Abstract
Staphylococcus aureus is a bacterium that causes various diseases in humans. Cold plasma is found to be an alternative to eliminate S. aureus. Most studies of cold plasma on S. aureus mainly focus on the physiochemical changes of the cells. So far, biological responses of S. aureus to cold plasma treatment under different treatment durations have not yet been evaluated. In this study, the results showed that the cold plasma was effective in eliminating S. aureus. At the initial exposure (1 min), the treated cells showed gene upregulations of stress proteins, antioxidants, nitrosative stress, and transporter proteins, but no significant change in other biological processes, such as cell membrane synthesis, DNA repairing, transcription, and translation. This indicated that the cells actively countered the damage from cold plasma. In contrast, during the prolonged treatment (3 and 5 min), biological processes related to central dogma were affected, including the DNA repairing mechanism, transcription, and translation. In addition, the majority of the genes related to cell membrane synthesis were downregulated, indicating that the treated cells could no longer sustain their cell integrity. In conclusion, this study elucidated how cold plasma inactivated S. aureus in a series of cold plasma exposures and highlighted the sequential transcriptomic responses of S. aureus.
1. Introduction
Staphylococcus aureus is a Gram-positive bacterium commonly found in human microbiota, such as on the skin and in the nasal vestibule [1]. Its ability to form biofilm and develop multidrug resistance makes many strains opportunistic pathogens or potential pathobionts [1]. These pathogenic strains can cause a range of ailments, including acne, impetigo, and abscesses, as well as life-threatening conditions, such as pneumonia, meningitis, osteomyelitis, endocarditis, toxic shock syndrome, bacteremia, and sepsis [1]. S. aureus is also a leading cause of wound infections after surgery and the well-known methicillin resistant or multidrug-resistant S. aureus (MRSA) is a global threat to medicine [1]. MRSA is defined as any strain of S. aureus that has developed to resist multiple antibiotics that are normally used to treat ordinary Staphylococcus infections [1].
Antibiotics are a common treatment for S. aureus infections, with options including penicillin, methicillin, nafcillin, oxacillin, dicloxacillin, flucloxacillin, kanamycin, gentamicin, streptomycin, vancomycin, and others [2]. However, S. aureus is constantly developing resistance to antibiotics. For instance, less than 2% of S. aureus isolates in the United Kingdom are susceptible to penicillin treatment [3]. MRSA occurs in 5–25% of cases in the United States and Europe, and up to 50% in Japan [4]. The effectiveness of antibiotics against S. aureus is decreasing globally. Alternative treatments, such as cell wall-anchored protein vaccines, nanoparticles, photochemical internalization, cell-penetrating peptides, antimicrobial peptides, and bacteriophage, have been suggested [5]. Disinfection methods, such as ultraviolet, hydrogen peroxide, and ozone, can also destroy S. aureus and prevent it from infecting the human body [6,7,8].
Cold plasma is defined as a type of plasma that operates at relatively low temperatures compared to thermal plasma, and is normally worked below 40 °C if it is used for biomedical applications [9]. It is an alternative disinfection method that can be used to eliminate various pathogens, such as Escherichia coli, Enterococcus faecalis, and others [10,11]. This method is useful in disinfecting wounds after medical surgery to prevent the aforementioned problem of wound infection by S. aureus [12]. Furthermore, it can also be pre-applied to burned skin to reduce the risk of bacterial infection or act as a post-treatment for microbial-infected skin [12]. Cold plasma consists of reactive oxygen/nitrogen species (ROS/RNS), ultraviolet radiation, charged molecules, and energized ions and atoms [13]. All these compositions can contribute to microbial inactivation, with ROS often identified as the main affecting species [13]. Cold plasma causes strong oxidative stress to the treated cells, causing damage through possible lipid peroxidation, enzyme inactivation, and DNA cleavage [13].
Studies on the effects of cold plasma on S. aureus are available elsewhere [13,14,15,16], and most of them mainly focus on the physiochemical changes of the cells. However, the transcriptomic responses of the cells under different treatment durations have not been evaluated so far. Hence, in this study, we evaluated both the antibacterial effects of cold plasma generated from a self-developed plasma jet device on S. aureus and the transcriptomics response of S. aureus under different durations of cold plasma treatment via RNA-sequencing, where the biological process changes in the treated cells were evaluated.
2. Materials and Methods
2.1. Cold Plasma Generator
The cold plasma generator used in this study was self-developed and built by the Suzhou Amazing Grace Medical Equipment Co., Ltd., China [17]. Figure 1 shows the schematic diagram of the developed device. The device consists of major components, such as an air pump, a plasma generating circuit and control circuit unit, a high voltage converter, and a power supply. In addition, a discharging chamber is included in the device, which is important for generating plasma and is formed from an air tubing, metal deflector, insulated enclosure, positive and negative poles, and barrier medium. With the help of a metal deflector, the working gas (atmospheric air) has more contact area per unit time in the channel, thus, having higher ionization efficiency. In this study, the device was optimized at a frequency of 120 kHz and a fixed duty ratio of 50%, when the input voltage was adjusted to approximately 12.8 V with the circuit current at 2.5 ± 0.2 A. Considering the duty ratio, the power of the cold plasma generator could be estimated at approximately 2.5 A × 12.8 V × 50% = 16 W. Nonetheless, the air pump was set to 5.0 ± 0.5 L/min flow rate.
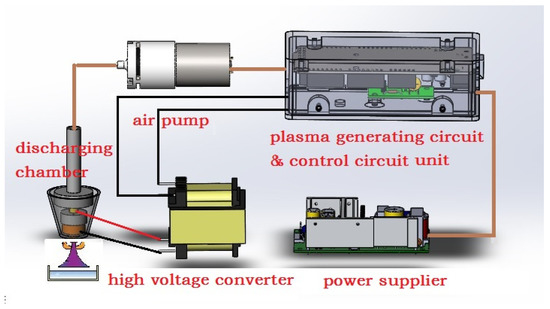
Figure 1.
Schematic diagram of cold plasma generator used in this study.
A fiber optic spectrometer (StarLine AvaSpec-ULS4096CL-EVO (Avantes, Apeldoorn, The Netherlands)) was used to generate the optical emission spectrum (OES) of the cold plasma to detect the effective species. Furthermore, the cold plasma generator was monitored in terms of its current usage, voltage, and each high voltage output pulse (plasma dose) across the different treatment times (1–30 min). The pH, electrical conductivity, and oxidation-reduction potential (ORP) of the cold plasma-treated water were also measured across the treatments. The cold plasma temperature at a distance of 3 cm and ozone generation by the device were detected to ensure the device’s safety and applicability in the biomedical field.
2.2. Cell Survivability
Staphylococcus aureus ATCC 6538 was acquired from the American Type Culture Collection (ATCC) in the form of a KWIK-STIKTM containing the lyophilized microorganism. The bacterium was revived following the manufacturer’s protocol and grown on Tryptic Soy Agar incubated at 37 °C for 16 h. A bacterial overnight-grown culture was prepared by inoculating bacterial colonies into 50 mL of Tryptic Soy Broth and incubated at 37 °C and 200 rpm for 16 h. The overnight culture was harvested by centrifugation (4 °C, 5000× g, 10 min), washed twice, and resuspended with 0.5% (w/v) saline water for further use.
To assess the effects of cold plasma on the solid agar surface, the spread plates of the culture were treated with cold plasma before incubation at 37 °C for 16 h. The distance between the nozzle and agar surface was fixed at 3 cm, the initial culture turbidity was set at OD600 nm of 0.5. Treatment durations ranging from 1–30 min were tested. The spread plate without cold plasma treatment served as the reference control, and the zone of inhibition of the treated plates was measured. The experiment was conducted in triplicate.
Cold plasma was also applied to the resuspension culture in liquid form. Briefly, a two mL culture was pipetted into a 5 mL Bijou bottle and subjected to different cold plasma treatment durations (1–30 min). The treated culture solution was then subjected to CFU plate counting via the serial dilution spread plate method. The resuspension culture without cold plasma treatment served as the control, and the CFU values were converted into cell survival rate and plotted into a graph. The experiment was conducted in triplicate.
