Abstract
In this paper, a new hybrid MpGA-CS is elaborated between multi-population genetic algorithm (MpGA) and cuckoo search (CS) metaheuristic. Developed MpGA-CS has been adapted and tested consequently for modelling of bacteria and yeast fermentation processes (FP), due to their great impact on different industrial areas. In parallel, classic MpGA, classic CS, and a new hybrid MpGA-CS have been separately applied for parameter identification of E. coli and S. cerevisiae FP models. For completeness, the newly elaborated MpGA-CS has been compared with two additional nature-inspired algorithms; namely, artificial bee colony algorithm (ABC) and water cycle algorithm (WCA). The comparison has been carried out based on numerical and statistical tests, such as ANOVA, Friedman, and Wilcoxon tests. The obtained results show that the hybrid metaheuristic MpGA-CS, presented herein for the first time, has been distinguished as the most reliable among the investigated algorithms to further save computational resources.
1. Introduction
As well known, one of the main advantages of metaheuristic techniques, when applied to a complicated optimization task with incomplete, imperfect or limited computation capacity, is that they may find a near-optimal solution for an acceptable computation time [1]. Metaheuristics can be classified in different ways, but one popular classification divides them into two classes—single-based and population-based [2]. The techniques from the first class modify and improve a single candidate solution, while population-based techniques operate over multiple candidate solutions. Some of the most powerful nature-inspired population-based metaheuristics are genetic algorithms (GA) [3], cuckoo search (CS) [4], bat algorithm [5], ant colony optimization [6], artificial bee colony optimization [7], etc. They have all been proven to be successful alternatives to the conventional optimization methods for solving a wide range of optimization problems, among them, parameter identification of fermentation process models [8,9].
Since each one of the aforementioned algorithms has numerous advantages and disadvantages, it became evident that focusing our attention on a single metaheuristic technique should be limited. This offers new ideas for combining the use of metaheuristic and other optimization techniques, or between metaheuristics themselves. These combinations (also known as hybrids) usually provide more efficient performance and higher flexibility when solving real-world large-scale problems [10,11]; namely, hybrid metaheuristic techniques utilize the strength of two or more algorithms, e.g., hybrids between cuckoo search and genetic algorithm [12,13], genetic algorithm and artificial neural network [14], cuckoo search and support vector machine [15], etc. As a result, they have become a powerful tool in the hands of investigators.
It is well-known that modelling and optimization of fermentation processes (FP) are difficult and challenging tasks due to their numerous specific peculiarities, such as nonlinearity, time-varying, and interdependent process variables, etc. [13,14,16,17,18,19]. The process models of yeasts and bacteria could have a complex structure [20], thus the user’s choice of an appropriate optimization method for model parameter identification is of crucial importance. In [13], the authors have focused their efforts on adaptation and application of a hybrid technique between simple genetic algorithm and cuckoo search for parameter identification of S. cerevisiae fed-batch fermentation process model. The hybrid algorithm SGA-CS is demonstrated to solve the task for parameter identification of yeast FP model better than SGA toward the value of optimization criterion, while maintaining the model accuracy of CS and even saving nearly 60% of CS CPU time. Therefore, the authors are motivated to propose a hybrid algorithm that saves time in computational resources, but maintains the quality of the obtained solution.
In this paper, a new hybrid technique between multi-population genetic algorithm (MpGA) and CS has been proposed. The authors have chosen MpGA and CS due to their substantial experience with both metaheuristic techniques, as well as due to the fact that metaheuristic techniques in general, and MpGA and CS in particular, are relevant nowadays, frequently preferred, and efficiently used for many optimization problems [21,22,23,24,25]. These algorithms continue to increasingly attract researchers’ interest, and their superiority in comparison with other metaheuristic methods is shown in many investigations, as considered above, which provokes the authors to choose MpGA and CS for their research. The authors expect that the combination of these two successful techniques might lead to a new hybrid technique, which performs better via the computational resources (e.g., computation time and memory) or reflects authors’ growing interest to improve the model(s) considered herein on fermentation processes of bacteria and yeast in terms of lower objective function value, which will lead to higher model accuracy.
A comparison between classic MpGA and classic CS, on the one hand, and MpGA-CS hybrid technique from another, on the other hand, has been performed. The three algorithms have been consequently applied for parameter identification of two fed-batch fermentation process models; namely, bacteria E. coli and yeast S. cerevisiae, in order to reveal the advantages and disadvantages of the newly proposed MpGA-CS hybrid technique.
Furthermore, the obtained results are compared with the already published ones in order to show the superiority of the proposed MpGA-CS hybrid metaheuristic algorithm. Two powerful metaheuristic algorithms are chosen—artificial bee colony and water cycle algorithm.
Karaboga has described the artificial bee colony (ABC) algorithm for numerical optimization problems on the basis of foraging behaviour of honey bees [7]. Recently, ABC has been successfully applied on multiple combinatorial optimization problems: Vehicle routing problem [26], optimal power sharing [27], nature-inspired cyber defense [28]. Moreover, it has been shown that the ABC algorithm provides better performance than most of the evolutionary computation-based optimization algorithms [29].
Eskandar et al. have proposed a promising metaheuristic approach called water cycle algorithm (WCA) [30]. The inspiration for WCA has been derived from the natural world, after observation of the entire hydrological cycle, including the flow of streams and rivers into the seas. Recently, WCA algorithm has been successfully applied to solve different problems, such as evaluation of the optimum parameter values of a proportional-integral-derivative (PID) controller for an automatic voltage regulator [31], solving an energy-efficient disassembly-line balancing problem [32] for distribution system reconfiguration [33], etc. In addition, it has often produced better results than the other algorithms, demonstrating its good optimization capabilities.
A widespread practice in computational intelligence for the evaluation of a newly proposed technique/method is to assess its performance by the application of statistical tests [34]. Typically, the experimental analysis is used to decide when one algorithm is considered better than another. Unfortunately, this comparison may not be trivial. It has become necessary to confirm whether a new proposed method offers a significant improvement over the existing methods for a given problem. Two classes of statistical procedures are developed to perform statistical analyses: Parametric and nonparametric. Parametric tests have been commonly used in the analysis of experiments in computational intelligence. They are based on assumptions which are most probably violated when analysing the performance of stochastic algorithms based on computational intelligence [35]. On the other hand, nonparametric statistical procedures are a practical tool when the previous assumptions cannot be satisfied [34].
A comparison of the performance of the proposed hybrid technique with the classic metaheuristic is carried out based on the application of a parametric statistical test, such as one-way analysis of variance (ANOVA) [36], and nonparametric statistical tests, such as Friedman [35] and Wilcoxon [37].
The main contributions of this study are as follows:
- (1)
- For the first time, a hybrid technique MpGA-CS combining the exploitation capability of MpGA and exploration power of CS is proposed.
- (2)
- MpGA-CS hybrid technique is adapted and successfully applied for parameter identification of two mathematical models of fermentation processes; namely, E. coli and S. cerevisiae.
- (3)
- The resulting hybrid reached the desired solution with the use of 8–10 times less computation resources; namely, computation time and memory, in comparison with the classic MpGA and CS.
- In the case of a simpler mathematical model of E. coli FP, an improvement of the model accuracy of up to 14.6% is achieved based on 8 times less objective function calculations.
- In the case of a more complex model of S. cerevisiae FP, the objective function calculations are decreased 10 times and the model accuracy is preserved, even slightly improved.
