Enhancing Biobased Volatile Fatty Acids Production from Olive Mill Solid Waste by Optimization of pH and Substrate to Inoculum Ratio
Abstract
1. Introduction
2. Material and Methods
2.1. Chemicals
2.2. Hydrothermally Pretreated Olive Mill Solid Waste
2.3. Statistical Analysis: Response Surface Methodology (RSM)
2.4. Batch Fermentation Experiments
2.5. Volatile Fatty Acids Quantification
2.6. Microbial Community Analysis
2.7. Kinetics of VFAs Production
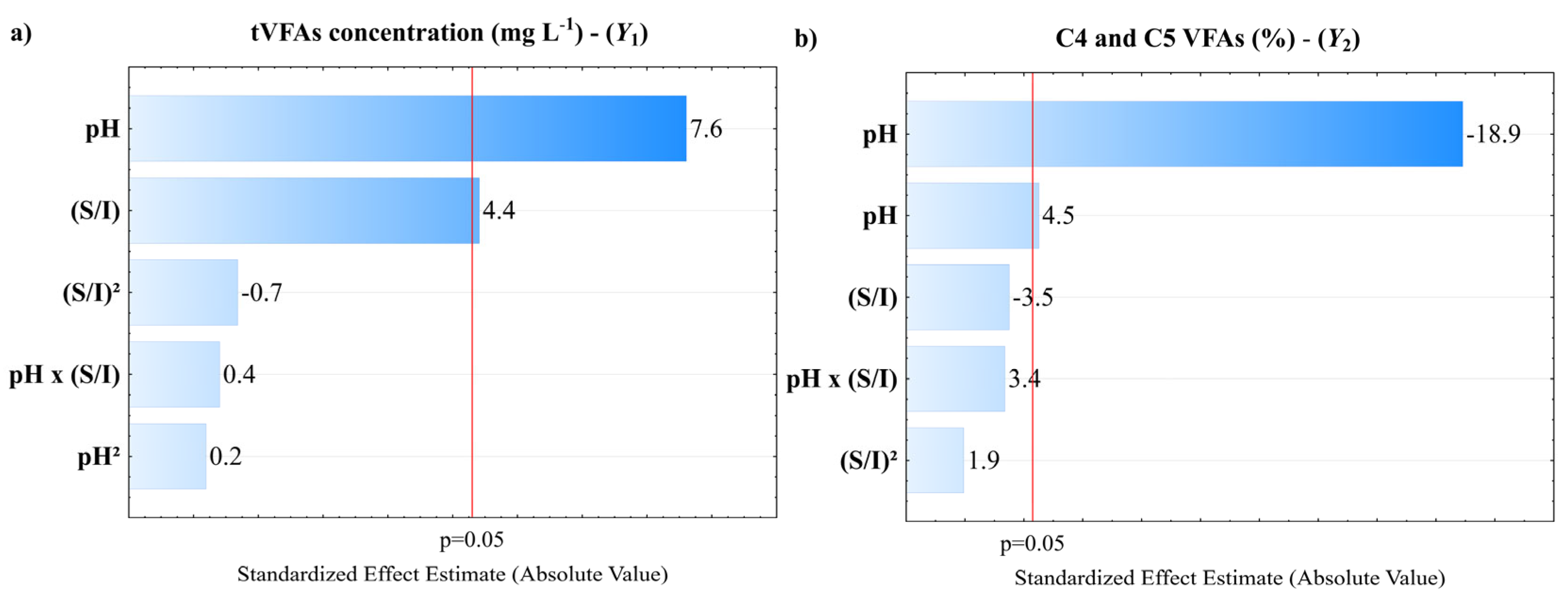
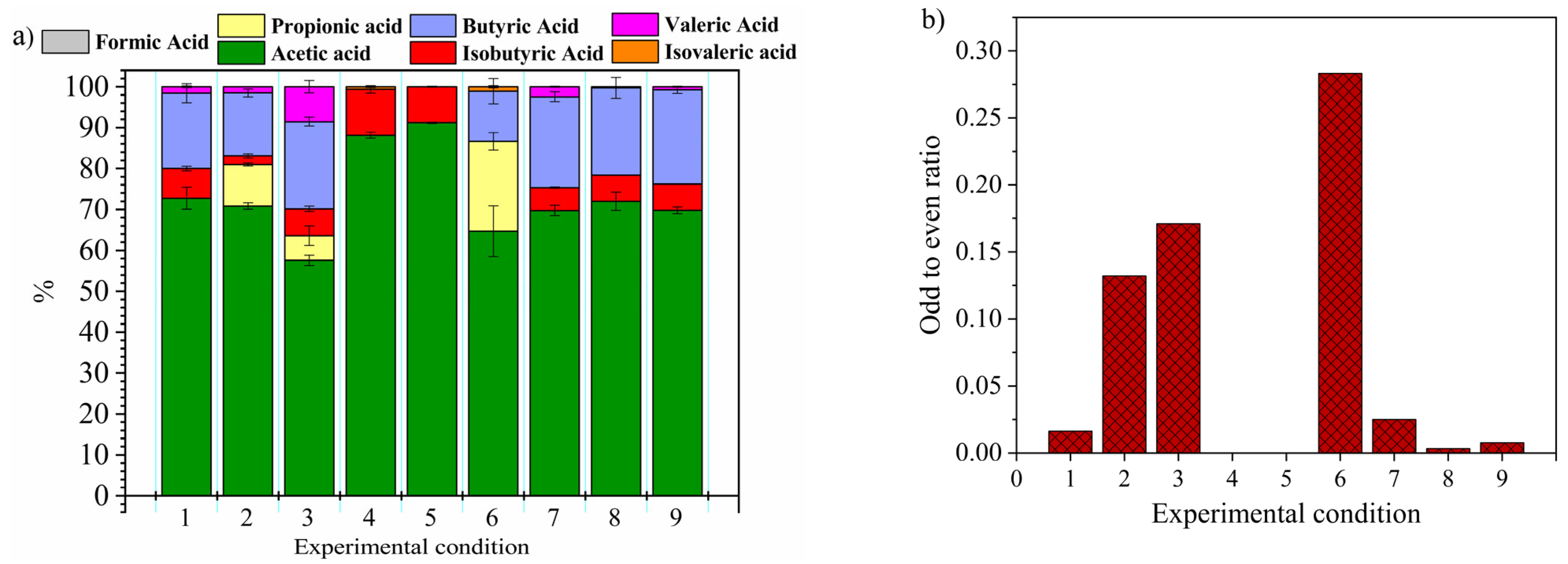
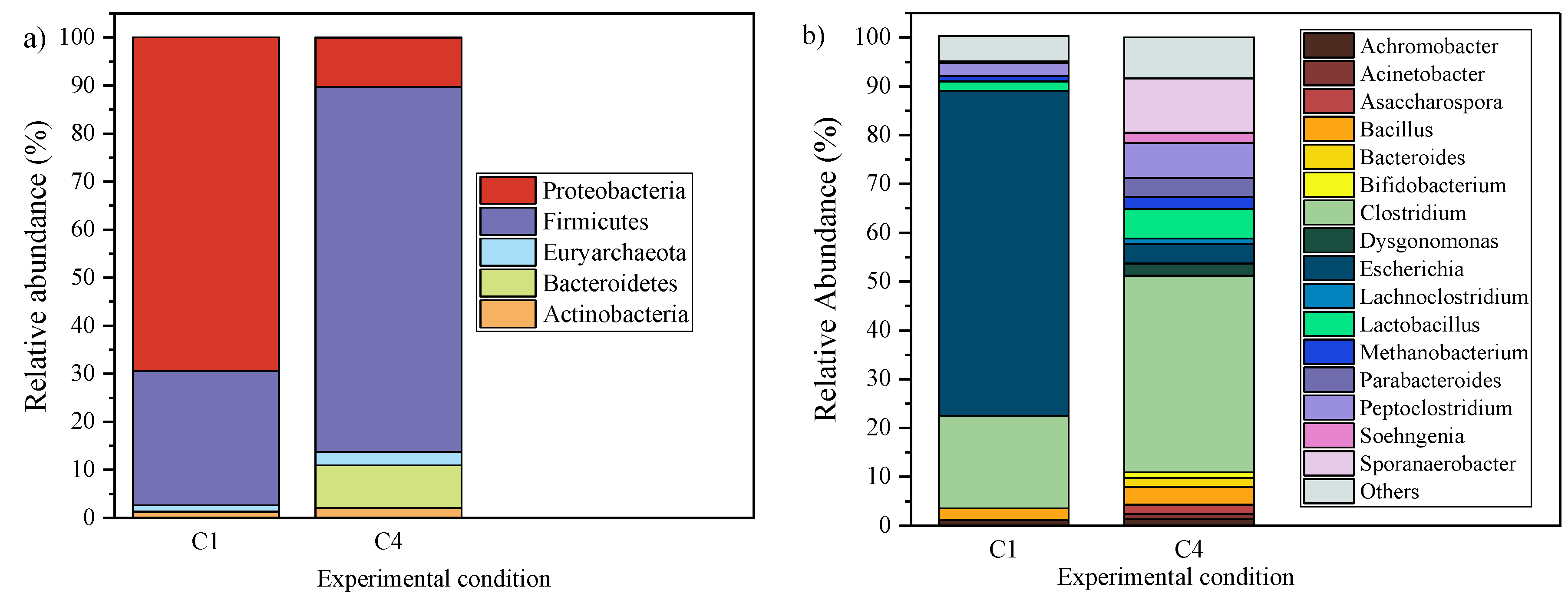
3. Results and Discussion
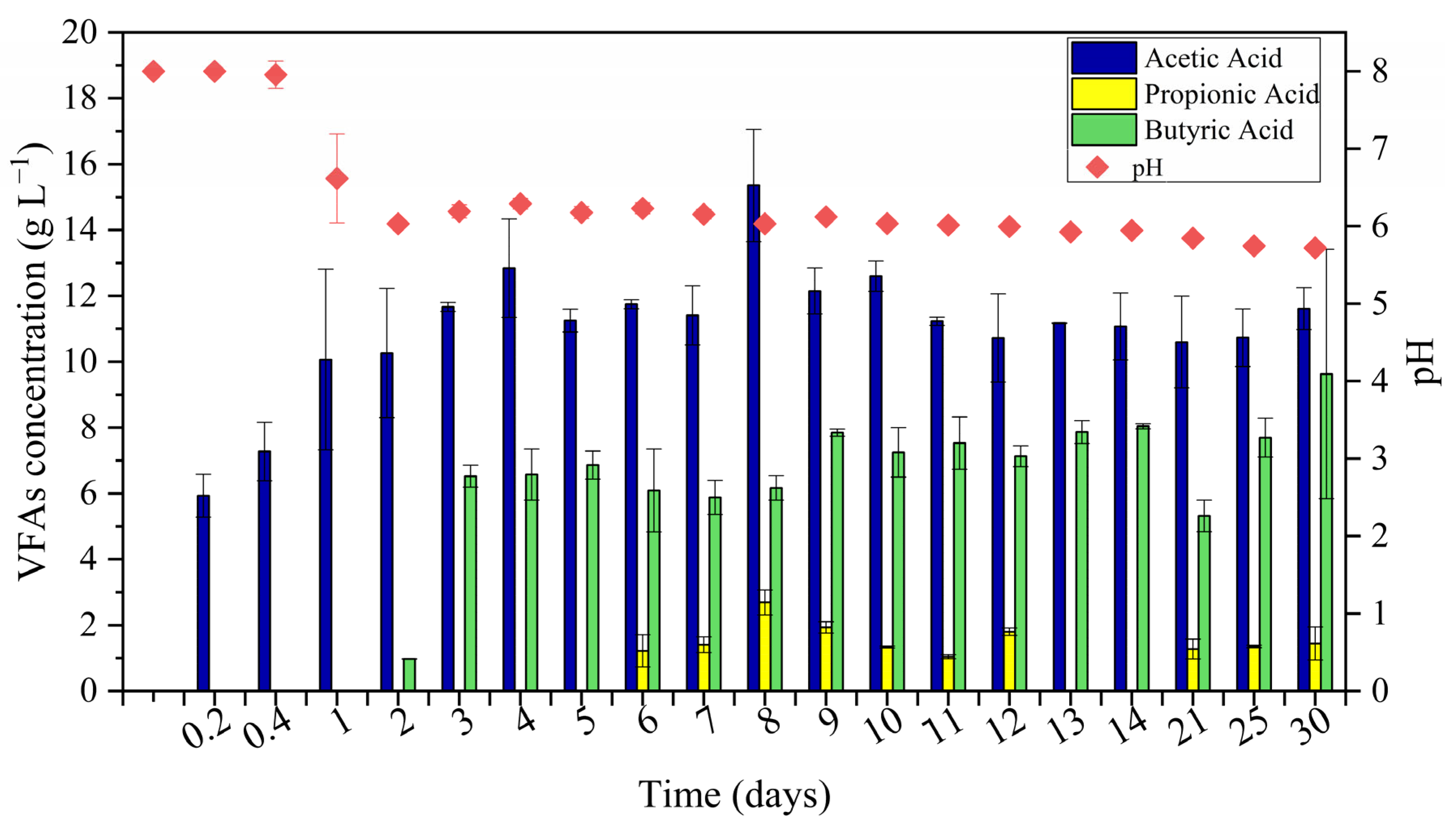
3.1. Fermentation Assays
3.2. VFAs Composition
3.3. Microbial Community
3.4. Simultaneous Optimization and Kinetic Studies
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- European Commission. Communication from the Commission to the European Parliament, the Council, the European Economic and Social Committee and the Committee of the Regions; European Commission: Brussels, Belgium, 2015. [Google Scholar]
- Atasoy, M.; Owusu-Agyeman, I.; Plaza, E.; Cetecioglu, Z. Bio-Based Volatile Fatty Acid Production and Recovery from Waste Streams: Current Status and Future Challenges. Bioresour. Technol. 2018, 268, 773–786. [Google Scholar] [CrossRef] [PubMed]
- Zacharof, M.; Lovitt, R. Complex Effluent Streams as a Potential Source of Volatile Fatty Acids. Waste Biomass Valorization 2013, 4, 557–581. [Google Scholar] [CrossRef]