2.3. Detection of Cell Lysate Composition
To detect the release of intracellular components, samples were collected from the resuspension cultures treated with cold plasma. The samples were centrifuged (4 °C, 5000× g, 10 min) and only the supernatants were used for detection. DNA and protein concentration (ng/µL) were measured by a NanoDrop® ND-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). The culture without cold plasma treatment was used as the reference control for the experiment. The experiment was conducted in triplicate.
2.4. Antioxidant Assays
The production of antioxidants by bacterial cells as a defense mechanism against the reactive oxygen and nitrogen species (ROS/RNS) generated from the cold plasma was detected using the ferric reducing antioxidant potential (FRAP) assay. The assay was conducted according to a method described by Benzie and Devaki [18], with slight modification. The FRAP reagent was produced by mixing acetate buffer (300 mM), 2,4,6-Tris(2-pyridyl)-s-triazine (10 mM) in HCl and FeCl3•6H2O (20 mM) in a 10:1:1 ratio. An amount of 500 µL of the sample solution was mixed with 500 µL of the FRAP reagent and incubated at room temperature for 30 min. The absorbance changes were measured at 593 nm and compared to a standard curve derived using FeSO4•7H2O solution. The FRAP values were expressed as mM FeSO4 equivalents and all assays were conducted in triplicate.
2.5. RNA Sequencing
The resuspension culture of Staphylococcus aureus ATCC 6538 that was treated with different durations (0, 1, 3, and 5 min) of cold plasma was harvested by centrifugation at 4 °C, 5000× g for 10 min. The supernatant was discarded, and the cell pellet was stored in 1× DNA/RNA shield solution (Zymo Research, Irvine, CA, USA) for further use. The cells were lysed using a mechanical lysis technique with a bead-beating method (2 × 3 min, 25 Hertz) in a Tissuelyser II (Qiagen, Germany). The cell lysate was subjected to total RNA extraction using a Quick-RNA Fungal/Bacterial MiniPrepTM kit with DNase I treatment (Zymo Research, Irvine, CA, USA). The extracted RNA was examined using 1% agarose gel electrophoresis, NanoDrop® ND-1000 for purity assessment, and Qubit® 3.0 fluorometer in conjunction with an RNA BR Assay Kit (Thermo Fisher Scientific, USA) for concentration determination. RNA integrity was determined using the TapeStation 2200 (Agilent, Santa Clara, CA, USA). Only RNA samples that passed the quality control were converted into cDNA and used to construct a sequencing library using an Illumina TruSeq Stranded Total RNA sample preparation kit, which included a ribosomal rRNA depletion step (Illumina, San Diego, CA, USA). The prepared libraries were checked using the TapeStation 2200 before sequencing with PE150 on a NovaSeq 6000 platform (Illumina, San Diego, CA, USA). Three biological replicates were conducted for each treatment, and a minimum of 10 million paired-end reads output was preserved for each sequencing.
2.6. Transcriptome Analysis
The raw data from the sequencer was processed using several software tools to ensure quality and accuracy. The raw data was first quality checked using Fastqc v0.11.9 [19], followed by adapter trimming and read filtering using BBDuk in BBTools v38.94 (parameters: ref = phix.fa, adapters.fa; trimq = 20; minlen = 100) [20]. The clean reads were then mapped to the reference genome of S. aureus ATCC 6538 (ASM202514v1) using HISAT2 v2.2.1 (parameters: --mp 6,2 --no-spliced-alignment) [21]. The gene expression quantification and profiling were performed using featureCounts in Subread v2.0.3 (parameters: -t gene -g gene_id -O -M -s 0 -p) [22], and differentially expressed genes (DEGs) were identified using DESeq2 v1.32.0 [23]. Unless otherwise specified, only genes with log2FC ≥ 0.5 and p value ≤ 0.05 were classified as DEGs and further analyzed. The consistency of biological replicates was evaluated via principal component analysis (PCA) in DESeq2. The functional annotations of the DEGs were assigned using EggNOG-mapper v2.1.9 with default parameters into clusters of orthologous groups (COGs) and KEGG orthology (KO numbers) [24]. Functional enrichment analyses were performed using ClusterProfiler [25]. All software was used with default parameters unless otherwise specified.
3. Results
3.1. Cold Plasma Properties
The cold plasma used in this study was generated from a self-developed in-house built device [17]. Its optical emission spectrum (OES) was obtained using a fiber optic spectrometer (StarLine AvaSpec-ULS4096CL-EVO) and the result was illustrated in Figure 2a. In brief, the spectrum showed emissions of species, such as NO lines (238.5 nm, 258.5 nm), O+ lines (272.3 nm, 328.2 nm, 436.8 nm, 464.9 nm), O lines (777.4 nm, 844.6 nm), N2 lines (333.9 nm, 401.4 nm), OH lines (306.4 nm, 307.5 nm), and N+ lines (343.7 nm, 399.5 nm). This indicated that the device-generated cold plasma consisted mainly of reactive oxygen species (ROS) and reactive nitrogen species (RNS), such as N2, O+, N+, OH, and NO.
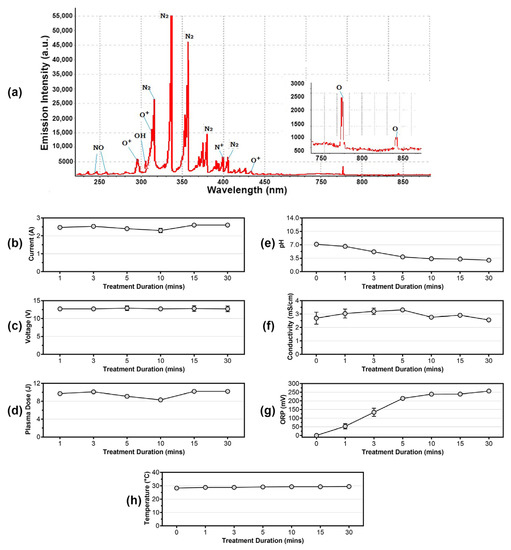
Figure 2.
(a) Optical emission spectrum (OES) of cold plasma. Measurement of (b) current, (c) voltage, and (d) plasma dose of the in-house built cold plasma device at different treatment durations. Detection of (e) pH changes, (f) electrical conduction, and (g) oxidation-reduction potential (ORP) of the cold plasma-treated solution. Measurement of (h) cold plasma temperature from 0 min to 30 min.
In addition, to monitor the stability of the device, the current, voltage, and each high voltage output pulse (plasma dose) in every discharging period were measured from 1 min to 30 min during the treatments. Based on the results, the current consumed by the device ranged from 2.30 A to 2.60 A (Figure 2b), voltage was considered stable at approximately 12.7–12.9 V (Figure 2c), and the generated plasma dose ranged from 8.3 J to 10.2 J, with an approximate average of 9.6 J (Figure 2d).
Based on studies, cold plasma-treated water will change in properties, such as pH, electrical conductivity, and oxidation-reduction potential (ORP) [26,27,28]. In this study, these parameters were measured after the 0.5% (w/v) saline distilled water was cold plasma-treated for different durations. As a result, the pH of the solution became more acidic as the treatment continued for a longer duration (Figure 2e). The electrical conductivity of the cold plasma-treated water fluctuated across the treatment durations, ranging from 2.57 to 3.31 mS/cm (Figure 2f), while the ORP of the treated water slowly increased from 0 mV (0 min) to 256 mV (30 min) (Figure 2g). Similar observations were recorded in other studies, in which the acidity was caused by the dissolving of acidic chemical species in cold plasma, and the increment of ORP was most likely due to the dissolved oxygen and other reactive particles in the water [26,27,28].
The temperature of the cold plasma remained at approximately 27.0–29.0 °C when the distance was fixed at 3 cm (Figure 2h). This is important for biomedical applications where cold plasma is directly applied on human tissue or skin. Nonetheless, during the experiment, the production of ozone by the device was approximately 0.12 ppm. This low ozone concentration renders the device safe for use in the medical field without worrying about the harm caused by excessive ozone exposure to the person-in-charge and to patients.
3.2. Cell Survivability
In this study, the effects of cold plasma on S. aureus cells was determined through both zone-of-inhibition diameter (on solid agar, Figure 3a,b) and cell survivability calculated via CFU plate count methods (in liquid suspension, Figure 3c) (cell survivability % = ). A zone-of-inhibition could only be observed when the treatment duration exceeded 5 min, indicating that cold plasma has a higher microbial inactivation rate with longer treatment. This finding was consistent with the results of cell survivability (Figure 3c), which showed that the survivability of S. aureus cells gradually decreased as the treatment duration of cold plasma on liquid culture increased. With treatments from 1–5 min, S. aureus could still retain a cell survivability over 40%, and complete cell death was achieved when the treatment reached 10 min or more.
