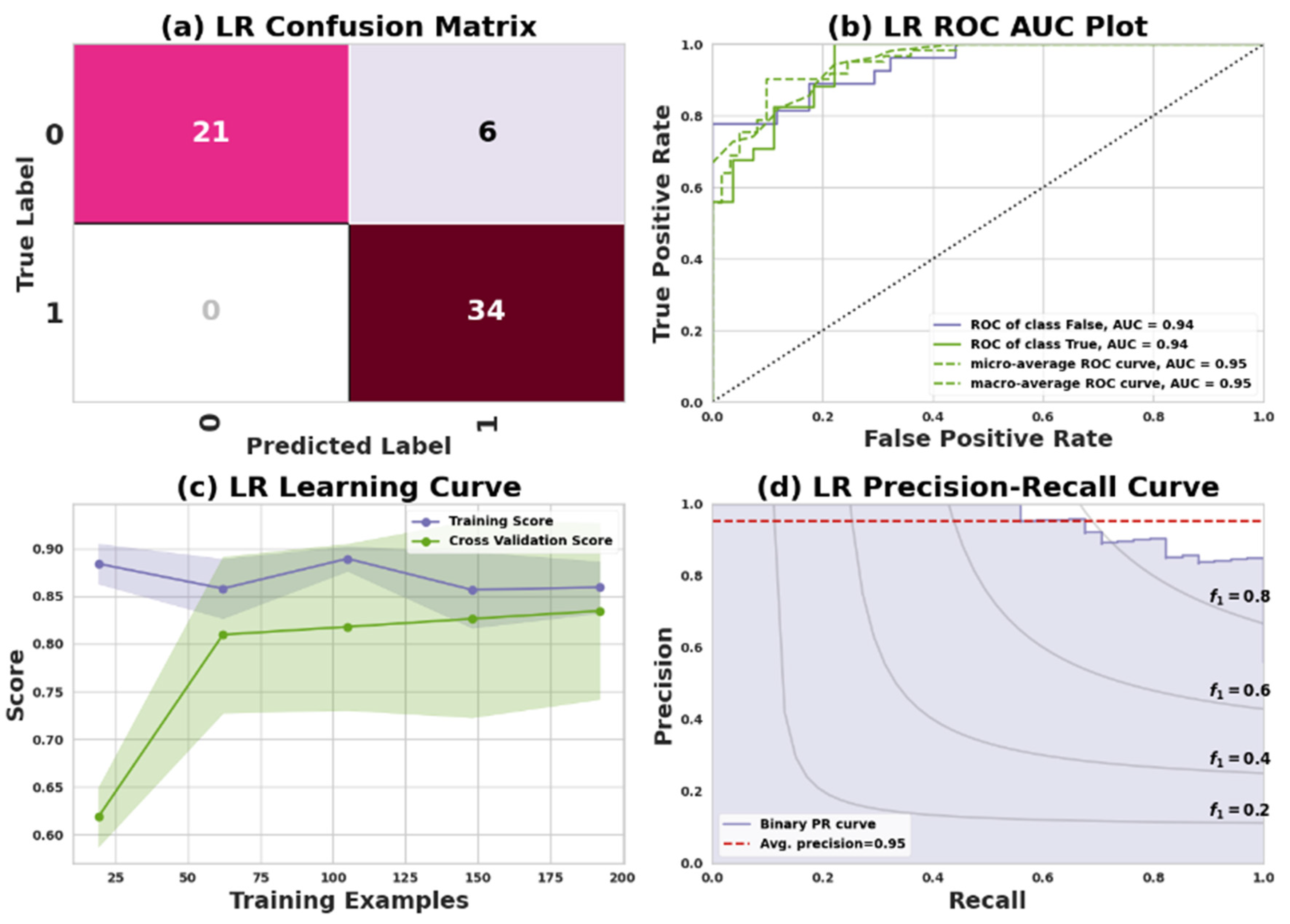
LR is an algorithm for predicting whether an observation belongs to one of two categories in ML. The LR classifiers predict the target class based on calculated logits (scores). A logistic function is used to convert probabilities into binary values that can be used to make predictions [
53]. The confusion matrix for the model reveals the following results for Dataset I and Dataset II: In Dataset I, the model accurately predicted 21 positive and 34 negative cases while making 6 incorrect predictions, all of which were false negatives and no false positives. In Dataset II, the model successfully predicted 75 positive and 88 negative cases, but it also made 21 incorrect predictions, comprising 13 false negatives and 8 false positives.
Table 8 illustrates the performance of a Logistic Regression (LR) classifier in predicting heart disease for two datasets: Dataset I (Cleveland) and Dataset II (IEEE Dataport). The evaluation metrics presented include precision, recall, F1 score, and support for both classes: 0 (no heart disease) and 1 (having heart disease).
In Dataset I (Cleveland), Class 0 has a precision of 100%, recall of 78%, F1 score of 88%, and 27 instances. Class 1 exhibits a precision of 85%, recall of 100%, F1 score of 92%, and 34 instances. With a total of 61 instances, the overall accuracy, macro average, and weighted average are 90%, 93%, and 92%, respectively.
For Dataset II (IEEE Dataport), Class 0 displays a precision of 90%, recall of 85%, F1 score of 88%, and 88 instances. Class 1 shows a precision of 87%, recall of 92%, F1 score of 89%, and 96 instances. The dataset, containing 184 instances, has an overall accuracy, macro average, and weighted average of 89%.
Figure 8 and
Figure 9 represent the LR model’s performance measuring plots on Dataset I and Dataset II.