- Chandrasekhar, K.; Kumar, S.; Lee, B.D.; Kim, S.H. Waste Based Hydrogen Production for Circular Bioeconomy: Current Status and Future Directions. Bioresour. Technol. 2020, 302, 122920. [Google Scholar] [CrossRef] [PubMed]
- Murali, N.; Srinivas, K.; Ahring, B.K. Biochemical Production and Separation of Carboxylic Acids for Biorefinery Applications. Fermentation 2017, 3, 22. [Google Scholar] [CrossRef]
- Wainaina, S.; Lukitawesa; Kumar Awasthi, M.; Taherzadeh, M. Bioengineering of Anaerobic Digestion for Volatile Fatty Acids, Hydrogen or Methane Production: A Critical Review. Bioengineered 2019, 10, 437–458. [Google Scholar] [CrossRef]
- Roig, A.; Cayuela, M.L.; Sa, M.A. An Overview on Olive Mill Wastes and Their Valorisation Methods. Waste Manag. 2006, 26, 960–969. [Google Scholar] [CrossRef]
- Rodríguez, G.; Lama, A.; Rodríguez, R.; Jiménez, A.; Guillén, R.; Fernández-Bolaños, J. Olive Stone an Attractive Source of Bioactive and Valuable Compounds. Bioresour. Technol. 2008, 99, 5261–5269. [Google Scholar] [CrossRef]
- Serrano, A.; Fermoso, F.G.; Rodríguez-Gutierrez, G.; Fernandez-Bolaños, J.; Borja, R. Biomethanization of Olive Mill Solid Waste after Phenols Recovery through Low-Temperature Thermal Pre-Treatment. Waste Manag. 2017, 61, 229–235. [Google Scholar] [CrossRef]