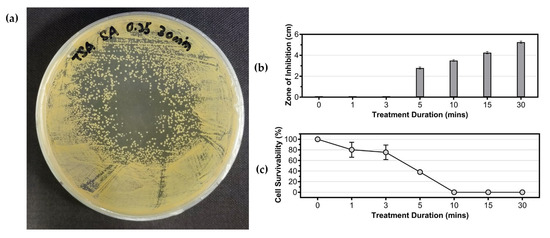
Figure 3.
Effects of cold plasma treatment on the growth of Staphylococcus aureus ATCC 6538. (a) Example of inhibition zone formation on the S. aureus spread plate; (b) effect of cold plasma treatment duration on zone of inhibition formed on agar plates; (c) cell survivability of S. aureus in liquid suspension after treatment with cold plasma.
3.3. Detection of Cell Lysate Composition
The cell membrane integrity of S. aureus was assessed via detection of intracellular components, such as DNA and protein, in the culture supernatant. As shown in Figure 4a, the concentration of DNA in the supernatant increased over the treatment duration, reaching a maximum of 10.30 ng/µL after 60 min of treatment. In contrast, the concentration of protein in the supernatant, as shown in Figure 4b, remained similar to the control (0 min) during treatment 1–15 min, but increased to 0.07 ng/µL and 0.13 ng/µL after 30 and 60 min of treatment, respectively. These findings suggest that the cell membrane integrity of S. aureus was compromised by cold plasma treatment, as indicated by the detection of both DNA and protein in the supernatant after longer treatment durations.
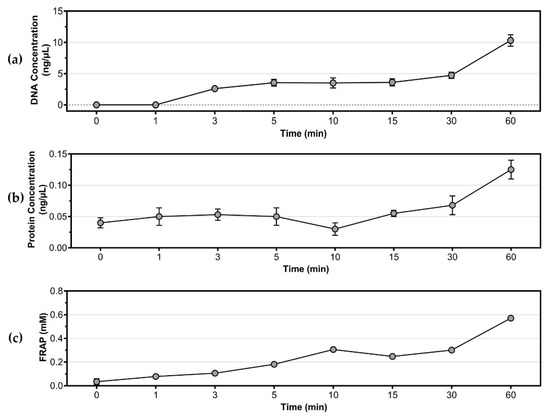
Figure 4.
Detection of intracellular components and antioxidants in culture supernatant. (a) Concentration of DNA; (b) concentration of protein; (c) total antioxidant activities of S. aureus as determined by FRAP assay (equivalent to mM FeSO4).
3.4. Antioxidant Assays
Cold plasma contains a high concentration of reactive oxygen species (ROS) and reactive nitrogen species (RNS) that can cause oxidative stress to bacteria. To counteract this stress, bacteria typically produce antioxidants. The total antioxidant level in cold plasma-treated S. aureus was measured by FRAP assay and is shown in Figure 4c. The results indicated that the antioxidant level gradually increased with the duration of cold plasma treatment, reaching a maximum of 0.57 mM FeSO4 equivalent at 60 min.
3.5. Differential Gene Expression on Cold Plasma-Treated Cell Culture
3.5.1. Technical Overview of RNA-Seq Data
Four experimental conditions were used for RNA sequencing, including a control experiment without cold plasma treatment (0 min, designated as T0) and three cold plasma treatments of 1, 3, and 5 min (designated as T1, T3, and T5, respectively). Three biological replicates were performed for each experimental condition to ensure the reproducibility and reliability of the transcriptome data. Table 1 provides general statistics of each sample. In general, each RNA sequencing generated approximately 3.32 Gb from approximately 11 million raw paired-end reads with a GC content of approximately 50.44%. Each sample had a sequencing quality of QPhred20 > 97% and QPhred30 > 93%. After adapter trimming and read filtering, each sample had approximately 8.1 million clean paired-end reads that were mapped to the reference genome of S. aureus ATCC 6538, with a mapping rate greater than 99%.

Table 1.
Summary of RNA sequencing data.
Figure 5a,b illustrates the biological replicate assessment of the RNA-sequencing data. The principal component analysis (PCA) plot in Figure 5a demonstrates that the three biological replicates of each treatment cluster close together, indicating high correlation among the replicates. Similarly, Figure 5b shows that the biological replicates from the same treatment tend to cluster together. Furthermore, when comparing across the four treatment groups, T0 and T1 form one cluster, while T3 and T5 form another, highlighting the high similarity within the two main clusters.
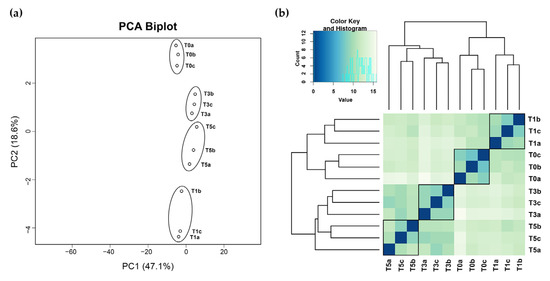
Figure 5.
Biological replicates assessment via (a) PCA plot and (b) heatmap and clustering of sample-to-sample distance.
3.5.2. Functional Enrichment Analyses of Differentially Expressed Genes (DEGs)
Differentially expressed genes (DEGs) analyses were performed by comparing the three treatment settings (T1, T3, and T5) with the control experiment (T0). Table 1 shows that the T1 treatment had 96 upregulated and 33 downregulated genes, T3 had 171 upregulated and 156 downregulated genes, while T5 had 204 upregulated and 142 downregulated genes, respectively. In total, the identified DEGs from each treatment group represented approximately 4.78–12.82% of the total genes (gene numbers = 2699) in the genome of S. aureus ATCC 6538.
Figure 6a,b shows the DEG enrichment analysis based on Clusters of Orthologous Groups (COGs) and Kyoto Encyclopedia of Genes and Genomes (KEGG) that assign genes to different microbial cell processes. Out of 497 identified DEGs, 444 and 312 DEGs were successfully assigned to COG categories and KO numbers, respectively. According to COGs classification (Figure 6a), the most affected DEGs after cold plasma treatment (T1, T3, and T5) were primarily categorized as “function unknown” (S), followed by biological processes of “inorganic ion transport and metabolism” (P), “transcription” (K), “energy production and conversion” (C), “amino acid transport and metabolism” (E), “cell wall/membrane/envelope biogenesis” (M), and “translation, ribosomal structures and biogenesis” (J).

Figure 6.
DEGs enrichment analyses based on (a) Clusters of Orthologous Groups and (b) Kyoto Encyclopedia of Genes and Genomes.
In terms of KEGG (Figure 6b), the metabolism of S. aureus was most affected by cold plasma treatment, with the highest number of DEGs found in metabolic processes such as “carbohydrate metabolism”, “amino acid metabolism”, and “metabolism of terpenoids and polyketides”. Some genes involved in “membrane transport” and “signal transduction”, which belong to the category of environmental information processing, were also differentially expressed. Pathways related to genetic information processing, such as “translation”, “replication and repair”, and “folding, sorting and degradation”, were also affected by cold plasma treatment, particularly in the T3 and T5 experiments.
3.5.3. In-Depth Comparative Analysis of DEGs Harbored from T1, T3, and T5 Experiments
A total of 12 DEGs related to stress proteins were identified in the three treatment settings (T1, T3, and T5), please refer to (a) in Table 2. The majority were upregulated, with nine out of the 12 stress protein-related genes being upregulated in T1, two up- and one downregulated in T3, and seven up- and one downregulated in T5. This disparity between treatments might be due to S. aureus’s response to the sudden exposure of cold plasma in the early treatment (T1), where more RNA encode for stress proteins were generated than in the other treatments (T3 and T5). Other research revealed each stress protein can be regulated by different stress conditions, such as Gls24 family stress response protein, which can be induced by aerated conditions [29] and copper stress [30], and GlsB/YeaQ/YmgE family stress response protein, which is possibly related to butanol [31] and salt tolerance [32].