4.5. Naive Bayes Classifier
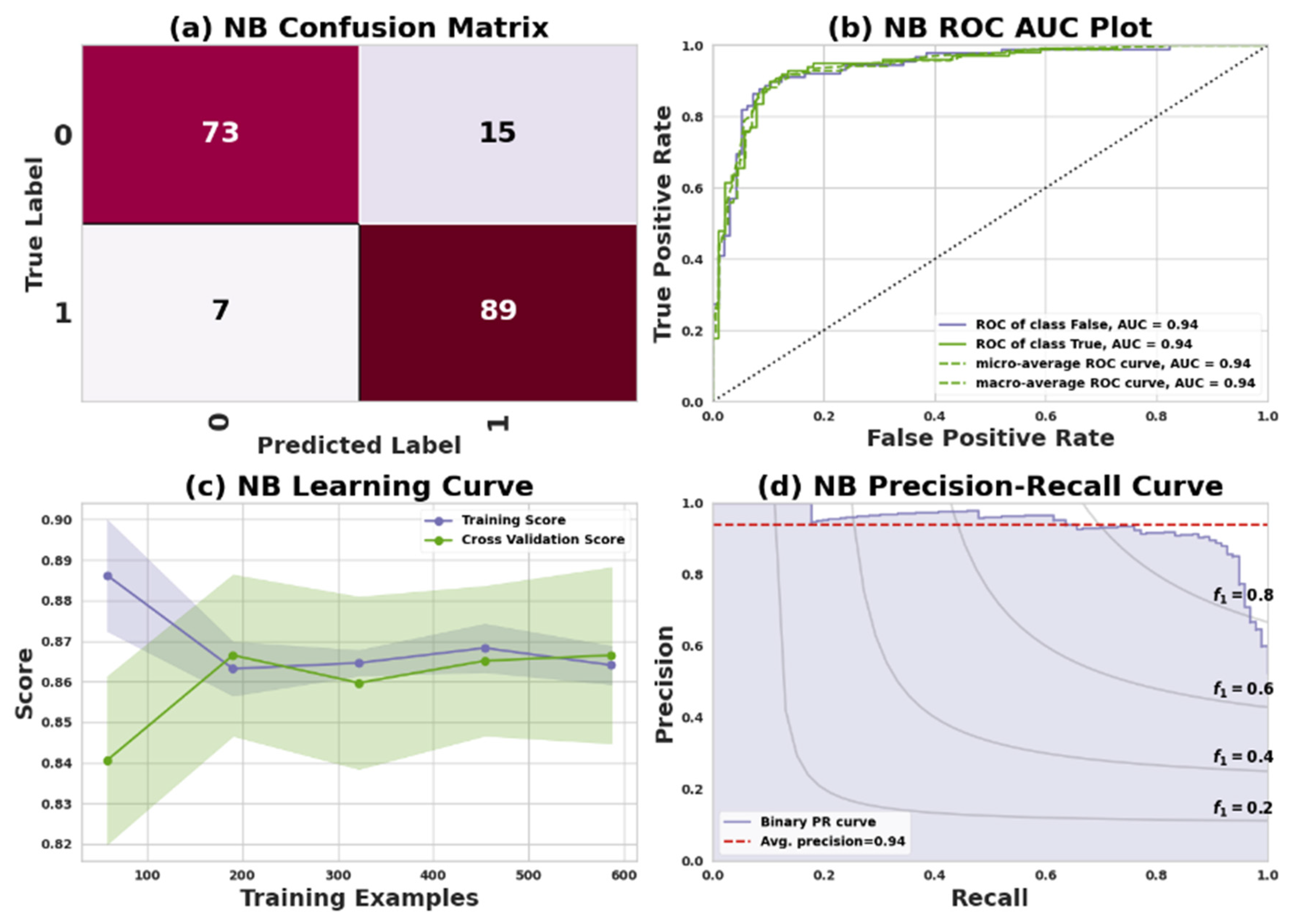
One of the most popular supervised ML algorithms for multi-classification problems is the NB algorithm. Several classification problems can be solved using the NB algorithm, which is based on the Bayes theorem. The basic concept of NB is to estimate the probability of each class we wish to reveal based on the probability of each feature being present in the data. According to Equation (10), naive models assume that the features of a model are independent of each other.
where the P(c/x) represents the posterior probability, which is the probability of a hypothesis (or class) given the observed data. The term p(x/c) denotes the likelihood, which is the probability of observing the data given the hypothesis (or class). The class prior to probability is denoted by p©, and it represents the probability of observing the hypothesis (or class) in the absence of any data. Finally, the predictor prior to probability, denoted by p(x), represents the probability of observing the data in the absence of any hypothesis (or class) [
54]. The NB algorithm assumes that each feature in the data has an independent condition on how the probability of an outcome will happen for each unique class of data in the dataset. For Dataset I, the confusion matrix reveals that the model accurately predicted 23 positive and 31 negative cases, while making 7 incorrect predictions, which include 4 false negatives and 3 false positives. In the case of Dataset II, the confusion matrix shows that the model successfully predicted 73 positive and 89 negative cases, but also made 22 incorrect predictions, comprising 15 false negatives and 7 false positives.
Table 9 presents the performance of a NB model in predicting heart disease for two datasets: Dataset I (Cleveland) and Dataset II (IEEE Dataport). Evaluation metrics include precision, recall, F1 score, and support for both classes: 0 (no heart disease) and 1 (having heart disease).
In Dataset I, Class 0 has a precision of 88%, recall of 85%, F1 score of 87%, and 27 instances. Class 1 exhibits a precision of 89%, recall of 91%, F1 score of 90%, and 34 instances. With 61 instances in total, the overall accuracy, macro average, and weighted average are 89%.
For Dataset II, Class 0 displays a precision of 88%, recall of 85%, F1 score of 87%, and 88 instances. Class 1 shows a precision of 89%, recall of 91%, F1 score of 90%, and 96 instances. With 184 instances, the overall accuracy, macro average, and weighted average are 89%.
Figure 10 and
Figure 11 represent the NB model’s performance measuring plots on Dataset I and Dataset II.
4.6. Gradient Boosting Classifier
A GB Classifier is an ML technique that uses an ensemble of weak models to produce a robust classifier. The algorithm sequentially trains individual models, each addressing the residual errors generated by the previous model. The final prediction is made by combining each model’s predictions, weighted according to their contribution [
55]. This technique can be applied to binary and multi-class classification problems and is often implemented using decision trees for weak learners. The goal is to minimize the loss function through the iterative process of model training and combining. For Dataset I, the confusion matrix indicates that the model accurately predicted 21 positive and 31 negative cases, while making 9 incorrect predictions, consisting of 6 false negatives and 3 false positives. In Dataset II, the confusion matrix reveals that the model successfully predicted 75 positive and 89 negative cases, but also made 20 incorrect predictions, which include 13 false negatives and 7 false positives.
Table 10 showcases the performance of a GB classifier in predicting heart disease for two datasets: Dataset I (Cleveland) and Dataset II (IEEE Dataport). It includes evaluation metrics such as precision, recall, F1 score, and support for both classes: 0 (no heart disease) and 1 (having heart disease).
For Dataset I (Cleveland), Class 0 has a precision of 88%, recall of 78%, F1 score of 82%, and 27 instances. Class 1 displays a precision of 84%, recall of 91%, F1 score of 87%, and 34 instances. The dataset, with 61 instances, has an overall accuracy, macro average, and weighted average of 85%.
In Dataset II (IEEE Dataport), Class 0 exhibits a precision of 91%, recall of 85%, F1 score of 88%, and 88 instances. Class 1 presents a precision of 87%, recall of 93%, F1 score of 90%, and 96 instances. With 184 instances, the overall accuracy, macro average, and weighted average are 89%.
Figure 12 and
Figure 13 represent the GB model’s performance measuring plots on Dataset I and Dataset II.