- (4)
- The mathematical models of E. coli and S. cerevisiae FP obtained by MpGA-CS achieved the highest accuracy to date, and could be used for process control and optimization in order to achieve an increasing productivity of FP toward enzymes, pharmaceuticals, or other important products.
- (5)
- The performance of the newly elaborated MpGA-CS is compared with the performance of CS, MpGA, ABC, and WCA using three statistical tests; namely, the parametric test ANOVA and the nonparametric tests Freidman and Wilcoxon. Statistical results show that there are statistical differences between the hybrid MpGA-CS and the other four investigated algorithms.
The rest of the paper is organized as follows: Section 2 describes the investigated E. coli and S. cerevisiae FP and their mathematical models. Next, the performances of MpGA, CS, ABC, WCA, and proposed hybrid technique MpGA-CS are presented. In Section 3, the obtained numerical results and some statistical analysis are described and discussed. Conclusions of the work are drawn in Section 4.
2. Materials and Methods
2.1. Escherichia coli and Saccharomyces cerevisiae Fermentation Processes
The well-studied and widespread representatives of bacteria and yeast microorganisms E. coli and S. cerevisiae, respectively are model organisms with a major impact on the industry and economics [38,39,40,41]. They are both widely used in genetics for the production of antibiotics and synthetics of other biomolecules, with a small number of examples from the latest results [42,43,44,45].
The experimental datasets of E. coli and S. cerevisiae are obtained after fed-batch FP is carried out in the Institute of Technical Chemistry, University of Hannover, Germany [20]. For completeness, specific conditions and mathematical models for each process will be provided below.
Online measurements of substrate (glucose) and offline measurements of biomass are available for both FP. Offline data for ethanol are available for S. cerevisiae fed-batch FP only.
A flow injection analysis (FIA) system was used for online glucose measurement using ACCU FM40 for a carrier with a flow rate at 1.7 mL·min−1. SciLog (Madison, WI, USA) pumps were used for a continuous sample stream at 0.5 mL·min−1 for E. coli fed-batch FP and 1.0 mL·min−1 for S. cerevisiae fed-batch FP, respectively. For E. coli FP, cell containing culture broth of 24 mL was injected into the carrier stream and mixed with an enzyme solution of 350 000 U·L−1 of 36 mL glucose oxidase (Fluka, Germany). In the case of S. cerevisiae fermentation processes, two injection valves (Knauer, Germany) injected 35 μL of glucose oxidase (Fluka, Germany) solution (100,000 U·L−1) and 18 μL of culture broth in fast succession in the carrier stream, in order that the culture broth was placed in front of the enzyme solution in the carrier stream. The software CAFCA (ANASYSCON, Germany) was applied for FIA system automation and glucose concentration determination. Measurement noise was reduced using the extended continuous-discrete Kalman filter.
Determination of biomass and ethanol concentration was carried out by 10 mL samples taken roughly every hour. The samples were immediately cooled down in ice water to prevent metabolic activity of the cells. The biomass concentration was analysed by dry weight analysis. The ethanol concentration was analysed in a gas chromatograph (Shimadzu GC-14B, Shimadzu, Germany).
Figure 1 presents the feeding profiles of E. coli and S. cerevisiae fed-batch FP, thus demonstrating the difference of control strategies in both FP.
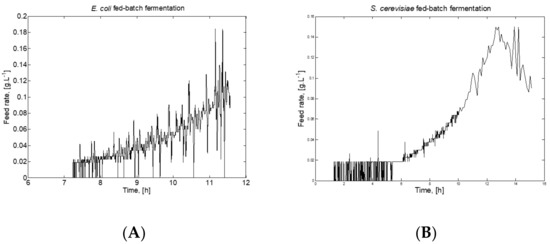
Figure 1.
Feeding profiles for (A) E. coli and (B) S. cerevisiae fed-batch FP.
Table 1 presents the FP models considered herein for fed-batch FP of E. coli and S. cerevisiae, supplemented with meanings of variables and vectors of identified parameters.

Table 1.
E. coli and S. cerevisiae fed-batch FP models and vectors of identified parameters.
The behaviour of two classic metaheuristics and the newly elaborated hybrid algorithm is initially studied by the simpler mathematical model E. coli fed-batch FP (three model parameters for identification, left column of Table 1), followed by the more complicated model of S. cerevisiae fed-batch FP (six model parameters for identification, right column of Table 1).
Optimization criterion (objective function) used for both processes is defined as the mean square deviation between the modelled and experimental FP data:
where ‖ ‖ is the -vector norm and . are modelled data and denote the experimental data in the case of E. coli fed-batch FP. are modelled data and denote the experimental data in the case of S. cerevisiae fed-batch FP, respectively.
2.2. Metaheuristic Algorithms
2.2.1. Multi-Population Genetic Algorithm
One of the widely applied metaheuristic techniques, inspired by nature processes, is GA [46]. GA uses a set of individuals called chromosomes to form the population. Each artificial chromosome is a solution of the problem and represents binary strings (genes) of certain length. Each gene stores information for the corresponding model parameter.
Simple GA [3] searches for a global optimal solution by implementing three fundamental genetic operators named selection, crossover, and mutation upon one population of individuals.
Selection. In selection operators, the selection is carried out according to the values of the objective function (fitness ) of each chromosome i in order to choose the chromosome that represents better possible solutions. Definition of the probability for selecting the individual i is:
where is the population size.
Reproduction. During the reproduction phase, crossover and mutation operators are employed in order to form a new offspring and respectively to prevent the algorithm from falling into a local optimum.
Fundamental theorem of genetic algorithms, the so-called reproductive scheme growth, which has an influence on the parameter performance of the GA algorithms is as follows:
where denotes the number of strings from generation t that belong to the schema S; is the average fitness of the schema S; is the average fitness of the population from generation t; is the probability of crossover; defining length is the distance between the first and last specific positions; is the order of the schema; and m is the length of the code.
MpGA simulates the evolution of the species in a way more similar to nature than SGA. While SGA works with one population, MpGA works with several populations, called subpopulations, which evolve independently for a certain number of generations. The MpGA starts generating k random subpopulations, each one containing n chromosomes. Then, the algorithm calculates the objective function and the fitness function of each chromosome and proceeds to create new populations through three main operators:
- ✓
- Selection: Parent chromosomes are selected from the subpopulations according to their fitness function.
- ✓
- Crossover: Selected parents are subjected to crossover in order to form a new generation.
- ✓
- Mutation: One or more values of a gene on a chromosome are changed to maintain genetic diversity in the next generation.
In this way, the new generation replaces the old subpopulations in the algorithm. After the isolation time, a certain number of individuals are distributed (migrate) between the subpopulations. The rate of migration and the structure of migration are responsible for the degree of genetic diversity [3]. The migration rate determines how many individuals will be distributed between the subpopulations. The structures of migration vary, but the most common applied migration strategy is unrestricted migration, where individuals can migrate from any subpopulation to another.
After migration, and if the final condition is satisfied, the MpGA stops and returns the "best" solution among the populations. Otherwise, the algorithm continues the execution until an algorithm termination criterion is reached, in this paper, when a predefined number of generations has been reached.
2.2.2. Cuckoo Search
The cunning and aggressive breeding behaviour of the cuckoo species has been implemented by Yang and Deb [4] in the cuckoo search metaheuristic algorithm for solving optimization problems. CS is a population-based metaheuristic algorithm, in which the eggs found in a nest are considered as a set of candidate solutions of the optimization problem, while the cuckoo egg is interpreted as a new possible solution. The ultimate goal of the method is to iteratively use these new and potentially better solutions in order to find the (sub)optimal solution to the problem. Cuckoo search is a (μ + λ), which is an evolution strategy in disguise according to the recent research [47]. Effectiveness and efficiency of CS are well established in practice [12,18,48,49,50].