Table 2.
List of DEGs related to various biological processes identified in T1, T3, and T5 treatments. The biological processes include (a) stress proteins, (b) antioxidants, (c) nitrosative stress, and (d) DNA repairing.
A total of six DEGs related to antioxidants were identified in cold plasma-treated S. aureus, all of which were upregulated (three in T1/T3/T5), please refer to (b) in Table 2. The involved antioxidant enzymes are glutathione peroxidase, superoxide dismutase, thioredoxin, and thiol peroxidase. Cold plasma consists of reactive nitrogen species (RNS), which can induce nitrosative stress response in microorganisms [33,34,35]. In this study, a nitrosative stress response by S. aureus upon exposure to cold plasma was observed, with a total of five DEGs related to nitrosative stress identified, please refer to (c) in Table 2. Four out of the five genes were upregulated in T1, three in T3, and four in T5, indicating that S. aureus responded equally high to RNS in all treatments.
In this study, 20 DEGs related to DNA repairing were identified. None of them were up- or downregulated in T1, but most of them were upregulated in T3 (20 genes) and T5 (17 genes). This difference is because the cold plasma had not yet caused damage to the bacterial intracellular DNA in the early treatment (T1); therefore, none of the genes were identified as DEGs in T1, please refer to (d) in Table 2. However, in the prolonged treatment (T3 and T5), the accumulative ROS, RNS, and other energized molecules and atoms surrounded the bacterial cells, damaging the DNA in the cells and triggering the respective repairing mechanisms.
In this study, besides the DEGs related to stress proteins, antioxidants, nitrosative stress, and DNA repair, other biological processes were also analyzed. As shown in Table 3, genes related to virulence factors, cell membrane/wall, transcription, and translation processes were identified. The virulence genes that encode for proteins, such as autolysin, adhesin, and hemolysin, play a significant role in making bacteria infectious [36,37,38]. In this study, 21 DEGs annotated as possible virulence factors of S. aureus were identified, and most of these virulence genes were downregulated after cold plasma treatment, please refer to (a) in Table 3. The results showed that cold plasma treatment may reduce the pathogenicity of S. aureus by decreasing the production of virulence factors.

Table 3.
List of DEGs related to various biological processes identified in T1, T3, and T5 treatments. The biological processes include (a) virulence factors, (b) cell wall and membrane, (c) transporters, (d) transcription, and (e) translation.
Regarding the cell membrane/wall-related genes, 18 DEGs were identified, please refer to (b) in Table 3. Only one gene was downregulated in T1, three up- and eight downregulated genes in T3, and five up- and eight downregulated genes in T5. This indicated that the effects of the cold plasma treatment on the cell membrane/wall were only apparent after 3 or 5 min of treatments.
The study also analyzed the transporter-related genes, with a total of 34 DEGs identified, please refer to (c) in Table 3. In T1, 13 were upregulated and one was downregulated, while in T3, 11 were upregulated and 11 were downregulated. In T5, there were 13 up- and 10 downregulated transporter genes. Most of these transporters are responsible for transporting inorganic ions (i.e., metal ions such as potassium, zinc, and sodium) and amino acids.
In terms of transcription and translation processes, a total of 30 and 15 DEGs were identified, respectively, please refer to (d) and (e) in Table 3. In T1, fewer DEGs were identified in the transcription (two up and two down) and translation (none of them are up or down) processes, indicating that the cold plasma treatment did not significantly affect these processes. In contrast, T3 had 19 (seven up and 12 down) and 11 (seven up and three down) DEGs identified in the transcription and translation processes. In T5, 22 (13 up and nine down) and 12 (10 up and two down) DEGs were identified, respectively. These results indicate that the cold plasma treatment impacted the transcription and translation processes in the cell starting from 3 min of treatment.
4. Discussion
Cold plasma is commonly utilized in the agricultural, food, and pharmaceutical industries for removing surface contaminants from products before packaging. It is also used in the medical field for disinfecting working surfaces and treating patients’ wounds. The effectiveness of cold plasma treatment is influenced by biotic and abiotic factors. Biotic factors refer to the types of targeted microorganisms, with studies finding cold plasma to be more effective against Gram-negative bacteria, followed by Gram-positive bacteria, and fungi [39,40,41]. The abiotic factors that affect the effectiveness of cold plasma include the design of the cold plasma device (i.e., plasma generation method, voltage or power applied, type of gas, and others) and treatment parameters (i.e., distance, duration, cell concentration, and others) [42]. The design of the cold plasma device used in this study had been previously reported and has advantages, such as lower power consumption, lower ultra-violet wavelength, lower temperature, and higher generation of reactive oxygen and nitrogen species [17]. The current study also established the minimum treatment time for cold plasma treatment of S. aureus, which were a duration of at least 5 min for culture on solid agar or 10 min for culture in liquid suspension.
Cold plasma treatment has been shown to cause cell death by lysis, which can be determined by detecting DNA and protein in the lysate [43]. In this study, DNA and protein leakage were observed, particularly with prolonged cold plasma treatment of 30 and 60 min, indicating the effectiveness of cold plasma in causing cell lysis. Similar findings have been observed in cold plasma treatment of E. coli [43], Pseudomonas aeruginosa [44], and Lentilactobacillus hilgardii [45]. However, the treatment duration required for the detection of DNA and protein is longer for Gram-positive bacteria, such as S. aureus, possibly due to the thicker cell wall. In E. coli, a significant amount of DNA and protein could be detected with treatment less than 40 s [43].
To investigate the biological process of sequential cell responses of cold plasma-treated S. aureus, the treatment experiments for transcriptome studies were conducted. Total RNA was extracted from each sample and subjected to RNA sequencing and differentially expressed genes (DEGs) determination. Stress proteins are a diverse group of evolutionary conserved proteins produced by both prokaryotes and eukaryotes when cells are exposed to stressful conditions, and are mainly used to protect the cells and other proteins from damage [46]. In this study, several genes (B4602_RS08710, B4602_RS03580, B4602_RS04355, B4602_RS08280, B4602_RS08950, B4602_RS09460, and B4602_RS09855) that encode for stress proteins were found to be upregulated, please refer to (a) in Table 2. B4602_RS08710 is a gene that encodes a universal stress protein, which belongs to the UspA family and can respond to many stressors, protecting the cell from DNA damage [47]. The expression of this protein is most likely due to oxidative stress caused by ROS/RNS or minimal ultraviolet wavelengths generated from cold plasma, which could damage the DNA of S. aureus. B4602_RS03580, a gene that encodes for another stress protein (CsbB), is an extracellular cell envelope stress protein that can cope with oxidative stress and is responsible for lipoteichoic acid glycosylation [48,49]. B4602_RS01845 encodes for a general stress protein containing a 17M-like domain, is induced by oxidative stress and has a non-specific protective mechanism for the cell [50]. The rest of the genes either encode for CsbD (B4602_RS04355 and B4602_RS08280) or YtxH (B4602_RS08950, B4602_RS09460, and B4602_RS09855) are currently with unknown function. The related reports on cold plasma-induced stress proteins were mainly from eukaryotic cells and discussed heat shock protein, another class of stress proteins [51,52]. However, heat shock proteins from S. aureus were not induced by cold plasma in this study, indicating that the microbial inactivation mechanism of cold plasma is not related to heat.
Reactive oxygen species (ROS) is the main species of cold plasma, contributing to a high oxidative stress environment for treated cells [13]. ROS includes species such as superoxide (•O2–), hydrogen peroxide (H2O2), triplet oxygen (O22•), hydroxyl radical (HO•), hydroxide ion (HO–), peroxide ion (O22–), and others. To counteract this oxidative stress, bacteria produce antioxidants. In this study, cold plasma-treated S. aureus expressed an equivalent number of antioxidants throughout the three treatment durations, please refer to (b) in Table 2. Superoxide dismutase (B4602_RS07925) was used to catalyze the dismutation of superoxide (O2–) into oxygen (O2) and hydrogen peroxide (H2O2), which was then catalyzed by glutathione peroxidase (B4602_RS06460) into H2O and O2, or by thioredoxin (B4602_RS08910 and B4602_RS13400) into H2O with the help of NADPH [53]. S. aureus also expressed an organic hydroperoxide resistance protein (B4602_RS04165), belonging to the OsmC/Ohr family, which is involved in the cellular defense against oxidative stress caused by exposure to O2– or elevated osmolarity [54]. Another thiol peroxidase (B4602_RS08725), containing a thioredoxin domain is believed to catalyze H2O2 into H2O. Nonetheless, the catalase, another possible antioxidant enzyme in S. aureus, was unresponsive. The genes related to staphyloxanthin biosynthesis governed by crtOPQMN operon were also not differentially expressed; staphyloxanthin is a carotenoid that contributes to golden yellow pigmentation in S. aureus and is a potent non-enzymatic antioxidant [55]. Despite this, the upregulation of enzymatic antioxidants as shown earlier indicated that the cells actively counteracted the oxidative stress. In addition, the antioxidant test using the FRAP assay (Figure 4c) confirmed that the treated S. aureus was constantly producing antioxidants throughout the cold plasma treatment.