4.7. AdaBoost Classifier
The AB is a ML algorithm designed for classification tasks. It combines multiple simple models, known as weak learners, to form a more robust overall classifier. The algorithm starts by training the first weak learner on the data and then calculates the error. Subsequently, misclassified samples are given greater weight, and the subsequent weak learner is trained on these samples with higher emphasis. This process is repeated several times. Each weak learner’s prediction is given a weight proportional to its accuracy before being combined to form the final prediction [
56]. The AB can be used for binary or multi-class classification problems, and weak learners often utilize decision trees. The algorithm adjusts the weight of the samples based on their classification performance, allowing it to focus on the samples that are challenging to classify. For Dataset I, the confusion matrix shows that the model accurately predicted 19 positive and 33 negative cases, while making 9 incorrect predictions, which include 8 false negatives and 1 false positive. In Dataset II, the confusion matrix indicates that the model successfully predicted 75 positive and 90 negative cases, but also made 19 incorrect predictions, consisting of 13 false negatives and 6 false positives.
Table 11 presents the performance of the AB classifier in predicting heart disease for two datasets: Dataset I (Cleveland) and Dataset II (IEEE Dataport). Evaluation metrics include precision, recall, F1 score, and support for both classes: 0 (no heart disease) and 1 (having heart disease).
In Dataset I (Cleveland), Class 0 has a precision of 95%, recall of 70%, F1 score of 81%, and 27 instances. Class 1 demonstrates a precision of 80%, recall of 97%, F1 score of 88%, and 34 instances. With 61 instances in total, the overall accuracy, macro average, and weighted average are 85%.
For Dataset II (IEEE Dataport), Class 0 shows a precision of 93%, recall of 85%, F1 score of 89%, and 88 instances. Class 1 displays a precision of 87%, recall of 94%, F1 score of 90%, and 96 instances. With 184 instances, the overall accuracy, macro average, and weighted average are 90%.
Figure 14 and
Figure 15 represent the AB model’s performance measuring plots on Dataset I and Dataset II.
4.8. Performance Measurement Analysis of Different Curves
For each classifier,