Three idealized rules are intrinsic to the standard CS algorithm [4]:
- Each cuckoo can only lay a single egg at a time in a random nest;
- The best nests containing high-quality eggs (solutions) are carried over to the next generation;
- The available host nests are fixed in number, and a host bird spots an egg laid by a cuckoo with probability ϵ (0, 1). In this case, the host bird can throw away the egg or abandon the nest altogether and build a new one.
The third assumption can be approximated by the parameter, which gives the possibility of a fraction of the n host nests to be replaced by new nests, containing new random eggs (solutions). In CS algorithm, the switching parameter controls the balanced combination between the local and global explorative random walk. Equations (6) and (7) represent the local random walk and global random walk, respectively, which is obtained using Lévy flights [4]:
In Equation (6), and are two different solutions, selected by random permutation, is the Heaviside function, is a uniform random number from the unit interval, S is the step size, and ⊗ denotes the Hadamard (entry wise) vector product.
In Equation (7), the step size scaling factor α > 0 is determined as follows:
2.2.3. Multi-Population Genetic Algorithm—Cuckoo Search Hybrid Technique
MpGA-CS hybrid algorithm is a combination of two powerful population-based metaheuristic techniques. MpGA-CS hybrid algorithm begins with MpGA exploring the search space in order to generate initial solutions for the CS algorithm. To generate good initial solutions for the CS algorithm, MpGA is run for a few generations with a small number of individuals in the population. Herein, the good exploitation of the MpGA is a benefit. Furthermore, CS uses an initial population obtained by MpGA optimal solutions, and explores search space to generate the best model parameters vector. The CS algorithm is run for a small number of iterations with a small number of nests. When starting with a good initial solution that is generated by MpGA, this allows for a better solution to be found while benefiting from good CS exploration of the parameter space.
The pseudo code of the newly elaborated MpGA-CS adapted for model parameters identification of the fed-batch FPs studied herein is presented in Appendix A.
The flow chart of MpGA-CS hybrid algorithm, according to presented pseudo code is given in Figure 2.
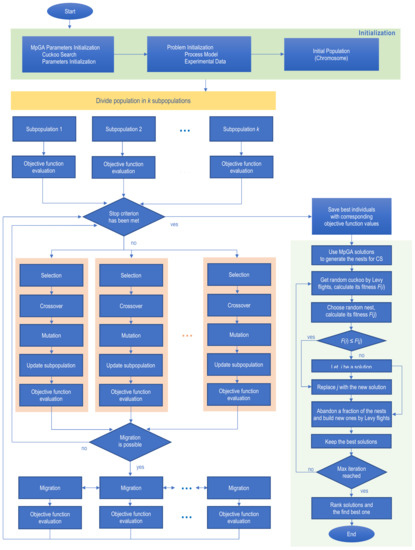
Figure 2.
Flow chart of MpGA-CS hybrid algorithm.
3. Results and Discussion
3.1. Tuning of the Metaheuristic Algorithm Parameters
The proposed metaheuristic algorithms have been executed on Intel® Core™i7-8700 CPU @ 3.20 GHz, 3192 MHz, 32 GB Memory (RAM), Windows 10 pro (64 bit) operating system. MATLAB and Simulink R2019a environment are used. MpGA and CS have been implemented in MATLAB environment based on the presented flow chart (Figure 2) and the provided pseudo codes (Appendix A). To achieve the advantages of both MpGA and CS algorithms, the hybrid technique MpGA-CS has been implemented in MATLAB code, as well. Mathematical models of E. coli and S. cerevisiae FP (Equations (1) and (2)) are modelled through Simulink implementation. Solver options are fixed-step size of 0.01 and ode4 (Runge-Kutta) with TIMESPAN = [6.69 11.57] in the case of E. coli FP, and with TIMESPAN = [1 14.95] in the case of S. cerevisiae FP.
All three algorithms have been applied for the two parameter identification problems of fed-batch FP model of E. coli and S. cerevisiae, respectively.
To achieve the best metaheuristic algorithm performance, some necessary adjustments of the MpGA, CS, and MpGA-CS parameters depending on the problem domain have been performed. The algorithm parameters have been tuned based on several pre-tests according to the particular parameter identification problem herein. The selected algorithm parameters for the tuning tests are based on the investigations and recommendations for GA tuning [51,52,53] and CS tuning [18,48], as well as on the substantial experience of previous authors [54,55,56]. To perform sensitivity analyses of the algorithm parameters, the following sets of values are tested for the optimization problems investigated herein:
MpGA parameters:
| population size | [15 20 30 50 100 150 200] |
| number of generations | [20 25 50 100 150 200] |
| generation gap | [0.80 0.85 0.90 0.95 0.96 0.97 0.98] |
| crossover probability | [0.65 0.7 0.75 0.80 0.85 0.9 0.95] |
| mutation probability | [0.01 0.03 0.05 0.07 0.1] |
| insertion probability | [0.70 0.75 0.80 0.85 0.90 0.95] |
| number of subpopulations | [3 5 7 9] |
| migration probability | [0.10 0.15 0.20 0.25 0.30] |
| isolation time | [10 15 20 25 30 35 40] |
CS parameters:
| number of nests | [5 10 15 20 25] |
| switching parameter | [0.10 0.13 0.15 0.17 0.20 0.23 0.25 0.27] |
| step size scaling factor | [1.00 1.50 2.00 2.5] |
MpGA has been performed using the following main functions: Selection function—roulette wheel selection; crossover function—extended intermediate recombination; mutation function—mutation operator of the Breeder genetic algorithm; fitness function—linear ranking; and reinsertion—fitness-based.
The set of optimal values used for the main parameters of the algorithms after a series of tuning procedures is provided in Table 2.

Table 2.
The optimal values of the main parameters of the metaheuristic algorithms.
MpGA requires a large number of parameters to be tuned, as can be seen from Table 2, while the workability of CS depends mainly on three parameters; namely, population size (number of nests), switching parameter pa (probability rate of replacement), and step size scaling factor α.
For parameter fine-tuning of the hybrid technique, MpGA maximum number of generations has been preliminary examined and 50 iterations have been set as quite appropriate for MpGA-CS. As can be seen from Table 2, MpGA-CS has been performed with the same number of subpopulations as in MpGA, but less isolation time (number of generations before migration) is considered in order to quickly reach an optimal solution. Or, you may consider the following reformulation:
Overall, as MpGA-CS disadvantage in comparison to classic MpGA and classic CS could be indicated bigger number of parameters required to be set before algorithm’s application.
In the case of parameter estimation of E. coli FP model, some of the MpGA-CS algorithm parameters are changed compared with the algorithm parameters of classic MpGA and CS. A slightly smaller value for (probability rate of crossover) and twice larger value of (probability rate of mutation) are chosen to achieve a better initial solution by MpGA. A small MpGA population of 25 chromosomes (population size) is run only for 50 generations with the chosen values of and , thus ensuring a construction of an initial solution for CS where the parameter space is explored in the best way. The higher rate of mutation gives the best possible decisions that are very distant from the current best to be chosen. Therefore, a better initial solution for CS is obtained in comparison with the optimal pc and pm values of the classic MpGA.