Reactive nitrogen species (RNS) is another group of energized ions and radicals that contribute to high oxidative stress to the treated cells. RNS includes species of dinitrogen trioxide (N2O3), nitric oxide (NO•), nitrite (NO2–), peroxynitrite (ONOO–), and others. This study observed the upregulation of genes related to nitrosative stress response of S. aureus in three treatment settings, as shown in (c) of Table 2. Most of the upregulated genes are found as part of the same gene cluster in the S. aureus genome and neighbor each other. This gene cluster includes a NarK/NasA family nitrate transporter (B4602_RS12580) involved in the transfer of nitrate ions (NO3–) into and out of the cell membrane. A nitrate respiration regulation regulator NreC (B4602_RS12585) is reported to regulate the transcription of relevant genes in the gene cluster. Nitrate reductase (B4602_RS12610, and B4602_RS12615) reduce nitrate (NO3–) to nitrite (NO2–) [56], and a nitrite reductase (B4602_RS12630) further reduces nitrite (NO2–) to ammonium (NH4+) [56]. This gene cluster also includes another seven genes that were not differentially expressed in either experiment.
The DNA repairing mechanism, also known as the SOS response, is a biological process that occurs in bacteria when DNA is damaged by external forces, such as ultraviolet radiation or oxidation caused by ROS, RNS, and antibiotics [57]. In this study, genes related to DNA repairing were only differentially expressed in T3 and T5 treatments, as shown in (d) of Table 2. T1 had zero DEGs, likely because the short duration of cold plasma exposure was not enough to cause damage or oxidation of the DNA. There was a total of 20 identified DEGs, mostly enzymatic-related, including nucleases, photolyase, polymerases, gyrases, helicase and recombinase. Nucleases, such as endonuclease III (B4602_RS07390), are responsible for removing and repairing damaged oxidized pyrimidine bases in the DNA [57], and excinuclease ABC (B4602_RS03910 and B4602_RS02585) repairs DNA damage caused by UV radiation [58]. Similarly, photolyase (B4602_RS03515) repairs UV-exposed DNA [58]. Polymerases (B4602_RS05610, B4602_RS08595, B4602_RS09940) play a role in DNA polymerization [59], refilling gaps in the DNA after damaged bases have been cleaved by nucleases. Gyrase (B4602_RS00030) and helicase (B4602_RS06055) help wind and unwind DNA during the DNA repairing process, while recombinase (B4602_RS06365) can facilitate DNA repair by recombination [59]. In addition to these enzymes, other proteins related to DNA repairing were observed, including nucleoid-associated proteins (B4602_RS02245 and B4602_RS14360) and DNA-binding proteins (B4602_RS03960, B4602_RS07395, and B4602_RS07495), which help maintain the structure and stability of the DNA [60]. Other proteins such as RecN (B4602_RS07745), work as regulators or are essential for the enzymatic activities described above, mediating the DNA recombination event by recombinase RecA (B4602_RS06365) [59]. These findings show the evidence of DNA damage caused by cold plasma at longer duration and the triggering of the DNA repairing mechanism in S. aureus.
The ROS and RNS produced by cold plasma have been shown to cause lipid peroxidation in the cell membrane of bacteria in various studies [13,14,33,34]. We were able to detect minor effects of cold plasma on the cell membrane/wall of S. aureus through DEG analysis. Eighteen DEGs were identified to be related to the cell membrane/wall, with most being unresponsive in T1 treatment due to the short duration of treatment, as shown in (b) of Table 3. However, in T3 and T5 treatments, a high proportion of these genes were downregulated. Some of these downregulated genes (B4602_RS03240, B4602_RS05125, B4602_RS06140, B4602_RS06915, B4602_RS06975) are involved in lipoteichoic acid and teichoic acid polymerization and attachment to the peptidoglycan cell wall of S. aureus, which are important for maintaining the integrity of the cell membrane/wall [61]. Other downregulated genes are responsible for cell wall/membrane remodeling during cell division (B4602_RS05445, B4602_RS05810, B4602_RS05820, B4602_RS05840, and B4602_RS07355). The downregulation of these genes suggests that the S. aureus may not be able to maintain its coccus shape during the cold plasma treatment and its cell division process may be hindered. On the other hand, DEGs related to peptidoglycan synthesis were not detected, indicating that the cell wall had not been heavily affected even after 5 min of cold plasma treatment and no repairing was required. This study highlights the potential of cold plasma to affect the biological process of S. aureus through its impact on the cell membrane/wall.
The virulence factors of microbial pathogens are cellular structures, molecules, and regulatory systems that play a role in colonizing and infecting the hosts, evading the host’s immune system, and causing diseases [62]. As S. aureus is an opportunistic pathogen that causes various diseases in humans, many of its potential virulence factors have been identified and studied by researchers [36,37,38,62,63,64]. Our study found that many genes of virulence factors in S. aureus were differentially expressed in all three treatments, as shown in (a) of Table 3. Most of them were downregulated, such as adhesins (B4602_RS02160 and B4602_RS02790), which help to anchor the cell to the host [37], and autolysins (B4602_RS02160 and B4602_RS03375), which break down peptidoglycan and play a role in cell wall turnover [36]. The complement convertase inhibitor Ecb (B4602_RS05670) and Efb (B4602_RS05690), which help the pathogen evade the host’s complement system to pathogens [65], were also downregulated, making the treated S. aureus more vulnerable to the host’s immune response. The downregulation of glycopeptide resistance-associated protein GraF (B4602_RS05255) and penicillin-binding protein 2 (B4602_RS07920) also showed that the treated S. aureus was now more vulnerable to antibiotics. The toxic peptides alpha-hemolysin (B4602_RS05715) and delta-hemolysin (B4602_RS14805), which can cause hemolytic effect in infected patients [38], were also downregulated. In summary, more virulence genes were downregulated after the cold plasma treatment, indicating a decrease in the virulence of S. aureus.
The gene expression of transporters, transcriptional regulators, and translation-related genes were also evaluated in this study, please refer to (c)–(e) in Table 3. The expression of metal ion transporters for potassium, zinc, iron, copper, and cadmium were mostly upregulated (B4602_RS10920, B4602_RS10925, B4602_RS11290, B4602_RS12970, B4602_RS13515, and B4602_RS14395). This upregulation could be due to the need for metal ions as cofactors for some of the antioxidants or enzymes [53], or as a result of the inner cell homeostasis of metal ions in response to the charges variation caused by ROS and RNS produced by the cold plasma [66]. Meanwhile, the regulation pattern of transporters responsible for amino acids (B4602_RS04230 and B4602_RS04880) and other substrates’ (B4602_RS00360, B4602_RS05390, and B4602_RS11840) transportation was not observed, and some transporters have unknown function. For the transcriptional regulation, T1 showed fewer DEGs compared to T3 and T5 treatments. Its DEGs were limited to B4602_RS04955 and B4602_RS05910, which were also downregulated in other treatments, and the upregulation of two unique genes (B4602_RS11285 and B4602_RS14390), which require metal ions for transcriptional regulation. T3 and T5 treatments had more DEGs related to transcriptional regulation, mostly downregulated, such as the YlbF/YmcA family competence regulator (B4602_RS09505) that controls the bacterial cell’s competency in taking up foreign DNA from the environment, LacI (B4602_RS10690) that regulates carbohydrate utilization genes, and AraC (B4602_RS12545) that regulates virulence genes in bacteria. T3 and T5 had more DEGs related to translation compared to T1 indicated that the translation process of S. aureus is affected by cold plasma treatment.