Table 12 shows the results obtained for both Dataset I and Dataset II. The performance is compared across different classifiers and datasets to understand which model performs better in each scenario. In general, the result indicates that the RF, LR, NB, and AB classifiers demonstrate higher ROC-AUC and precision-recall values, suggesting better overall performance compared to the other models. However, the GB classifier shows a perfect learning curve score of 100% for Dataset I, which implies it effectively learns from the training data. The ROC-AUC curve, learning curve, and precision-recall curves for both datasets are illustrated in
Figure 4,
Figure 5,
Figure 6,
Figure 7,
Figure 8,
Figure 9,
Figure 10,
Figure 11,
Figure 12,
Figure 13,
Figure 14 and
Figure 15. This visualization allows for a comprehensive comparison of classifier performance across the two datasets, considering multiple evaluation metrics.
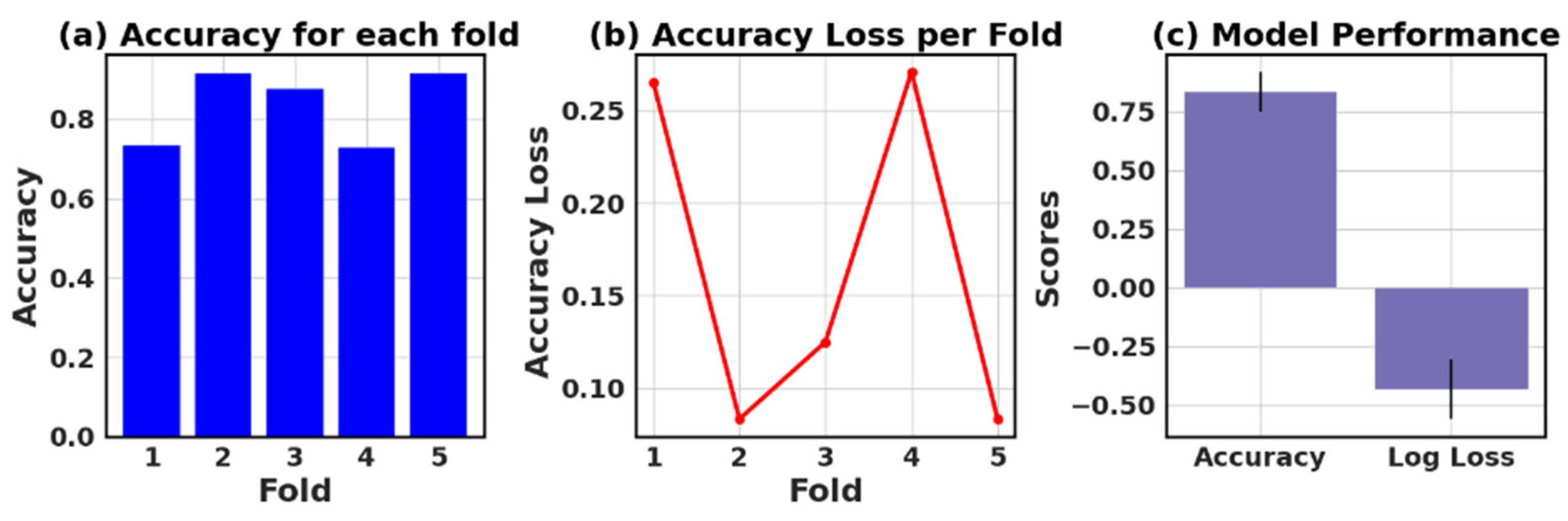
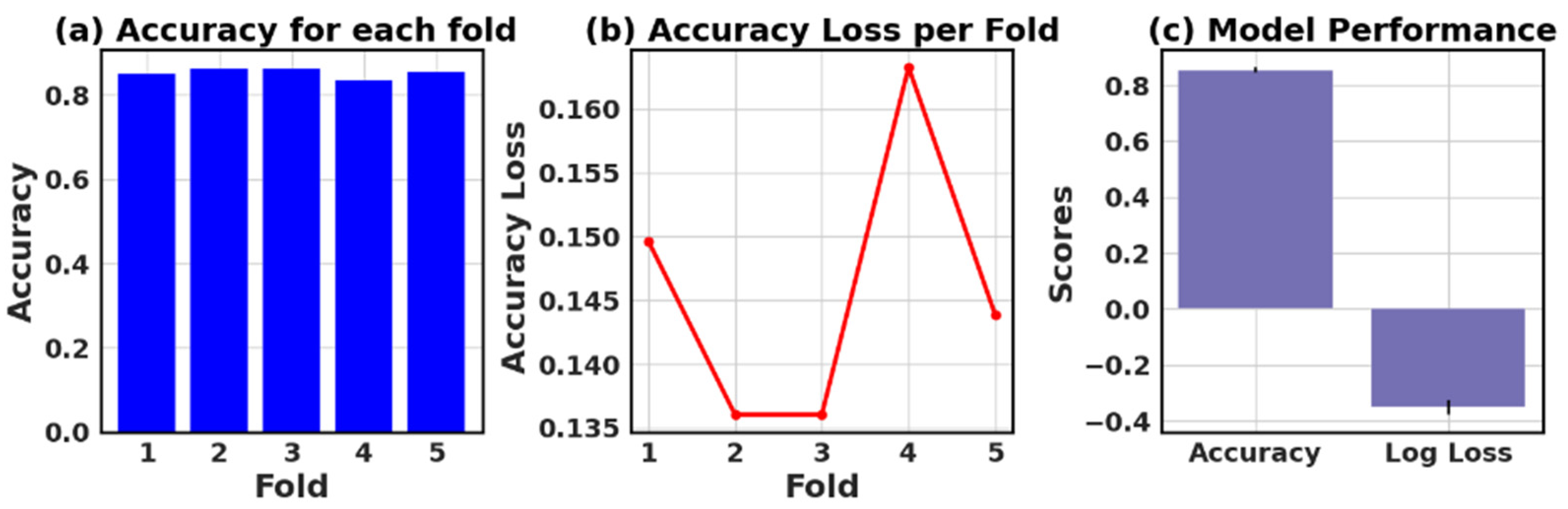
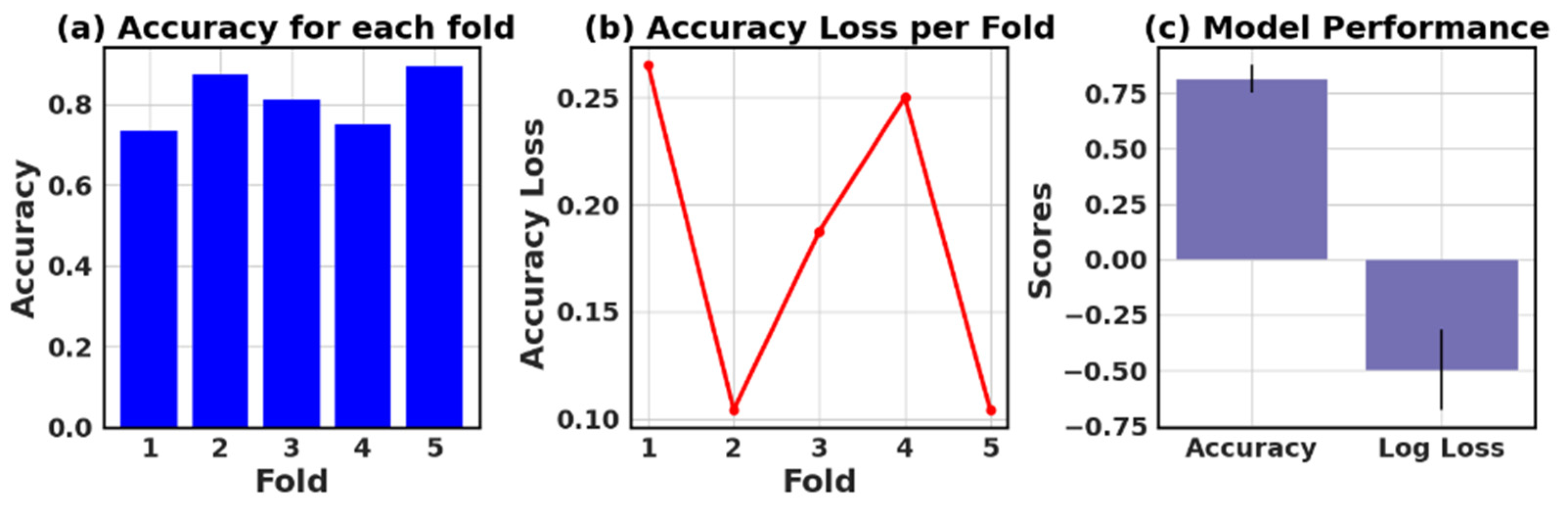
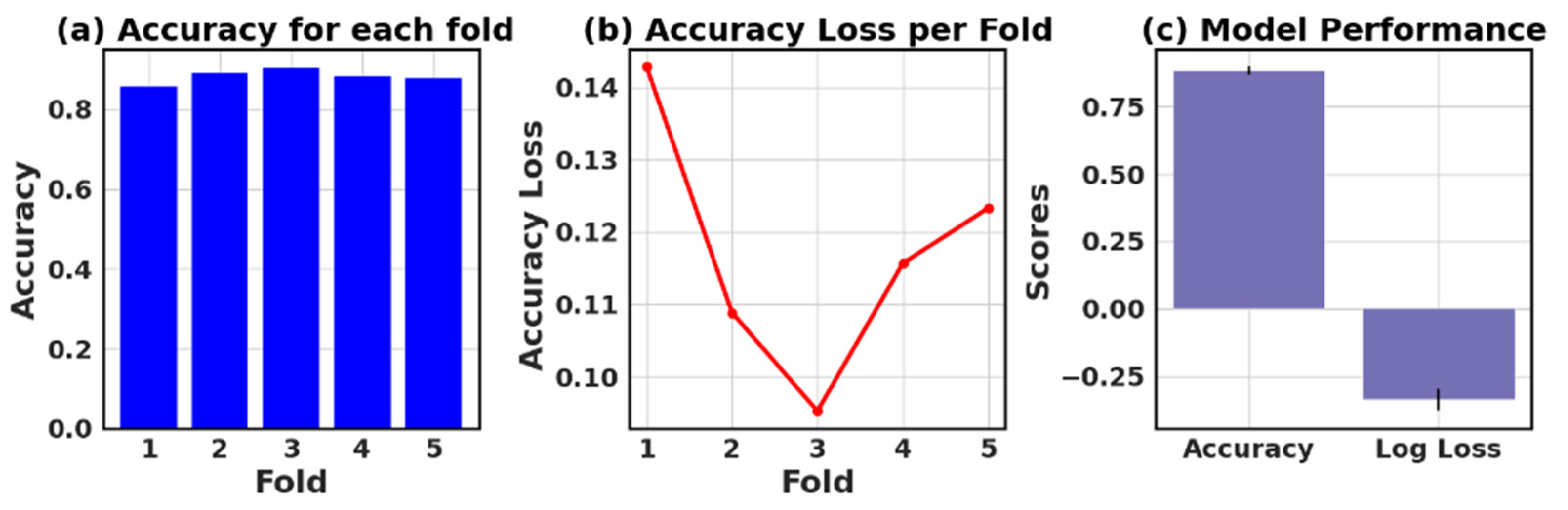
4.9. Assessing the Accuracy and Accuracy Loss of Each Fold: Measurement and Performance Evaluation
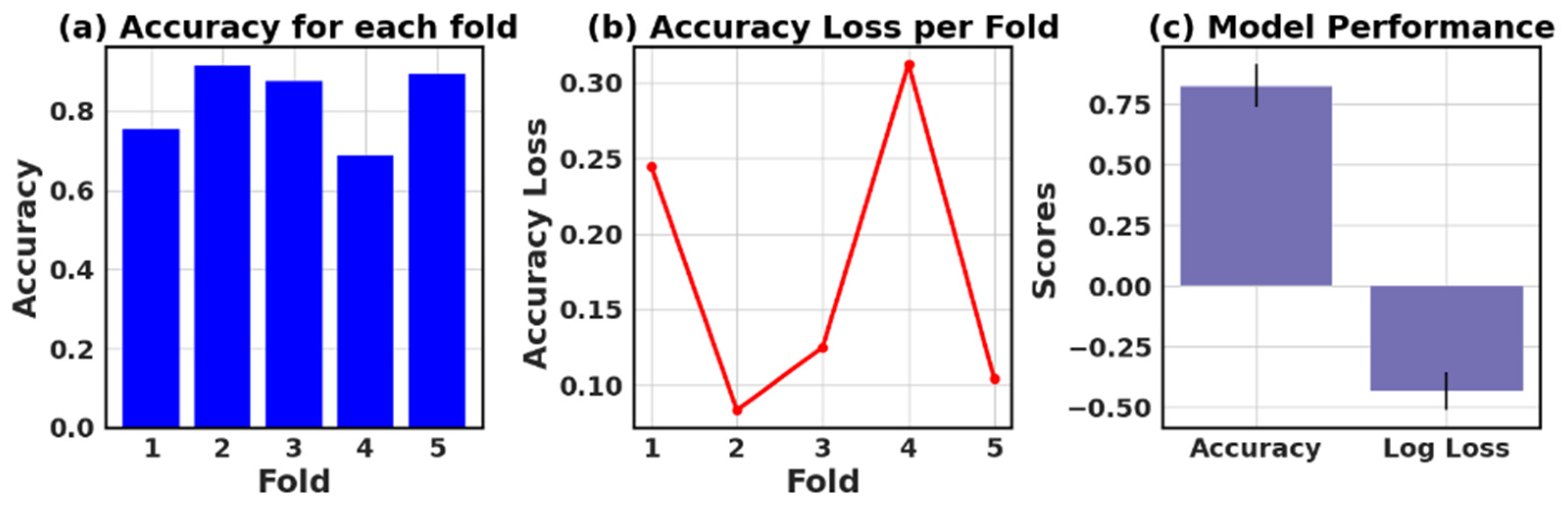
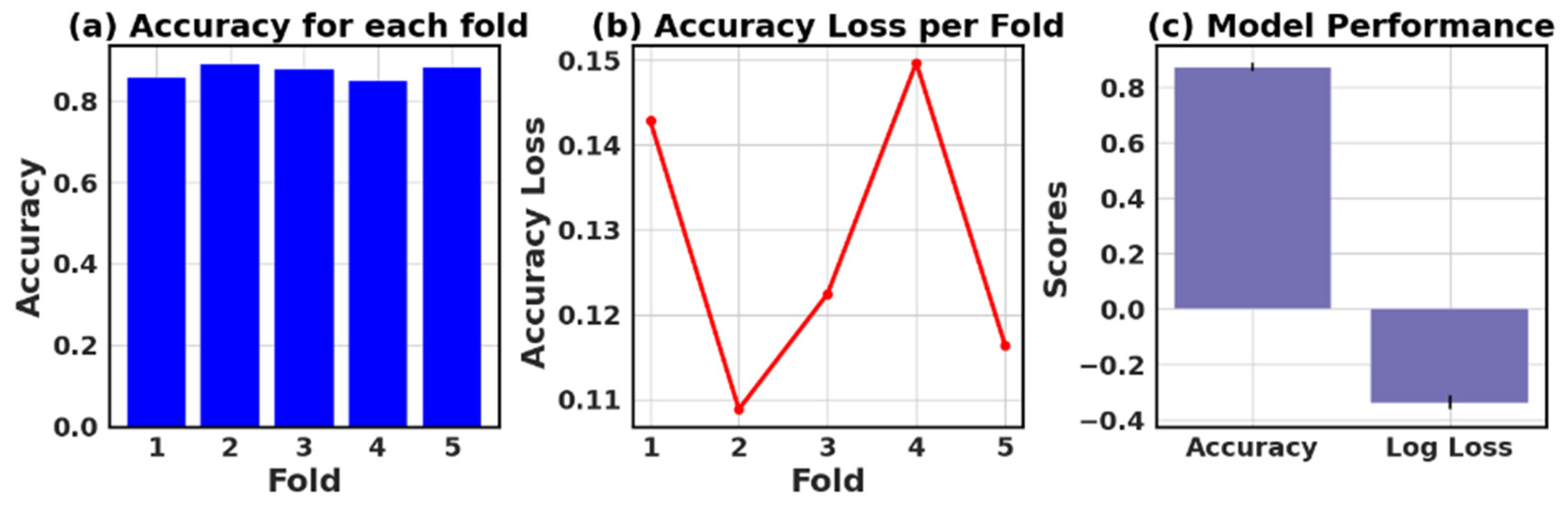
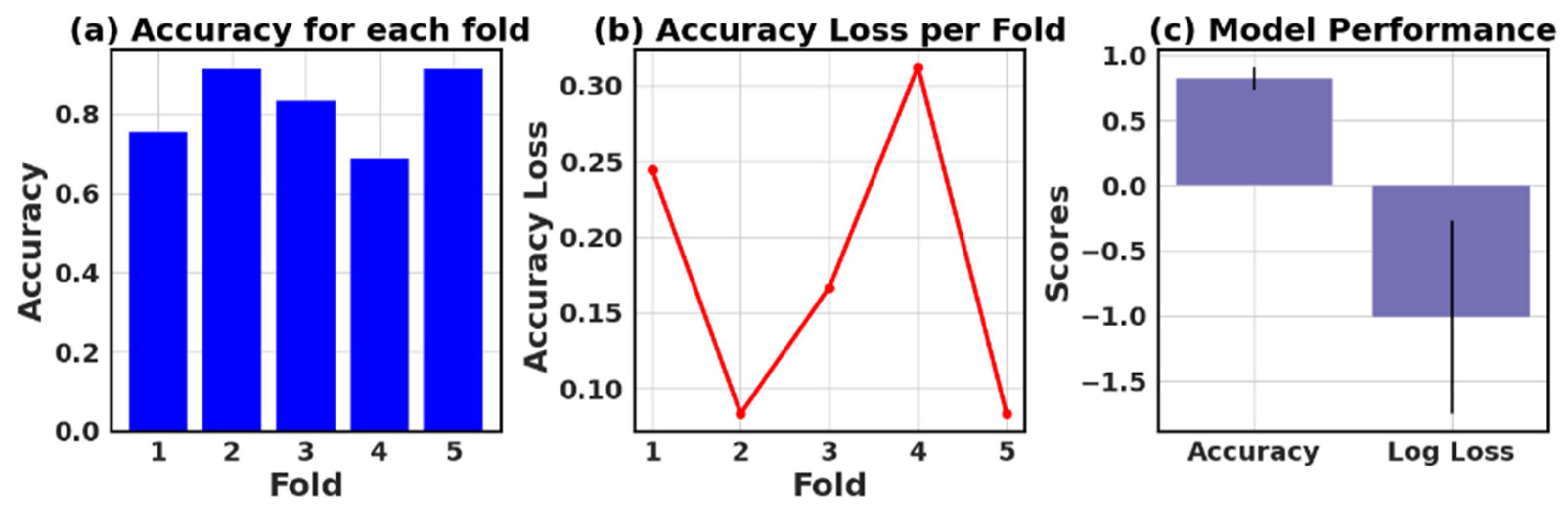
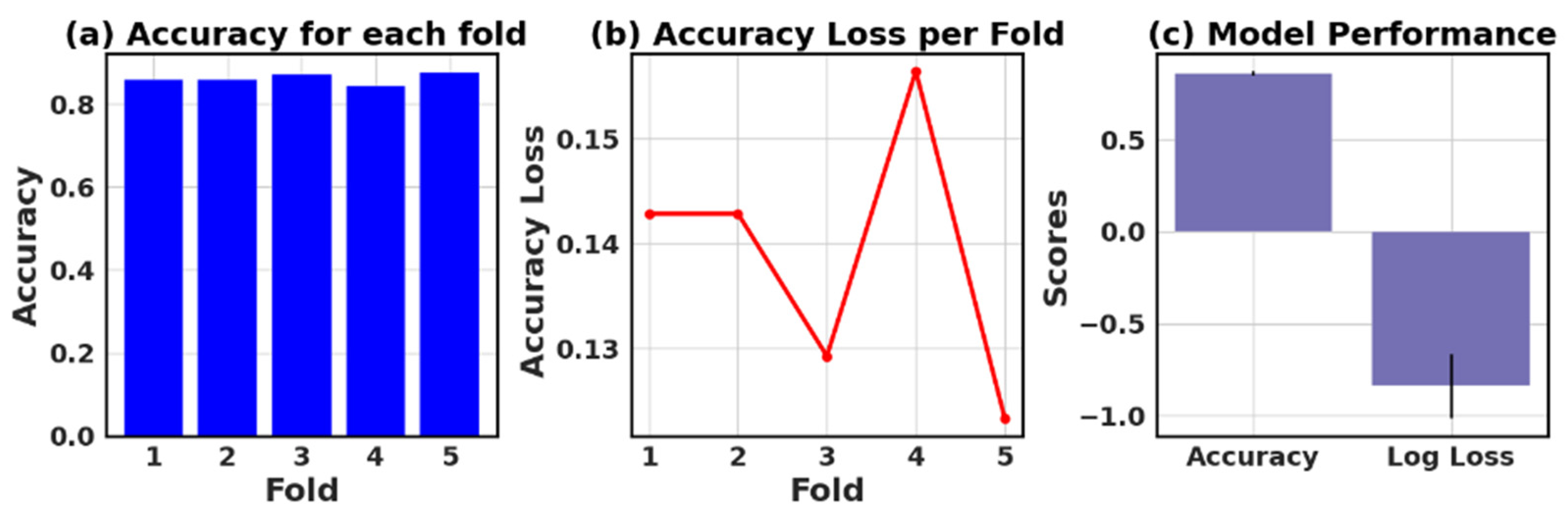
The loss and accuracy values for each fold provide an estimate of how well the model is performing on different subsets of the data.