- Yarimtepe, C.; Oz, N.; Ince, O. Volatile Fatty Acid Production Dynamics during the Acidification of Pretreated Olive Mill Wastewater. Bioresour. Technol. 2017, 241, 936–944. [Google Scholar] [CrossRef]
- Rincón, B.; Borja, R. Assessment of the Anaerobic Acidogenesis of Wet Olive Cake from a Two-Phase Olive Oil Mill. J. Environ. Sci. Health 2012, 47, 1439–1445. [Google Scholar] [CrossRef]
- Cabrera, F.; Serrano, A.; Torres, Á.; Guillermo, R.-G.; Jeison, D.; Fermoso, F. The Accumulation of Volatile Fatty Acids and Phenols through a pH- Controlled Fermentation of Olive Mill Solid Waste. Sci. Total Environ. 2019, 657, 1501–1507. [Google Scholar] [CrossRef]
- Da Fonseca, Y.A.; Silva, N.C.S.; De Camargos, A.B.; Silva, S.D.Q.; Wandurraga, H.J.L.; Gurgel, L.V.A.; Baêta, B.E.L. Influence of Hydrothermal Pretreatment Conditions, Typology of Anaerobic Digestion System, and Microbial Profile in the Production of Volatile Fatty Acids from Olive Mill Solid Waste. J. Environ. Chem. Eng. 2021, 9, 105055. [Google Scholar] [CrossRef]
- Angelidaki, I.; Alves, M.; Bolzonella, D.; Borzacconi, L.; Campos, J.L.; Guwy, A.J.; Kalyuzhnyi, S.; Jenicek, P.; Van Lier, J.B. Defining the Biomethane Potential (BMP) of Solid Organic Wastes and Energy Crops: A Proposed Protocol for Batch Assays. Water Sci. Technol. 2009, 59, 927–934. [Google Scholar] [CrossRef]