In the case of parameter estimation of S. cerevisiae FP model, some changes have been applied to the hybrid MpGA-CS algorithm parameters in comparison with the algorithm parameters used for the running of classic MpGA and CS. Once again, smaller MpGA population with 20 chromosomes (even smaller than in the case of parameter estimation of E. coli FP model) are set for 50 generations. Herein, the chosen values of pc and pm have been left unchanged since they have been proven to be optimal for the more complex task of parameter identification of S. cerevisiae FP model with parameters’ vector with six parameters. The solution obtained after MpGA ensures an initial solution for CS, relying on its proven feature to explore the parameter space in the best way.
3.2. Parameter Identification of E. coli Fermentation Process Model
A series of parameter identification procedures of the model (Equation (1)), using tuned classic MpGA and CS as well as a hybrid MpGA-CS, have been performed. Due to the stochastic nature of the algorithms, a meaningful statistical analysis requires each metaheuristic to run for at least 30 times.
The model parameters’ vector; namely, , is estimated based on the objective function J (Equation (3)).
Based on [54,55,57], each parameter of the model in Equation (1) has been coded in a specific range (lower bound (Lb) ≤ parameter ≤ upper bound (Ub)), as follows:
The obtained best model parameters’ estimates with their standard deviation (SD) and the corresponding value of the objective function are shown in Table 3.

Table 3.
Parameter estimates of E. coli FP model (Equation (1)).
In addition, the obtained results are compared with the already published ones for the same problem in order to show the superiority of the proposed MpGA-CS hybrid metaheuristic algorithm. In [57], several differently tuned ABC algorithms are applied to the parameter identification of an E. coli fed-batch FP. For comparison, the best results obtained in [57] are listed in Table 3 and the results obtained in [58] from the application of WCA are demonstrated, as well.
The parameter estimates presented in Table 3 correspond to their physical meaning [19,59,60] and thus are adequate. The obtained SD values indicate the good performance of the applied algorithms.
A graphical comparison can clearly show whether the model follows the real experimental data, i.e., to establish the presence or absence of systematic deviations between the model predictions and the real measurements. Moreover, this quantitative measure is an important evidence for the adequacy of the obtained models. The model predictions of the state variables and, based on an estimated set of MpGA, CS, ABC, WCA, and MpGA-CS model parameters, are compared with the experimental data of E. coli fed-batch FP in Figure 3.
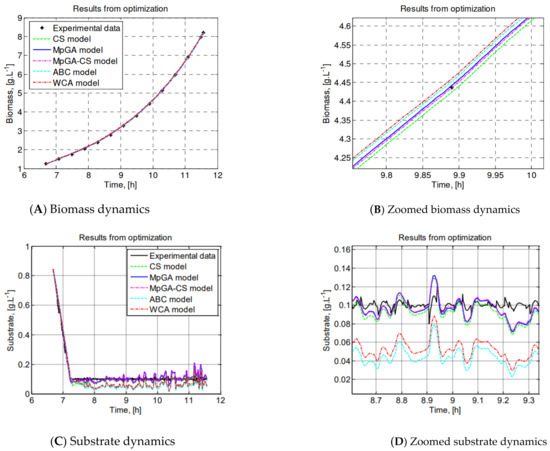
Figure 3.
Graphical comparison of the results from parameter identification of E. coli FP model.
The resulting model obtained by hybrid MpGA-CS has the lowest J value (J = 2.7444). To show the superior performance of the hybrid MpGA-CS, a zoomed view of the graphical results is presented. The graphical comparisons show that the model describes the experimental data in a superb way, following the trend of the process dynamics. The model that shows the best fit to the real data is obtained by the proposed hybrid algorithm.
The performance of the considered metaheuristic algorithms has been statistically evaluated by comparing the observed average value, SD, and the median of the estimated model parameters and the obtained objective function value J. To present summary statistics, box plot diagrams are shown in Figure 4.
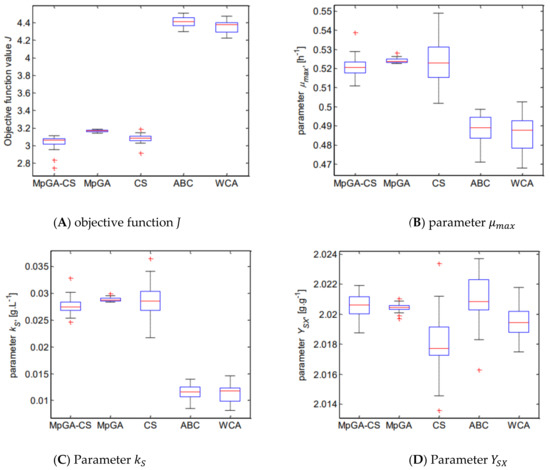
Figure 4.
Box plot for the results from parameter identification of E. coli FP model.
The results clearly show that the proposed hybrid algorithm has a performance based on the advantages of both metaheuristics, precisely the exploration of the CS algorithm and the better exploitation of the MpGA. Moreover, MpGA-CS outperforms both ABC and WCA algorithms. The hybrid algorithm found a different solution for the model parameters, in which a model with higher accuracy is obtained as compared with the models obtained by ABC and WCA.
Studies show that CS is potentially significantly more efficient than GA in balancing exploration search strategies [49,50]. Selection according to fitness is the source of exploitation, mutation and crossover operators are the sources of exploration [61,62]. According to [63], recombination serves as a very helpful role at exploitation when the necessary diversity is ensured by some means. In GA, this diversity is achieved through higher mutation rates as used in the proposed hybrid in the case of E. coli modelling.
The performance of the MpGA-CS hybrid is compared with the performance of CS, MpGA, ABC, and WCA based on the three statistical tests—parametric test ANOVA and nonparametric tests Freidman and Wilcoxon. The tests are run in MATLAB using functions ‘anova1’, ‘ranksum’, and ‘friedman’ on the obtained results of objective function J over 30 runs of each algorithm. The results are presented in the Table 4.

Table 4.
Parameter identification of E. coli results from statistical tests.
The presented results reveal that there are statistical differences between the MpGA-CS hybrid and competing algorithms—CS, MpGA, ABC, and WCA. Since the mean J value of MpGA-CS is lowest in comparison with the other obtained mean values, it may be concluded that the proposed hybrid metaheuristic algorithm outperforms all of the considered metaheuristic algorithms in the case of parameter identification of E. coli FP model presented with Equation (1).
To show the underlying frequency distribution of the results obtained by the classic MpGA and CS and the proposed hybrid MpGA-CS, histograms are presented in Figure 5 and Figure 6, respectively for the objective function and for model parameter estimates.
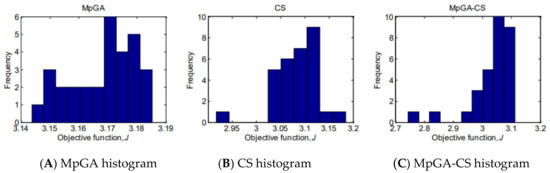
Figure 5.
Histograms of the results for the objective function values from parameter identification of E. coli FP model (30 runs).
The observed distribution of the objective function values produced by MpGA algorithm (Figure 5) is a plateau or multimodal distribution. CS and MpGA-CS produce results with a left-skewed distribution. Considering the model parameter estimates obtained by MpGA-CS hybrid algorithm (Figure 6), the results are quite closer to the normal distribution, in contrast to the results obtained by MpGA and CS. The advantage of hybridizing of MpGA algorithm with CS algorithm over the considered classic algorithms is that the multi-modality or skewed distribution could be avoided.