Figure 16,
Figure 17,
Figure 18,
Figure 19,
Figure 20,
Figure 21,
Figure 22,
Figure 23,
Figure 24,
Figure 25,
Figure 26 and
Figure 27 show six models’ five-fold accuracy and loss value plots for Datasets I and II.
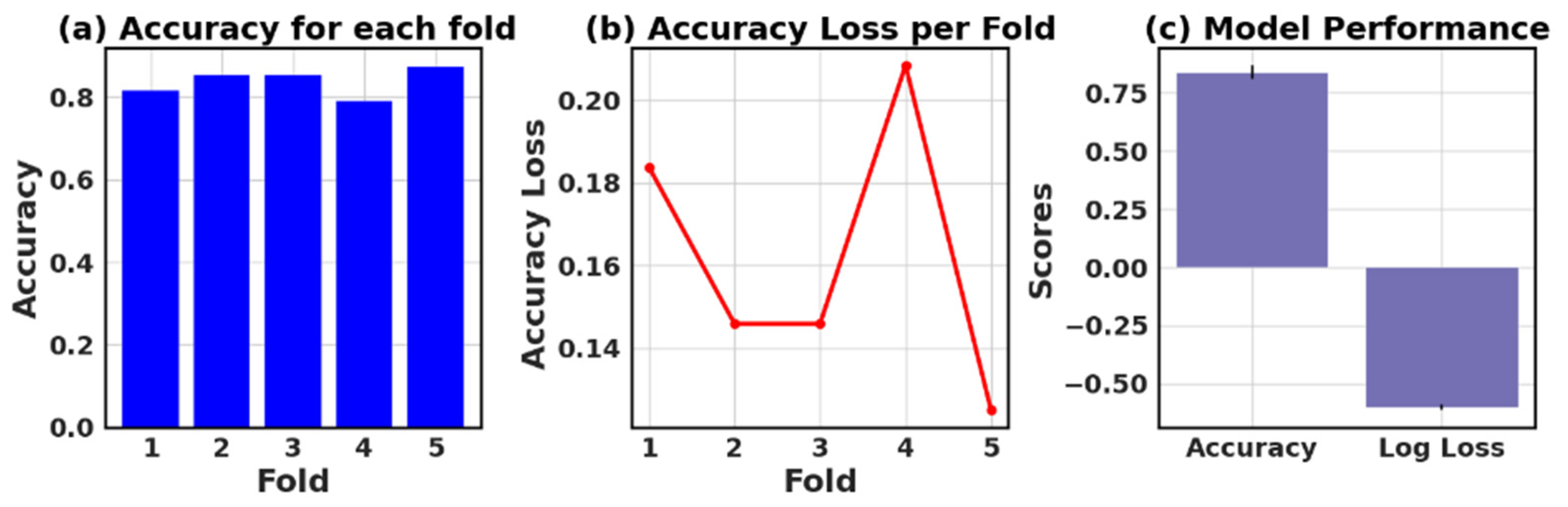
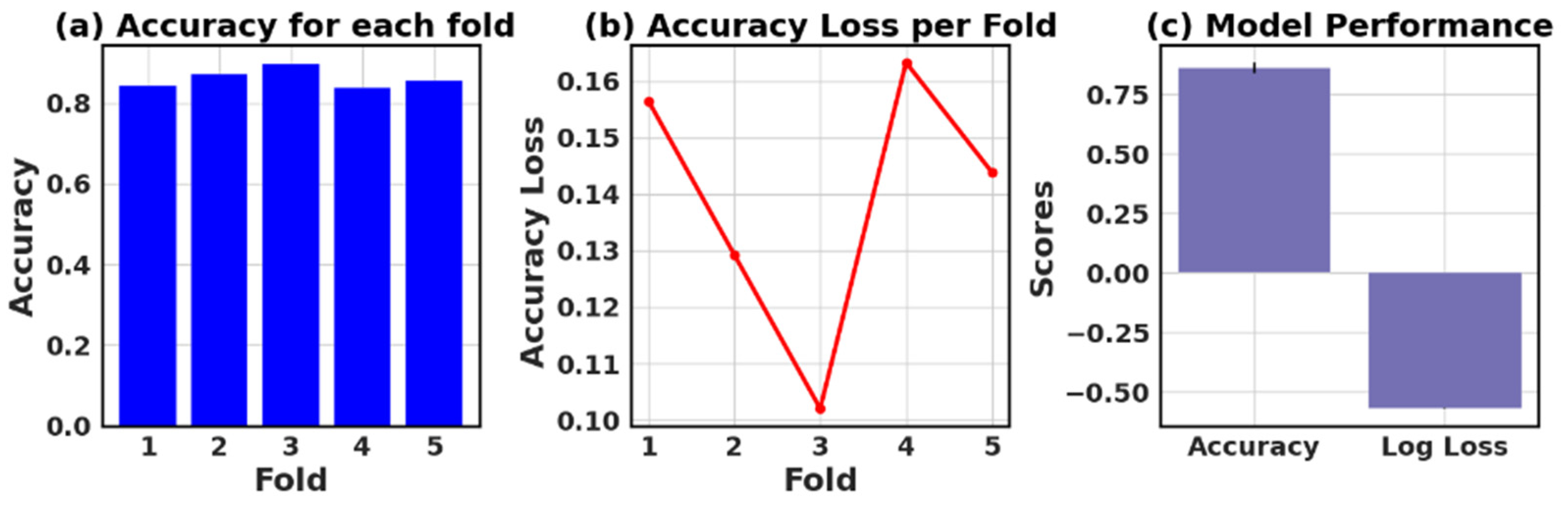
Table 13 presents the values of accuracy, accuracy loss of each fold, and mean and standard deviation values of six models.
In this study, the performance of various ML models, including RF, KNN, LR, NB, GB, and AB, was evaluated on two different datasets (I and II). The models’ performance was assessed using five-fold cross-validation, and three metrics were reported: accuracy, loss (1—accuracy), and negative log loss.
The results indicate that, for Dataset I, the RF model achieved the highest mean accuracy (0.826) with a standard deviation of 0.089, followed closely by the NB and LR models with mean accuracies of 0.839 and 0.834, respectively. On the other hand, the KNN model had the lowest negative log loss (−1.009) with the largest standard deviation (0.740), which could suggest overfitting or instability in model performance across different folds.