- Li, Y.; Chen, Z.; Peng, Y.; Huang, W.; Liu, J.; Mironov, V.; Zhang, S. Deeper Insights into the Effects of Substrate to Inoculum Ratio Selection on the Relationship of Kinetic Parameters, Microbial Communities, and Key Metabolic Pathways during the Anaerobic Digestion of Food Waste. Water Res. 2022, 217, 118440. [Google Scholar] [CrossRef]
- Elbeshbishy, E.; Nakhla, G.; Hafez, H. Biochemical Methane Potential (BMP) of Food Waste and Primary Sludge: Influence of Inoculum Pre-Incubation and Inoculum Source. Bioresour. Technol. 2012, 110, 18–25. [Google Scholar] [CrossRef]
- APHA. Standard Methods for the Examination of Water and Wastewater; American Public Health Association: Washington, DC, USA, 2005. [Google Scholar]
- Derringer, G.; Suich, R. Simultaneous Optimization of Several Response Variables. J. Qual. Technol. 1980, 12, 214–219. [Google Scholar] [CrossRef]
- Lima, D.; Adarme, O.; Baêta, B.; Gurgel, L.; Aquino, S. Influence of Different Thermal Pretreatments and Inoculum Selection on the Biomethanation of Sugarcane Bagasse by Solid-State Anaerobic Digestion: A Kinetic Analysis. Ind. Crop. Prod. 2018, 111, 684–693. [Google Scholar] [CrossRef]
- Baêta, B.; Lima, D.; Filho, J.; Adarme, O.; Gurgel, L.; Aquino, S. Evaluation of Hydrogen and Methane Production from Sugarcane Bagasse Hemicellulose Hydrolysates by Two-Stage Anaerobic Digestion Process. Bioresour. Technol. 2016, 218, 436–446. [Google Scholar] [CrossRef]