Previous transcriptomic studies on microorganisms treated with cold plasma have been conducted [13,14,15,16,67,68,69,70], where the studies mainly focused on the bacterial cell response at a single timepoint cold plasma treatment (treated vs. untreated). The Bacillus subtilis exposed to cold plasma treatment (15 min) exhibited transcriptional changes in cell membrane related processes, transcriptional regulators and oxidative stress response [70]. Cold plasma treatment of E. coli (15 min) showed DEGs related to cellular metabolism, transporters, and ribosomal proteins [67]. Our results for prolonged treatment (T3 and T5) demonstrated consistency to the findings of the single timepoint cold plasma treatment using other bacterial cells (Bacillus subtilis and E. coli).
Collectively, S. aureus showed differences in its cell biological response when exposed to cold plasma for different duration (Figure 7). In brief, at the initial treatment (T1), genes that encode for proteins related to stress, antioxidants, and transporter proteins were upregulated. No significant changes were observed in the cell membrane/wall synthesis, DNA repair mechanisms, transcription, and translation biological processes when T1 was compared to the control experiment (T0). When compared to the T1 treatment, T3 and T5 demonstrated that more DEGs related to transcription, translation and DNA repairing were identified, indicating that the biological processes of central dogma in S. aureus were affected by cold plasma treatment. In addition, lesser genes related to the production of stress proteins were upregulated in T3 and T5 compared to T1. The majority of the genes related to cell membrane/wall synthesis were downregulated in T3 and T5, indicating that S. aureus can no longer sustain its cell integrity under prolonged cold plasma treatment. Transporter proteins were also affected by cold plasma during prolonged treatment.

Figure 7.
Sequential responses in biological processes of S. aureus revealed by comparative analysis of the three treatment settings (T1, T3 and T5).
5. Conclusions
This study demonstrated the antimicrobial efficiency of the cold plasma generated from a self-developed plasma jet device. The transcriptome analysis showed different DEGs in the biological processes of S. aureus when different durations of cold plasma were applied, which can be interpreted as the sequential response of the bacteria against the cold plasma. Briefly, the initial response of S. aureus to cold plasma involves biological processes such as stress proteins, antioxidants production, and nitrosative stress response. Then, it was followed by affection on biological processes in DNA repair, transcription, translation, and cell membrane/wall synthesis, which can be found in the prolonged cold plasma treatment. Other transcriptome studies in cold plasma-treated bacteria were focused on single timepoint treatment, while the comparative study across different treatment duration was first attempted in this study. In future research, the sequential response of other bacteria can be conducted by using a similar approach. Furthermore, proteomics and metabolomics response of cold plasma treated bacterial cells could be explored.
Author Contributions
Conceptualization, K.J.L., X.Z. and C.S.C.; methodology, K.J.L., X.Z. and C.S.C.; software, K.J.L.; validation, K.J.L., X.Z. and C.S.C.; formal analysis, K.J.L.; investigation, K.J.L., K.L.C. and M.C.Y.T.; resources, X.Z., X.C., D.R., J.C., Z.C. and C.S.C.; data curation, K.J.L.; writing—original draft preparation, K.J.L.; writing—review and editing, X.Z., D.R., J.C. and C.S.C.; visualization, K.J.L.; supervision, K.J.L., X.Z. and C.S.C.; project administration, K.J.L., X.Z. and C.S.C.; funding acquisition, X.Z. and C.S.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Professional Development Research University Grant (grant number: 06E00) and External Grant (4B779) from Universiti Teknologi Malaysia. This study was also supported by the Gusu Innovation and Entrepreneurship Leading Talents Project (ZXL2018192), and the Suzhou 2020 Novel Coronavirus Emergency Prevention and Control Technology Project, and the 2020 Jiangsu Province Industry-University-Research Cooperation Project (BY2020506). K.J.L. is a Researcher at the Universiti Teknologi Malaysia under the Post-Doctoral Fellowship Scheme for the Project: “Transcriptome Analysis of Low Temperature Air Plasma Jet-Treated Microorganisms”.
Data Availability Statement
The raw data for RNA sequencing of Staphylococcus aureus ATCC 6538 in this study had been deposited in NCBI sequence Read Archive (SRA) with accession numbers SRR22386641–SRR22386652. The data can also be accessed via BioProject accession number PRJNA904504 and BioSample accession numbers SAMN31843833–SAMN31843844.
Acknowledgments
K.L.C. and M.C.Y.T. would like to thank Universiti Teknologi Malaysia for providing them the Zamalah Scholarship for their Ph.D. studies.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Silva-Santana, G.; Cabral-Oliviera, G.; Oliveira, D.R.; Nogueira, B.A.; Pereira-Ribeiro, P.M.A.; Mattos-Guaraldi, A.L. Staphylococcus aureus Biofilms: An Opportunistic Pathogen with Multidrug Resistance. Rev. Res. Med. Microbiol. 2020, 32, 12–21. [Google Scholar] [CrossRef]
- Guo, Y.; Song, G.; Sun, M.; Wang, J.; Wang, Y. Prevalence and Therapies of Antibiotic-Resistance in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed]
- Stein, E. Microbiologically Safe Foods; ED-Tech Press: Waltham Abbey, UK, 2018; ISBN 978-1-83947-354-8. [Google Scholar]
- Azuma, T.; Murakami, M.; Sonoda, Y.; Ozaki, A.; Hayashi, T. Occurrence and Quantitative Microbial Risk Assessment of Methicillin-Resistant Staphylococcus aureus (MRSA) in a Sub-Catchment of the Yodo River Basin, Japan. Antibiotics 2022, 11, 1355. [Google Scholar] [CrossRef]
- Li, J.; Wen, Q.; Gu, F.; An, L.; Yu, T. Non-Antibiotic Strategies for Prevention and Treatment of Internalized Staphylococcus aureus. Front. Microbiol. 2022, 13, 974984. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.; Small, D.A.; Toghrol, F.; Bentley, W.E. Global Transcriptome Analysis of Staphylococcus aureus Response to Hydrogen Peroxide. J. Bacteriol. 2006, 188, 1648–1659. [Google Scholar] [CrossRef]
- Song, M.; Zeng, Q.; Xiang, Y.; Gao, L.; Huang, J.; Huang, J.; Wu, K.; Lu, J. The Antibacterial Effect of Topical Ozone on the Treatment of MRSA Skin Infection. Mol. Med. Rep. 2018, 17, 2449–2455. [Google Scholar] [CrossRef]
- Kaiki, Y.; Kitagawa, H.; Hara, T.; Nomura, T.; Omori, K.; Shigemoto, N.; Takahashi, S.; Ohge, H. Methicillin-Resistant Staphylococcus aureus Contamination of Hospital-Use-Only Mobile Phones and Efficacy of 222-Nm Ultraviolet Disinfection. Am. J. Infect. Control 2021, 49, 800–803. [Google Scholar] [CrossRef]
- Laroussi, M. Cold Plasma in Medicine and Healthcare: The New Frontier in Low Temperature Plasma Applications. Front. Phys. 2020, 8, 74. [Google Scholar] [CrossRef]
- Ziuzina, D.; Han, L.; Cullen, P.J.; Bourke, P. Cold Plasma Inactivation of Internalised Bacteria and Biofilms for Salmonella enterica Serovar Typhimurium, Listeria monocytogenes and Escherichia coli. Int. J. Food Microbiol. 2015, 210, 53–61. [Google Scholar] [CrossRef]
- Xu, D.; Zhang, X.; Zhang, J.; Feng, R.; Wang, S.; Yang, Y. Metabolomics of Pseudomonas aeruginosa Treated by Atmospheric-Pressure Cold Plasma. Appl. Sci. 2021, 11, 10527. [Google Scholar] [CrossRef]
- Nasir, N.M.; Lee, B.; Yap, S.S.; Thong, K.; Yap, S.L. Cold Plasma Inactivation of Chronic Wound Bacteria. Arch. Biochem. Biophys. 2016, 605, 76–85. [Google Scholar] [CrossRef]
- Han, L.; Patil, S.; Boehm, D.; Milosavljević, V.; Cullen, P.J.; Bourke, P. Mechanisms of Inactivation by High-Voltage Atmospheric Cold Plasma Differ for Escherichia Coli and Staphylococcus aureus. Appl. Environ. Microbiol. 2016, 82, 450–458. [Google Scholar] [CrossRef]
- Liao, X.; Xiang, Q.; Liu, D.; Chen, S.; Ye, X.; Ding, T. Lethal and Sublethal Effect of a Dielectric Barrier Discharge Atmospheric Cold Plasma on Staphylococcus aureus. J. Food Prot. 2017, 80, 928–932. [Google Scholar] [CrossRef]
- Liao, X.; Cullen, P.J.; Liu, D.; Muhammad, A.I.; Chen, S.; Ye, X.; Wang, J.; Ding, T. Combating Staphylococcus aureus and Its Methicillin Resistance Gene (MecA) with Cold Plasma. Sci. Total Environ. 2018, 645, 1287–1295. [Google Scholar] [CrossRef]
- Liao, X.; Liu, D.; Ding, T. Nonthermal Plasma Induces the Viable-but-Nonculturable State in Staphylococcus aureus via Metabolic Suppression and the Oxidative Stress Response. Appl. Environ. Microbiol. 2020, 86, e02216–e02219. [Google Scholar] [CrossRef]
- Zhang, X.; Liew, K.J.; Chong, C.S.; Cai, X.; Chang, Z.; Jia, H.; Liu, P.; He, H.; Liu, W.; Li, Y. Low-Temperature Air Plasma Jet for Inactivation of Bacteria (S. aureus and E. coli) and Fungi (C. albicans and T. rubrum). Acta Phys. Pol. A 2023, 143, 12–18. [Google Scholar] [CrossRef]
- Benzie, I.F.; Devaki, M. The Ferric Reducing/Antioxidant Power (FRAP) Assay for Non-enzymatic Antioxidant Capacity: Concepts, Procedures, Limitations and Applications. In Measurement of Antioxidant Activity & Capacity: Recent Trends and Applications; Apak, R., Capanoglu, E., Shahidi, F., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2018; Chapter 5; pp. 77–106. [Google Scholar]
- Andrews, S. FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 1 September 2022).