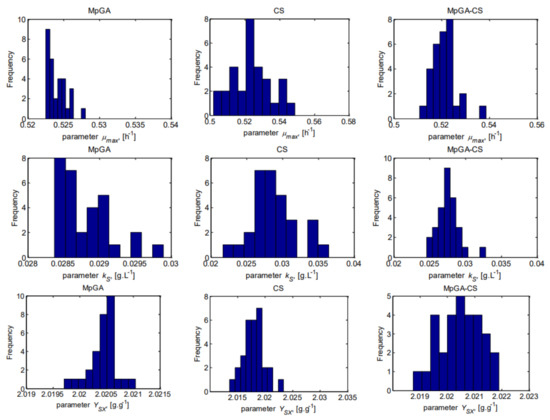
Figure 6.
Histograms of the results for model parameter estimates from parameter identification of E. coli FP model (30 runs).
Finally, the results obtained for the objective function J (Table 3) should be discussed in terms of computer memory, and thus computational time. Rather than performing a calculation of the objective function 50,000 times in MpGA (100 generations, 100 chromosomes in 5 populations), the hybrid MpGA-CS performed 6250 calculations of the objective function (50 generations, 25 chromosomes in 5 populations) and achieved a solution with greater accuracy compared with the solutions of the two algorithms—classic MpGA and CS. We will appreciate if you change “a model even with higher accuracy” with “a model with even higher accuracy”
3.3. Parameter Identification of S. cerevisiae Fermentation Process Model
By analogy with the parameter identification procedure of E. coli FP model, the classic MpGA, classic CS, and hybrid MpGA-CS tuned herein have been consequently applied to S. cerevisiae FP model (Equation (2)). Once again, due to the stochastic nature of the algorithms, 30 independent runs have been carried out for each metaheuristic.
Six model parameters have been estimated and the corresponding model parameters’ vector is .
Based on [20], each parameter of the model Equation (2) has been coded between the lower bound and upper bound, as follows:
Table 5 presents the best achieved values of model parameter estimates with their SD for the model Equation (2) after 30 runs, as well as the corresponding value of the objective function J.

Table 5.
Model parameter estimates of S. cerevisiae FP model (Equation (2)).
Additionally, the obtained results are compared with those produced by ABC [64] and WCA [58] algorithms, in order to show the advantages of the proposed MpGA-CS hybrid algorithm.
As can be seen from Table 5, the values of the optimization criterion J obtained by the MpGA, CS, ABC, WCA, and the hybrid MpGA-CS investigated herein are quite similar. The obtained results demonstrate that WCA outperforms MpGA, CS, and ABC, but the newly proposed hybrid MpGA-CS yields the best results with the lowest value J = 1.342.
As presented in Table 5, the parameter estimates maintain the physical meaning of model parameters [65,66,67]. The obtained SD values again show that all of the algorithms have good performance for the solved task.
The workability of MpGA-CS hybrid algorithm for solving complex problems, such as the parameter identification of S. cerevisiae FP model is demonstrated in Figure 7, where the results from experimental and model predicted data, respectively, for biomass, substrate, and ethanol have been graphically presented.
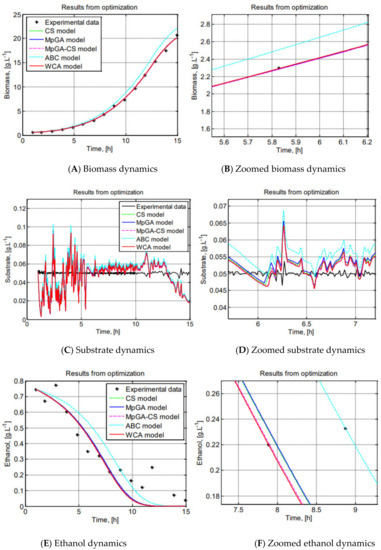
Figure 7.
Graphical comparison of the results from parameter identification of S. cerevisiae FP model.
Once again, the performance of the five algorithms is statistically evaluated by comparison of the observed average value, standard deviation, and median of the estimated model parameters and the objective function value J. To visualize summary statistics for S. cerevisiae parameter identification, box plot diagrams are shown in Figure 8.
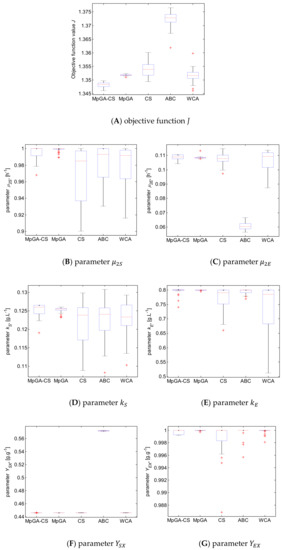
Figure 8.
Box plot for the results from parameter identification of S. cerevisiae FP model.
In analogy with the case of E. coli FP, the performance of MpGA-CS hybrid is again compared with the performance of CS, MpGA, ABC, and WCA based on the aforementioned statistical tests. The results are presented in Table 6.

Table 6.
Parameter identification of S. cerevisiae results from statistical tests.
In the case of S. cerevisiae, the presented results reveal again that there are statistical differences between the MpGA-CS hybrid and other algorithms performed herein—CS, MpGA, ABC, and WCA. The mean J value of MpGA-CS is lowest again in comparison with the mean values obtained by other metaheuristics. Therefore, it may be concluded again that the proposed MpGA-CS hybrid outperforms the other considered metaheuristic algorithms.
In analogy with the E. coli FP model parameter identification procedure, Figure 9 and Figure 10 present histograms that show the underlying frequency distribution of the results obtained by the compared algorithms, respectively for the objective function and model parameter estimates.
Once again, the results definitely demonstrate the successful combination of MpGA and CS population-based metaheuristics. The MpGA-CS hybrid incorporates the advantages of MpGA, namely, the better convergence, and CS, namely, the improved accuracy of the solution.
The results presented in Figure 9 show that the classic CS and hybrid MpGA-CS produce a multimodal type distribution for the objective function, while the results obtained by MpGA produce a right-skewed distribution. Concerning the results presented in Figure 10, some similarities might be outlined in the distributions of model parameter estimates between MpGA and hybrid MpGA-CS (e.g., for parameters and ), while for parameters and , the distribution of MpGA-CS is closer to those in CS. The distributions of when applying CS and MpGA-CS are very close to the normal distribution.
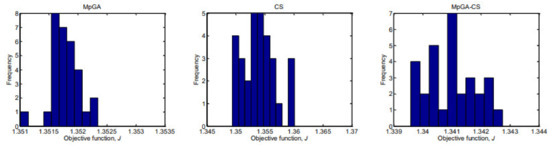
Figure 9.
Histograms of the results of objective function values from parameter identification of S. cerevisiae FP model (30 runs).
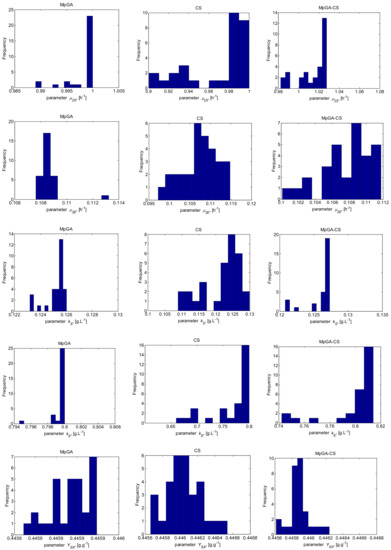
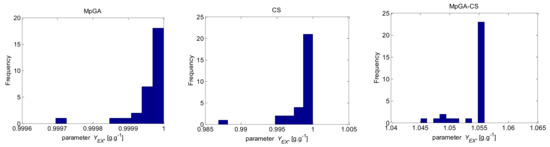
Figure 10.