- Iglesias-Iglesias, R.; Campanaro, S.; Treu, L.; Kennes, C.; Veiga, M.C. Valorization of Sewage Sludge for Volatile Fatty Acids Production and Role of Microbiome on Acidogenic Fermentation. Bioresour. Technol. 2019, 291, 121817. [Google Scholar] [CrossRef]
- Adarme, O.; Eduardo Lobo Baêta, B.; Cardoso Torres, M.; Camilo Otalora Tapiero, F.; Vinicius Alves Gurgel, L.; de Queiroz Silva, S.; Francisco de Aquino, S. Biogas Production by Anaerobic Co-Digestion of Sugarcane Biorefinery Byproducts: Comparative Analyses of Performance and Microbial Community in Novel Single-and Two-Stage Systems. Bioresour. Technol. 2022, 354, 127185. [Google Scholar] [CrossRef]
- Christoff, A.; Fernanda, A.; Sereia, R.; Rodrigues, D.; Lucio, R.; De Moraes, V.; Felipe, L.; De Oliveira, V. Bacterial Identification through Accurate Library Preparation and High-Throughput Sequencing. Neoprospecta Microbiome Technol. 2017, 25, 1–5. [Google Scholar]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive Functional Profiling of Microbial Communities Using 16S RRNA Marker Gene Sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Zhao, X.; Li, W.; Chen, H.; Zhu, X.; Zhu, H.; Zhang, P. Responses of Short-Chain Fatty Acids Production to the Addition of Various Biocarriers to Sludge Anaerobic Fermentation. Bioresour. Technol. 2020, 304, 122989. [Google Scholar] [CrossRef] [PubMed]
- Morais, N.W.S.; Coelho, M.M.H.; Silva, A.D.S.E.; Pereira, E.L.; Leitão, R.C.; dos Santos, A.B. Kinetic Modeling of Anaerobic Carboxylic Acid Production from Swine Wastewater. Bioresour. Technol. 2020, 297, 122520. [Google Scholar] [CrossRef] [PubMed]