- Bushnell, B. BBTools—DOE Joint Genome Institute. Available online: https://jgi.doe.gov/data-and-tools/software-tools/bbtools/ (accessed on 1 September 2022).
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-Based Genome Alignment and Genotyping with HISAT2 and HISAT-Genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L. ClusterProfiler 4.0: A Universal Enrichment Tool for Interpreting Omics Data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Soni, A.; Choi, J.; Brightwell, G. Plasma-Activated Water (PAW) as a Disinfection Technology for Bacterial Inactivation with a Focus on Fruit and Vegetables. Foods 2021, 10, 166. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Zhou, X.; Yang, W.; Zhang, Y.; Ye, Z.; Hu, S.; Ye, C.; Li, Y.; Lan, Y.; Shen, J. In Vitro Antimicrobial Effects and Mechanism of Air Plasma-activated Water on Staphylococcus aureus Biofilm. Plasma Process. Polym. 2020, 17, 1900270. [Google Scholar] [CrossRef]
- Zhao, Y.; Patange, A.; Sun, D.; Tiwari, B. Plasma-activated Water: Physicochemical Properties, Microbial Inactivation Mechanisms, Factors Influencing Antimicrobial Effectiveness, and Applications in the Food Industry. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3951–3979. [Google Scholar] [CrossRef]
- Cretenet, M.; Le Gall, G.; Wegmann, U.; Even, S.; Shearman, C.; Stentz, R.; Jeanson, S. Early Adaptation to Oxygen Is Key to the Industrially Important Traits of Lactococcus Lactis Ssp. Cremoris during Milk Fermentation. BMC Genom. 2014, 15, 1–15. [Google Scholar] [CrossRef]
- Stoyanov, J.V.; Mancini, S.; Lu, Z.H.; Mourlane, F.; Poulsen, K.R.; Wimmer, R.; Solioz, M. The Stress Response Protein Gls24 Is Induced by Copper and Interacts with the CopZ Copper Chaperone of Enterococcus hirae. FEMS Microbiol. Lett. 2010, 302, 69–75. [Google Scholar] [CrossRef]
- Goyal, L.; Jalan, N.K.; Khanna, S. Butanol Tolerant Bacteria: Isolation and Characterization of Butanol Tolerant Staphylococcus sciuri sp. J. Biotech Res. 2019, 10, 68–77. [Google Scholar]
- Jo, E.; Hwang, S.; Cha, J. Transcriptome Analysis of Halotolerant Staphylococcus saprophyticus Isolated from Korean Fermented Shrimp. Foods 2022, 11, 524. [Google Scholar] [CrossRef]
- Ercan, U.; Sen, B.; Brooks, A.; Joshi, S. Escherichia coli Cellular Responses to Exposure to Atmospheric-pressure Dielectric Barrier Discharge Plasma-treated N-acetylcysteine Solution. J. Appl. Microbiol. 2018, 125, 383–397. [Google Scholar] [CrossRef]
- Patange, A.; O’Byrne, C.; Boehm, D.; Cullen, P.; Keener, K.; Bourke, P. The Effect of Atmospheric Cold Plasma on Bacterial Stress Responses and Virulence Using Listeria monocytogenes Knockout Mutants. Front. Microbiol. 2019, 10, 2841. [Google Scholar] [CrossRef]
- Gilmore, B.F.; Flynn, P.B.; O’Brien, S.; Hickok, N.; Freeman, T.; Bourke, P. Cold Plasmas for Biofilm Control: Opportunities and Challenges. Trends Biotechnol. 2018, 36, 627–638. [Google Scholar] [CrossRef]
- Heilmann, C.; Hartleib, J.; Hussain, M.S.; Peters, G. The Multifunctional Staphylococcus aureus Autolysin Aaa Mediates Adherence to Immobilized Fibrinogen and Fibronectin. Infect. Immun. 2005, 73, 4793–4802. [Google Scholar] [CrossRef]
- Bartlett, A.H.; Hulten, K.G. Staphylococcus aureus Pathogenesis: Secretion Systems, Adhesins, and Invasins. Pediatr. Infect. Dis. J. 2010, 29, 860–861. [Google Scholar] [CrossRef]
- Vandenesch, F.; Lina, G.; Henry, T. Staphylococcus aureus Hemolysins, Bi-Component Leukocidins, and Cytolytic Peptides: A Redundant Arsenal of Membrane-Damaging Virulence Factors? Front. Cell. Infect. Microbiol. 2012, 2, 12. [Google Scholar] [CrossRef]
- Mai-Prochnow, A.; Murphy, A.B.; McLean, K.M.; Kong, M.G.; Ostrikov, K.K. Atmospheric Pressure Plasmas: Infection Control and Bacterial Responses. Int. J. Antimicrob. Agents 2014, 43, 508–517. [Google Scholar] [CrossRef]
- González-González, C.R.; Labo-Popoola, O.; Delgado-Pando, G.; Theodoridou, K.; Doran, O.; Stratakos, A.C. The Effect of Cold Atmospheric Plasma and Linalool Nanoemulsions against Escherichia coli O157: H7 and Salmonella on Ready-to-Eat Chicken Meat. LWT 2021, 149, 111898. [Google Scholar] [CrossRef]
- Qian, J.; Ma, L.; Yan, W.; Zhuang, H.; Huang, M.; Zhang, J.; Wang, J. Inactivation Kinetics and Cell Envelope Damages of Foodborne Pathogens Listeria monocytogenes and Salmonella enteritidis Treated with Cold Plasma. Food Microbiol. 2022, 101, 103891. [Google Scholar] [CrossRef]
- Coutinho, N.M.; Silveira, M.R.; Rocha, R.S.; Moraes, J.; Ferreira, M.V.S.; Pimentel, T.C.; Freitas, M.Q.; Silva, M.C.; Raices, R.S.L.; Ranadheera, C.S. Cold Plasma Processing of Milk and Dairy Products. Trends Food Sci. Technol. 2018, 74, 56–68. [Google Scholar] [CrossRef]
- Ahmed, M.W.; Naqvi, S.M.; Qasim, I.; Noreen, Z.; Shafiq, M.; Bukhari, H. Degradation of Multidrug-Resistant E. coli by Low Pressure Plasma. Int. J. Food Prop. 2021, 24, 1289–1299. [Google Scholar] [CrossRef]
- Ziuzina, D.; Boehm, D.; Patil, S.; Cullen, P.; Bourke, P. Cold Plasma Inactivation of Bacterial Biofilms and Reduction of Quorum Sensing Regulated Virulence Factors. PLoS ONE 2015, 10, e0138209. [Google Scholar] [CrossRef]
- Niedźwiedź, I.; Juzwa, W.; Skrzypiec, K.; Skrzypek, T.; Waśko, A.; Kwiatkowski, M.; Pawłat, J.; Polak-Berecka, M. Morphological and Physiological Changes in Lentilactobacillus hilgardii Cells after Cold Plasma Treatment. Sci. Rep. 2020, 10, 18882. [Google Scholar] [CrossRef] [PubMed]