Histograms of the results for six model parameter estimates from parameter identification of S. cerevisiae FP model (30 runs).
Finally, the results obtained for the objective function J (Table 5) have been discussed again in terms of computer memory and computational time. Herein, rather than performing 50,000 calculations of the objective function in MpGA (as explained above 100 generations, 100 chromosomes in 5 populations), the hybrid MpGA-CS performed 5000 calculations of the objective function (50 generations, 20 chromosomes in 5 populations) and achieved a solution with greater accuracy compared with the solutions of the classic MpGA and CS. Therefore, with an objective function performance of 10 times less the number of calculations by MpGA-CS toward MpGA, a model with even higher accuracy (although less than 1% improvement) has been obtained.
4. Conclusions
In this paper, a hybrid metaheuristic technique MpGA-CS implementing the advantages of multi-population genetic algorithm and cuckoo search has been elaborated herein for the first time. MpGA-CS has been tested for parameter identification of two non-linear dynamic fed-batch FP models. Gradually, the investigation starts applying MpGA-CS for simple E. coli fed-batch FP model. Next, to verify the results, a more complicated FP model of S. cerevisiae is considered. Thoroughly conducted comparison between MpGA-CS hybrid and the competing metaheuristics MpGA, CS, ABC, and WCA, has been carried out, following the application of each algorithm for parameter identification of E. coli and S. cerevisiae FP models. In addition, ANOVA, Friedman, and Wilcoxon tests have been performed and showed statistical differences between the MpGA-CS hybrid and other metaheuristics considered herein. Since the mean J value of MpGA-CS is lowest in both FP models in comparison with the other obtained mean values from the other metaheuristics, it may be concluded that the proposed MpGA-CS hybrid outperforms all algorithms. From eight (in the case of E. coli) to ten times (in the case of S. cerevisiae) less number of calculations of the objective function by MpGA-CS toward MpGA, models with higher accuracy (less than 1% improvement in the case of S. cerevisiae, but up to 14.6% improvement in the case of E. coli) have been obtained. As shown, the results unambiguously demonstrate that the MpGA-CS developed herein outperforms MpGA, CS, ABC, and WCA toward the optimization criterion value in both FP models of bacteria and yeasts, while significantly saves CPU resources in terms of computer memory and computational time.
Author Contributions
Conceptualization and methodology, M.A., O.R. and T.P.; software implementation, M.A., O.R., P.V. and T.P.; validation, M.A., O.R. and T.P.; investigation, M.A., O.R., P.V. and T.P.; writing—original draft preparation, M.A., O.R., P.V. and T.P.; writing—review and editing, M.A., O.R., P.V. and T.P.; visualization, O.R., P.V. and T.P.; supervision, O.R. and T.P. All authors have read and agreed to the published version of the manuscript. They contributed equally.
Funding
This research received no external funding.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A
| Algorithm A1 Pseudo Code of MpGS-CS Hybrid Algorithm |
| Begin Objective function f(x), x = (x1, …, xd)T Define the MpGA parameters Define the CS parameters [Begin MpGA] [Start] Generate k random subpopulations each of which with n chromosomes [Objective function] Evaluate the objective function of each chromosome n in the subpopulations. Evaluate the fitness function of each chromosome n in the subpopulations [New population] Create a new population by repeating the following steps: [Selection] Select parent chromosomes from the subpopulation according to their fitness function [Crossover] Cross over the parents to form new offspring with a crossover probability [Mutation] Mutate new offspring at each locus with a mutation probability [Accepting] Place new offspring in the new population [Replace] Use newly generated population for the further run of the algorithm [Migration] Migration of individuals between the subpopulations after following isolation time [Test] If the end condition is satisfied, stop and return the best solution in current population, else move to Loop step [Loop] Go to Step Objective function Evaluate P(i) fitness [end Loop] Rank the chromosomes and find the current best n chromosomes [End begin MpGA] [Begin CS] [Start] Initial population of n host nests = Final best MpGA n solutions [Create new solution] Get a cuckoo (i) randomly and create a new solution by Lévy flights Evaluate its quality or fitness value Fi Choose a nest among n randomly (j) If the fitness value of the cuckoo is better than the one of the nest, replace j with the new solution i [Abandon] Abandon a fraction of the worse nests [Generate new solutions] Build new solutions (nest) Keep the best solution, i.e., the nests with highest quality solutions Rank the solutions and find the current best [Test] If the end condition is satisfied or stop criteria is reached return the best solution, else move to Create new solution step [End begin CS] Postprocess and visualize the results End |
References
- Chopard, B.; Tomassini, M. Performance and limitations of metaheuristics. In An Introduction to Metaheuristics for Optimization; Springer International Publishing: Berlin/Heidelberg, Germany, 2018; pp. 191–203. [Google Scholar]
- Gogna, A.; Tayal, A. Metaheuristics: Review and application. J. Exp. Theor. Artif. Intell. 2013, 25, 503–526. [Google Scholar] [CrossRef]
- Goldberg, D. Genetic Algorithms in Search, Optimization and Machine Learning, 1st ed.; Addison-Wesley Professional: Hoboken, NJ, USA, 1989. [Google Scholar]
- Yang, X.-S.; Deb, S. Cuckoo search via levy flights. In Proceedings of the World Congress on Nature and Biologically Inspired Computing, Coimbatore, India, 9–11 December 2009; IEEE: Piscataway, NJ, USA, 2009; pp. 210–214. [Google Scholar]
- Yang, X.-S. A new metaheuristic bat-inspired algorithm. Stud. Comp. Int. 2010, 284, 65–74. [Google Scholar]
- Dorigo, M.; Stutzle, T. Ant Colony Optimization; MIT Press: Cambridge, UK, 2004. [Google Scholar]
- Karaboga, D. An Idea Based on Honey Bee Swarm for Numerical Optimization; Technical Report-TR06; Erciyes University, Engineering Faculty, Computer Engineering Department: Kayseri, Turkey, 2005. [Google Scholar]