For Dataset II, the GB showed the best performance with a mean accuracy of 0.887 and a standard deviation of 0.016. The other models, including KNN, LR, and NB, also demonstrated relatively high mean accuracies, ranging between 0.854 and 0.866. The negative log loss values were more stable for this dataset, with the AB model having the most consistent performance, indicated by a mean negative log loss of −0.567 and a standard deviation of 0.002. Further, this study reveals that selecting the best model requires careful consideration of the evaluation metrics and their respective standard deviations.
6. Comparative Study
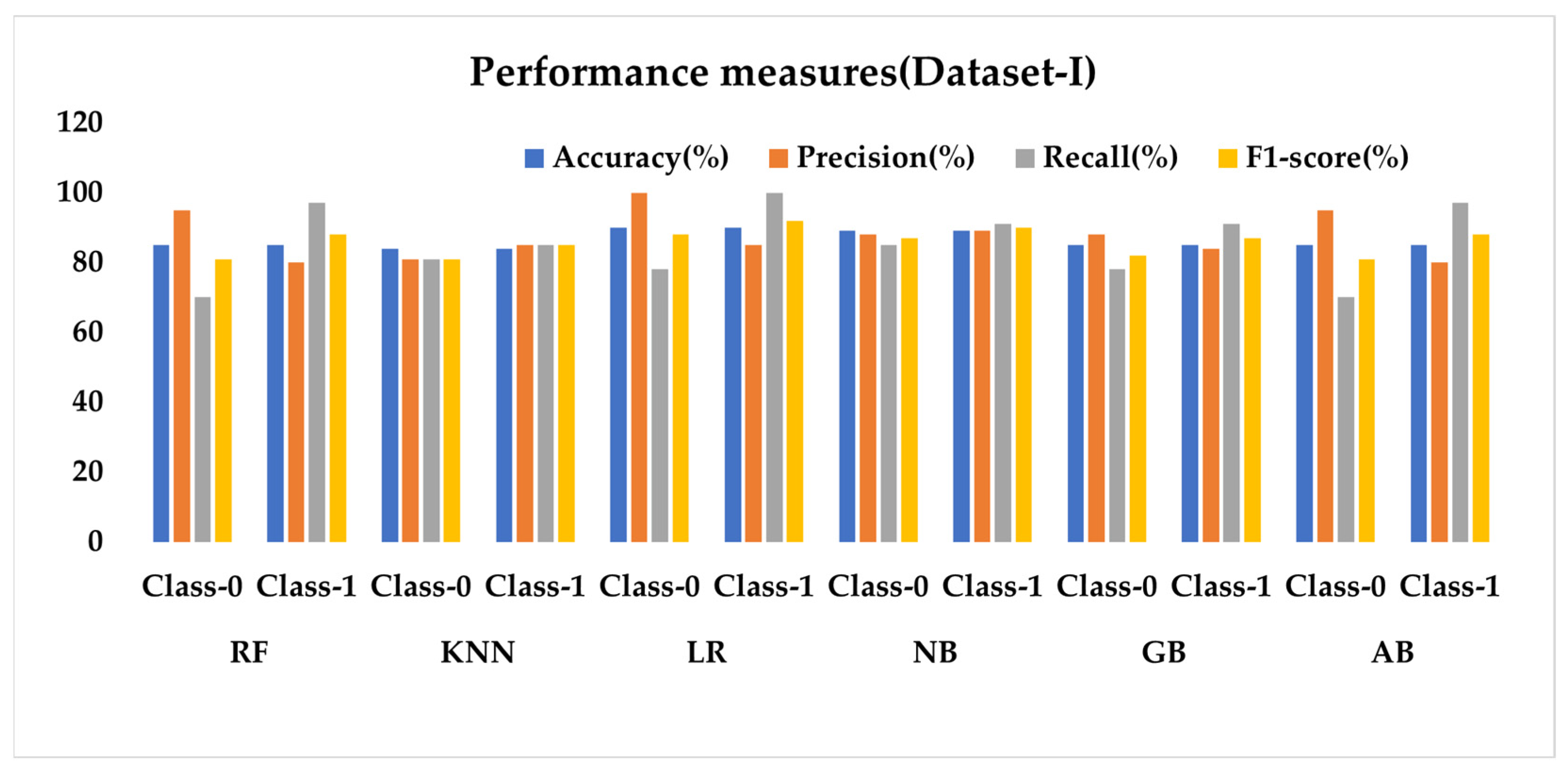
Figure 29 presents a performance analysis of six ML classifiers applied to Dataset I. The results show that LR achieves the highest accuracy of 90% among the classifiers, with notable precision and recall values for both classes. Other classifiers, such as RF, KNN, NB, GB, and AB display varying levels of performance across the different metrics. Upon examining the results, it is evident that the classifiers exhibit different strengths and weaknesses. For instance, while RF and AB have high precision for Class 0, they show lower recall values for the same class. Conversely, LR demonstrates remarkable precision for Class 0 and recall for Class 1.