- Wan, Q.; Song, D.; Li, H.; He, M. Stress Proteins: The Biological Functions in Virus Infection, Present and Challenges for Target-Based Antiviral Drug Development. Signal Transduct. Target. Ther. 2020, 5, 125. [Google Scholar] [CrossRef] [PubMed]
- Siegele, D.A. Universal Stress Proteins in Escherichia coli. J. Bacteriol. 2005, 187, 6253–6254. [Google Scholar] [CrossRef] [PubMed]
- Inoue, H.; Suzuki, D.; Asai, K. A Putative Bactoprenol Glycosyltransferase, CsbB, in Bacillus subtilis Activates SigM in the Absence of Co-Transcribed YfhO. Biochem. Biophys. Res. Commun. 2013, 436, 6–11. [Google Scholar] [CrossRef]
- Rismondo, J.; Percy, M.G.; Gründling, A. Discovery of Genes Required for Lipoteichoic Acid Glycosylation Predicts Two Distinct Mechanisms for Wall Teichoic Acid Glycosylation. J. Biol. Chem. 2018, 293, 3293–3306. [Google Scholar] [CrossRef]
- Völker, U.; Engelmann, S.; Maul, B.; Riethdorf, S.; Völker, A.; Schmid, R.; Mach, H.; Hecker, M. Analysis of the Induction of General Stress Proteins of Bacillus subtilis. Microbiology 1994, 140, 741–752. [Google Scholar] [CrossRef]
- Bekeschus, S.; Lippert, M.; Diepold, K.; Chiosis, G.; Seufferlein, T.; Azoitei, N. Physical Plasma-Triggered ROS Induces Tumor Cell Death upon Cleavage of HSP90 Chaperone. Sci. Rep. 2019, 9, 4112. [Google Scholar] [CrossRef]
- Holubová, Ľ.; Švubová, R.; Slováková, Ľ.; Bokor, B.; Chobotová Kročková, V.; Renčko, J.; Uhrin, F.; Medvecká, V.; Zahoranová, A.; Gálová, E. Cold Atmospheric Pressure Plasma Treatment of Maize Grains—Induction of Growth, Enzyme Activities and Heat Shock Proteins. Int. J. Mol. Sci. 2021, 22, 8509. [Google Scholar] [CrossRef]
- Nimse, S.B.; Pal, D. Free Radicals, Natural Antioxidants, and Their Reaction Mechanisms. RSC Adv. 2015, 5, 27986–28006. [Google Scholar] [CrossRef]
- Park, S.-C.; Pham, B.P.; Jia, B.; Lee, S.; Yu, R.; Han, S.W.; Yang, J.-K.; Hahm, K.-S.; Cheong, G.-W. Structural and Functional Characterization of Osmotically Inducible Protein C (OsmC) from Thermococcus kodakaraensis KOD1. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2008, 1784, 783–788. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, H.; Zhou, H.; Hu, Z.; Shang, W.; Rao, Y.; Peng, H.; Zheng, Y.; Hu, Q.; Zhang, R. Protective Effect of the Golden Staphyloxanthin Biosynthesis Pathway on Staphylococcus aureus under Cold Atmospheric Plasma Treatment. Appl. Environ. Microbiol. 2020, 86, e01998–e19. [Google Scholar] [CrossRef]
- Yılmaz, H.; İbici, H.N.; Erdoğan, E.M.; Türedi, Z.; Ergenekon, P.; Özkan, M. Nitrite Is Reduced by Nitrite Reductase NirB without Small Subunit NirD in Escherichia coli. J. Biosci. Bioeng. 2022, 134, 393–398. [Google Scholar] [CrossRef]
- Wozniak, K.J.; Simmons, L.A. Bacterial DNA Excision Repair Pathways. Nat. Rev. Microbiol. 2022, 20, 465–477. [Google Scholar] [CrossRef]
- Haney, A.M.; Sanfilippo, J.E.; Garczarek, L.; Partensky, F.; Kehoe, D.M. Multiple Photolyases Protect the Marine Cyanobacterium Synechococcus from Ultraviolet Radiation. mBio 2022, 13, e01511–e01522. [Google Scholar] [CrossRef]
- Mercolino, J.; Lo Sciuto, A.; Spinnato, M.C.; Rampioni, G.; Imperi, F. RecA and Specialized Error-Prone DNA Polymerases Are Not Required for Mutagenesis and Antibiotic Resistance Induced by Fluoroquinolones in Pseudomonas aeruginosa. Antibiotics 2022, 11, 325. [Google Scholar] [CrossRef]
- Hołówka, J.; Zakrzewska-Czerwińska, J. Nucleoid Associated Proteins: The Small Organizers That Help to Cope with Stress. Front. Microbiol. 2020, 11, 590. [Google Scholar] [CrossRef]
- Xia, G.; Kohler, T.; Peschel, A. The Wall Teichoic Acid and Lipoteichoic Acid Polymers of Staphylococcus aureus. Int. J. Med. Microbiol. 2010, 300, 148–154. [Google Scholar] [CrossRef]
- Al-Mebairik, N.F.; El-Kersh, T.A.; Al-Sheikh, Y.A.; Marie, M.A.M. A Review of Virulence Factors, Pathogenesis, and Antibiotic Resistance in Staphylococcus aureus. Rev. Med. Microbiol. 2016, 27, 50–56. [Google Scholar] [CrossRef]
- Cheung, G.Y.; Bae, J.S.; Otto, M. Pathogenicity and Virulence of Staphylococcus aureus. Virulence 2021, 12, 547–569. [Google Scholar] [CrossRef]
- Jurado-Martín, I.; Sainz-Mejías, M.; McClean, S. Pseudomonas aeruginosa: An Audacious Pathogen with an Adaptable Arsenal of Virulence Factors. Int. J. Mol. Sci. 2021, 22, 3128. [Google Scholar] [CrossRef]
- Jongerius, I.; Köhl, J.; Pandey, M.K.; Ruyken, M.; van Kessel, K.P.; van Strijp, J.A.; Rooijakkers, S.H. Staphylococcal Complement Evasion by Various Convertase-Blocking Molecules. J. Exp. Med. 2007, 204, 2461–2471. [Google Scholar] [CrossRef] [PubMed]
- Kiselyov, K.; Muallem, S. ROS and Intracellular Ion Channels. Cell Calcium 2016, 60, 108–114. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.G.; Yost, A.; Joshi, S.S.; Addya, S.; Ehrlich, G.; Brooks, A. Microarray Analysis of Transcriptomic Response of Escherichia coli to Nonthermal Plasma-Treated PBS Solution. Adv. Biosci. Biotechnol. 2015, 6, 49. [Google Scholar] [CrossRef]
- Mols, M.; Mastwijk, H.; Nierop Groot, M.; Abee, T. Physiological and Transcriptional Response of Bacillus cereus Treated with Low-temperature Nitrogen Gas Plasma. J. Appl. Microbiol. 2013, 115, 689–702. [Google Scholar] [CrossRef]
- Azam, M.W.; Khan, A.U. Updates on the Pathogenicity Status of Pseudomonas aeruginosa. Drug Discov. Today 2019, 24, 350–359. [Google Scholar] [CrossRef]
- Winter, T.; Winter, J.; Polak, M.; Kusch, K.; Mäder, U.; Sietmann, R.; Ehlbeck, J.; van Hijum, S.; Weltmann, K.; Hecker, M. Characterization of the Global Impact of Low Temperature Gas Plasma on Vegetative Microorganisms. Proteomics 2011, 11, 3518–3530. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).