- Angelova, M.; Tzonkov, S.; Pencheva, T. Genetic algorithms based parameter identification of yeast fed-batch cultivation. In Numerical Methods and Applications: NMA 2010; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2011; Volume 6046, pp. 224–231. [Google Scholar]
- Angelova, M.; Roeva, O.; Pencheva, T. Cuckoo search algorithm for parameter identification of fermentation process model. In Numerical Methods and Applications: NMA 2018; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2019; Volume 11189, pp. 39–47. [Google Scholar]
- El-Ghazali, T. (Ed.) Hybrid Metaheuristics; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Wang, G.; Guo, L. A Novel Hybrid Bat Algorithm with Harmony Search for Global Numerical Optimization. J. Appl. Math. 2013, 2013, 696491. [Google Scholar] [CrossRef]
- Lim, W.; Kanagaraj, G.; Ponnambalam, S. A Hybrid Cuckoo Search-genetic Algorithm for Hole-making Sequence Optimization. J. Int. Manufact. 2014, 27, 417–429. [Google Scholar]
- Angelova, M.; Vassilev, P.; Pencheva, T. Genetic Algorithm and Cuckoo Search Hybrid Technique for Parameter Identification of Fermentation Process Model. Int. J. Bioaut. 2020, 24, 277–288. [Google Scholar]
- De Menezes, L.H.S.; Carneiro, L.L.; de Carvalho Tavares, I.M.; Santos, P.H.; das Chagas, T.P.; Mendes, A.A.; da Silva, E.G.P.; Franco, M.; de Oliveira, J.R. Artificial Neural Network Hybridized with a Genetic Algorithm for Optimization of Lipase Production from Penicillium roqueforti ATCC 10110 in Solid-State Fermentation. Biocat. Agricult. Biotechnol. 2021, 31, 101885. [Google Scholar]
- Wang, Y.; Liu, Q.; Yang, Y.; Wang, L.; Song, X.; Zhao, X. Prognostic Staging of Esophageal Cancer Based on Prognosis Index and Cuckoo Search Algorithm-Support Vector Machine. Biomed. Sign. Proc. Contr. 2023, 79, 104207. [Google Scholar]
- Pan, N.; Wang, H.; Tian, Y.; Chorukova, E.; Simeonov, I.; Christov, N. Comparison Study of Dynamic Models for One-stage and Two-stage Anaerobic Digestion Processes. IFAC-PapersOnLine 2022, 55, 667–672. [Google Scholar]
- Chorukova, E.; Hubenov, V.; Gocheva, Y.; Simeonov, I. Two-Phase Anaerobic Digestion of Corn Steep Liquor in Pilot Scale Biogas Plant with Automatic Control System with Simultaneous Hydrogen and Methane Production. Appl. Sci. 2022, 12, 6274. [Google Scholar]
- Khoja, I.; Ladhari, T.; M’sahli, F.; Sakly, A. Cuckoo search approach for parameter identification of an activated sludge process. Comp. Comput. Intell. Neurosci. 2018, 2018, 3476851. [Google Scholar] [CrossRef]
- Anane, E.; Neubauer, P.; Bournazou, M.N.C. Modelling Overflow Metabolism in Escherichia coli by Acetate Cycling. Biochem. Eng. J. 2017, 125, 23–30. [Google Scholar]
- Pencheva, T.; Roeva, O.; Hristozov, I. Functional State Approach to Fermentation Processes Modelling; Prof. Marin Drinov Academic Publishing House: Sofia, Bulgaria, 2006. [Google Scholar]
- Guo, C.; Yang, Z.; Wu, X.; Tan, T.; Zhao, K. Application of an Adaptive Multi-Population Parallel Genetic Algorithm with Constraints in Electromagnetic Tomography with Incomplete Projections. Appl. Sci. 2019, 9, 2611. [Google Scholar] [CrossRef]
- Park, J.; Park, M.-W.; Kim, D.-W.; Lee, J. Multi-Population Genetic Algorithm for Multilabel Feature Selection Based on Label Complementary Communication. Entropy 2020, 22, 876. [Google Scholar] [CrossRef]
- Zhang, W.; He, H.; Zhang, S. A Novel Multi-stage Hybrid Model with Enhanced Multi-population Niche Genetic Algorithm: An Application in Credit Scoring. Expert Syst. Appl. 2019, 121, 221–232. [Google Scholar]
- Turan-Karaoğlan, S.; Göktürkler, G. Cuckoo Search Algorithm for Model Parameter Estimation from Self-Potential Data. J. Appl. Geophys. 2021, 194, 104461. [Google Scholar]
- Zhang, X.; Li, Y.; Fan, Y. Regularization Cuckoo Search Algorithm for Multi-Parameter Optimization of the Multi-Laminated Controlled Release System. Axioms 2022, 11, 500. [Google Scholar] [CrossRef]
- Nagy, Z.; Werner-Stark, Á.; Dulai, T. An Artificial Bee Colony Algorithm for Static and Dynamic Capacitated Arc Routing Problems. Mathematics 2022, 10, 2205. [Google Scholar] [CrossRef]
- Ullah, K.; Jiang, Q.; Geng, G.; Rahim, S.; Khan, R.A. Optimal Power Sharing in Microgrids Using the Artificial Bee Colony Algorithm. Energies 2022, 15, 1067. [Google Scholar] [CrossRef]
- Ganguli, C.; Shandilya, S.K.; Nehrey, M.; Havryliuk, M. Adaptive Artificial Bee Colony Algorithm for Nature-Inspired Cyber Defense. Systems 2023, 11, 27. [Google Scholar] [CrossRef]
- Karaboga, D.; Gorkemli, B. Solving traveling salesman problem by using combinatorial artificial bee colony algorithms. Int. J. Artif. Intell. Tools 2019, 28, 1950004. [Google Scholar]
- Eskandar, H.; Sadollah, A.; Bahreinineja, A.; Shukor, M. Water cycle algorithm—A novel metaheuristic optimization method for solving constrained engineering optimization problems. Comput. Struct 2012, 110–111, 151–166. [Google Scholar]
- Pachauri, N. Water cycle algorithm-based PID controller for AVR. Int. J. Comput. Math. Electr. Electron. Eng. 2020, 39, 551–567. [Google Scholar]
- Zhang, X.; Yuan, J.; Chen, X.; Zhang, X.; Zhan, C.; Fathollahi-Fard, A.M.; Wang, C.; Liu, Z.; Wu, J. Development of an Improved Water Cycle Algorithm for Solving an Energy-Efficient Disassembly-Line Balancing Problem. Processes 2022, 10, 1908. [Google Scholar] [CrossRef]
- Alwash, S.; Ibrahim, S.; Abed, A.M. Distribution System Reconfiguration with Soft Open Point for Power Loss Reduction in Distribution Systems Based on Hybrid Water Cycle Algorithm. Energies 2023, 16, 199. [Google Scholar] [CrossRef]
- Derrac, J.; García, C.; Molina, D.; Herrera, F. A practical tutorial on the use of nonparametric statistical tests as a methodology for comparing evolutionary and swarm intelligence algorithms. Swarm Evol. Comput. 2011, 1, 3–18. [Google Scholar] [CrossRef]
- García, S.; Molina, D.; Lozano, M.; Herrera, F. A study on the use of nonparametric tests for analyzing the evolutionary algorithms’ behaviour: A case study on the CEC’2005 special session on real parameter optimization. J. Heuristics 2009, 15, 617–644. [Google Scholar]
- Fisher, R.A. Statistical Methods and Scientific Inference, 2nd ed.; Hafner Publishing Co.: New York, NY, USA, 1959. [Google Scholar]
- García, S.; Fernández, A.; Luengo, J.; Herrera, F. Advanced nonparametric tests for multiple comparisons in the design of experiments in computational intelligence and data mining: Experimental analysis of power. Inf. Sci. 2010, 180, 2044–2064. [Google Scholar]