Figure 30 represents the performance analysis for six ML classifiers. In this, the analysis reveals that the classifiers demonstrate relatively similar performance on Dataset II, with the AB classifier achieving the highest accuracy of 90%. Precision, recall, and F1 score values are also consistent across the classifiers. However, there are some differences in performance, such as RF having a higher precision for Class 0 and a lower recall for the same class.
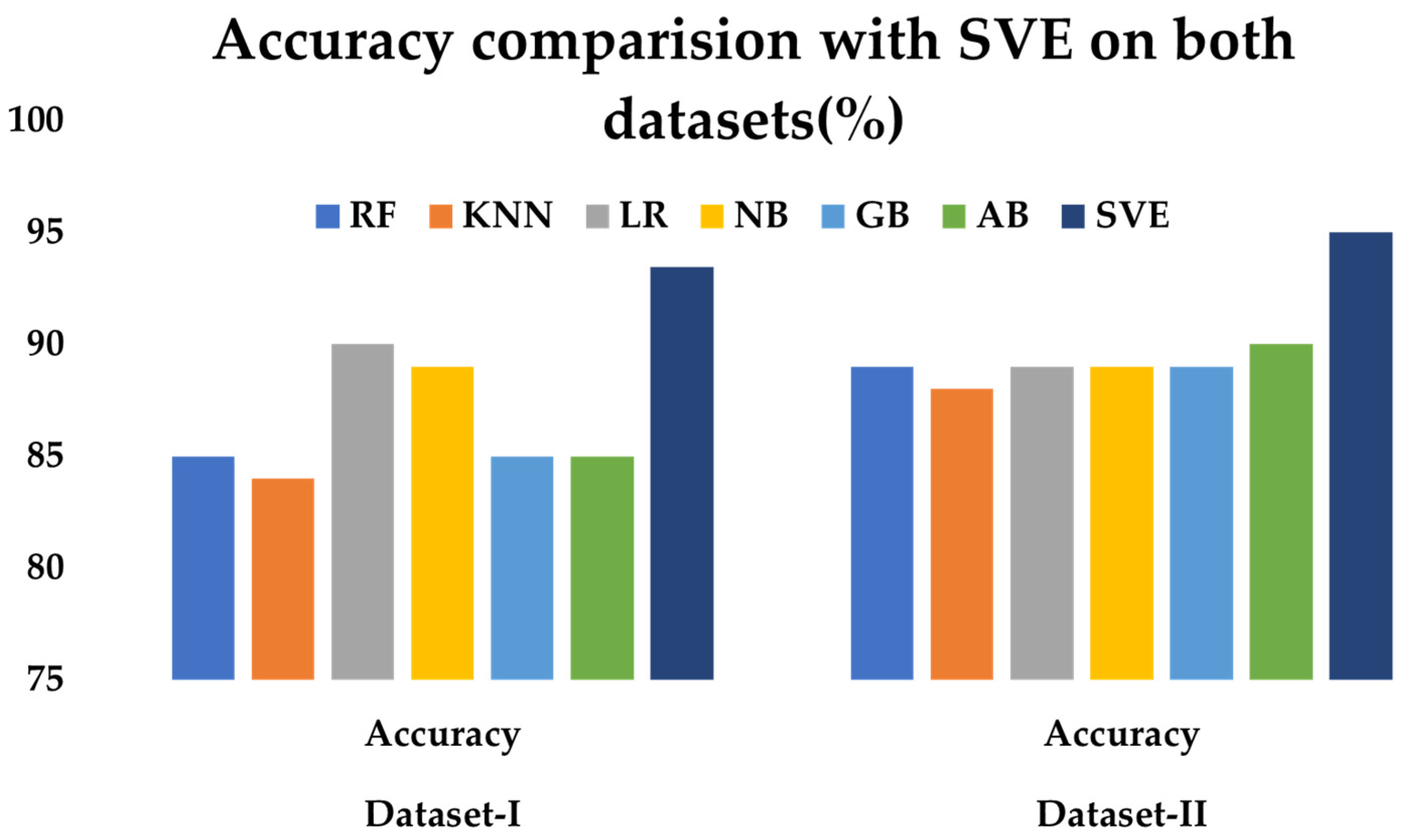
Upon analyzing
Figure 31, it is evident that the SVE classifier consistently outperforms the individual ML classifiers in both datasets, achieving 93.44% accuracy on Dataset I and 95% on Dataset II. As it is considered from individual classifiers, it has observed the maximum accuracy is only 90%, which is obtained from AB classifier on both the data sets. It is also notable that the performance of all classifiers improves from Dataset I to Dataset II. The SVE classifier effectively combines the strengths of the six individual classifiers, leading to enhanced accuracy in both datasets. This demonstrates the potential of ensemble methods for improved performance in heart disease prediction tasks.
Table 14 and
Table 15 compare the previous researcher’s accuracy and the proposed work result accuracy on Dataset I and Dataset II. Compared to the previous work results, our proposed model produced more accuracy.
The limitation of this model is that it is based on a limited amount of patient data, which only include 303 and 1190 patients in the datasets. Future work includes more patient data, the application of the feature selection method, and the development of a deep learning-based system for early heart disease detection. Additionally, utilizing medical IoT devices and sensors for the simultaneous collection of clinical parameters such as ECG, blood oxygen level, and body temperature can further improve the performance of the proposed system.