- Costa, C.E.; Romaní, A.; Teixeira, J.A.; Domingues, L. Resveratrol production for the valorisation of lactose-rich wastes by engineered industrial Saccharomyces cerevisiae. Bioresour. Technol. 2022, 359, 127463. [Google Scholar]
- Xu, L.; Wang, D.; Chen, J.; Li, B.; Li, Q.; Liu, P.; Qin, Y.; Dai, Z.; Fan, F.; Zhang, X. Metabolic engineering of Saccharomyces cerevisiae for gram-scale diosgenin production. Metab. Eng. 2022, 70, 115–128. [Google Scholar]
- Wang, X.; Chen, J.; Zhang, J.; Zhou, Y.; Zhang, Y.; Wang, F.; Li, X. Engineering Escherichia coli for production of geraniol by systematic synthetic biology approaches and laboratory-evolved fusion tags. Metab. Eng. 2021, 66, 60–67. [Google Scholar]
- Rinaldi, M.A.; Ferraz, C.A.; Scrutton, N.S. Alternative metabolic pathways and strategies to high-titre terpenoid production in E. coli. Nat. Prod. Rep. 2022, 39, 90–118. [Google Scholar]
- Ganjave, S.D.; Dodia, H.; Sunder, A.V.; Madhu, S.; Wangikar, P.P. High cell density cultivation of E. coli in shake flasks for the production of recombinant proteins. Biotechnol. Rep. 2022, 33, e00694. [Google Scholar]
- Last, D.; Hasan, M.; Rothenburger, L.; Braga, D.; Lackner, G. High-yield production of coenzyme F420 in Escherichia coli by fluorescence-based screening of multi-dimensional gene expression space. Metab. Eng. 2022, 73, 158–167. [Google Scholar]
- Kumari, P.; Sharma, J.; Singh, A.K.; Pandey, A.K.; Yusuf, F.; Kumar, S.; Gaur, N.A. Tailored designing of a diploid S. cerevisiae natural isolate for increased production of fatty acid ethyl ester. Chem. Eng. J. 2023, 453, 139852. [Google Scholar]
- Schlabitz, C.; Lehn, D.N.; de Souza, C.F.V. A review of Saccharomyces cerevisiae and the applications of its byproducts in dairy cattle feed: Trends in the use of residual brewer’s yeast. J. Clean. Prod. 2021, 332, 130059. [Google Scholar]
- Katoch, S.; Chauhan, S.S.; Kumar, V. A Review on Genetic Algorithm: Past, Present, and Future. Multimed. Tools Appl. 2021, 80, 8091–8126. [Google Scholar] [CrossRef]
- Christian, L.; Camacho-Villalón, D.M.; Stützle, T. An analysis of why cuckoo search does not bring any novel ideas to optimization. Comput. Oper. Res 2022, 142, 105747. [Google Scholar] [CrossRef]
- Liu, L.; Liu, X.; Wang, N.; Zou, P. Modified cuckoo search algorithm with variational parameters and logistic map. Algorithms 2018, 11, 30. [Google Scholar]
- Yang, X.-S.; Deb, S. Engineering optimisation by cuckoo search. Int. J. Math. Mod. Num. Opt. 2010, 1, 330–343. [Google Scholar]
- Li, J.; Xiao, D.-d.; Lei, H.; Zhang, T.; Tian, T. Using Cuckoo Search Algorithm with Q-Learning and Genetic Operation to Solve the Problem of Logistics Distribution Center Location. Mathematics 2020, 8, 149. [Google Scholar] [CrossRef]
- Mosayebi, M.; Sodhi, M. Tuning genetic algorithm parameters using design of experiments. In Proceedings of the 2020 Genetic and Evolutionary Computation Conference Companion, Cancun, Mexico, 8–12 July 2020; pp. 1937–1944. [Google Scholar]
- Sipper, M.; Fu, W.; Ahuja, K.; Moore, J.H. Investigating the parameter space of evolutionary algorithms. BioData Min. 2018, 11, 2. [Google Scholar] [CrossRef]
- Vlasov, A.; Khomchenko, A.; Faizliev, A.; Mironov, S.; Grigoriev, A. Parameter tuning of a genetic algorithm for finding central vertices in graphs. J. Phys. 2021, 1784, 012009. [Google Scholar]
- Roeva, O.; Atanassova, V. Cuckoo search algorithm for model parameter identification. Int. J. Bioautom. 2016, 20, 483–492. [Google Scholar]
- Slavov, T.; Roeva, O. Genetic Algorithm Tuning of PID Controller in Smith Predictor for Glucose Concentration Control. Int. J. Bioautom. 2011, 15, 101–114. [Google Scholar]
- Angelova, M.; Pencheva, T. Tuning Genetic Algorithm Parameters to Improve Convergence Time. Int. J. Chem. Eng. 2011, 7, 646917. [Google Scholar] [CrossRef]
- Roeva, O. Application of Artificial Bee Colony Algorithm for Model Parameter Identification. In Innovative Computing, Optimization and Its Applications; Zelinka, I., Vasant, P., Duy, V., Dao, T., Eds.; Studies in Computational Intelligence; Springer: Cham, Switzerland, 2018; Volume 741, pp. 285–303. [Google Scholar]
- Roeva, O.; Angelova, M.; Zoteva, D.; Pencheva, T. Water Cycle Algorithm for Modelling of Fermentation Processes. Processes 2020, 8, 920. [Google Scholar] [CrossRef]
- Chen, R.; John, J.; Rode, B.; Hitzmann, B.; Gerardy-Schahn, R.; Kasper, C.; Scheper, T. Comparison of Polysialic Acid Production in Escherichia coli K1 During Batch Cultivation and Fed-batch Cultivation Applying Two Different Control Strategies. J. Biotechnol. 2011, 154, 222–229. [Google Scholar]
- Vital, M.; Hammes, F.; Egli, T. Competition of Escherichia coli O157 with a Drinking Water Bacterial Community at low Nutrient Concentrations. Water Res. 2012, 46, 6279–6290. [Google Scholar]
- Črepinšek, M.; Liu, S.H.; Mernik, M. Exploration and exploitation in evolutionary algorithms: A survey. ACM Comp. Surv. 2013, 45, 1–35. [Google Scholar]
- Hussain, A.; Muhammad, Y.S. Trade-off between exploration and exploitation with genetic algorithm using a novel selection operator. Complex Intell. Syst. 2020, 6, 1–14. [Google Scholar] [CrossRef]
- Corus, D.; Oliveto, P.S. On the benefits of populations for the exploitation speed of standard steady-state genetic algorithms. In Proceedings of the Genetic and Evolutionary Computation Conference, Prague, Czech Republic, 13–17 July 2019; pp. 1452–1460. [Google Scholar]
- Angelova, M.; Roeva, O.; Pencheva, T. Artificial Bee Colony Algorithm for Parameter Identification of Fermentation Process Model. In Applied Physics, System Science and Computers III: APSAC 2018; Lecture Notes in Electrical Engineering; Springer: Cham, Switzerland, 2019; Volume 574, pp. 317–323. [Google Scholar]
- Dlangamandla, N.; Ntwampe, S.K.O.; Angadam, J.O.; Chidi, B.S.; Mewa-Ngongang, M. Kinetic Parameters of Saccharomyces cerevisiae Alcohols Production Using Nepenthes mirabilis Pod Digestive Fluids-Mixed Agro-Waste Hydrolysates. Fermentation 2019, 5, 10. [Google Scholar]
- Mukhtar, K.; Asgher, M.; Afghan, S.; Hussain, K.; Zia-ul-Hussnain, S. Comparative Study on Two Commercial Strains of Saccharomyces cerevisiae for Optimum Ethanol Production on Industrial Scale. BioMed Res. Int. 2010, 2010, 419586. [Google Scholar]
- Ahmad, F.; Jameel, A.T.; Kamarudin, M.H.; Mel, M. Study of Growth Kinetic and Modeling of Ethanol Production by Saccharomyces cerevisae. Afr. J. Biotechnol. 2011, 16, 18842–18846. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).