- Akaike, H.A.I. A New Look at the Statistical Model Identification. IEEE Trans. Automat. Contr. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Motulsky, H.; Christopoulos, A. Fitting Models to Biological Data Using Linear and Nonlinear Regression; GraphPad Software: San Diego, CA, USA, 2003. [Google Scholar]
- Aghapour Aktij, S.; Zirehpour, A.; Mollahosseini, A.; Taherzadeh, M.; Tiraferri, A.; Rahimpour, A. Feasibility of Membrane Processes for the Recovery and Purification of Bio-Based Volatile Fatty Acids: A Comprehensive Review. J. Ind. Eng. Chem. 2020, 81, 24–40. [Google Scholar] [CrossRef]
- Reyhanitash, E.; Kersten, S.R.A.; Schuur, B. Recovery of Volatile Fatty Acids from Fermented Wastewater by Adsorption. ACS Sustain. Chem. Eng. 2017, 5, 9176–9184. [Google Scholar] [CrossRef]
- Sarkar, O.; Rova, U.; Christakopoulos, P.; Matsakas, L. Influence of Initial Uncontrolled pH on Acidogenic Fermentation of Brewery Spent Grains to Biohydrogen and Volatile Fatty Acids Production: Optimization and Scale-Up. Bioresour. Technol. 2021, 319, 124233. [Google Scholar] [CrossRef]
- Dahiya, S.; Sarkar, O.; Swamy, Y.V.; Venkata Mohan, S. Acidogenic Fermentation of Food Waste for Volatile Fatty Acid Production with Co-Generation of Biohydrogen. Bioresour. Technol. 2015, 182, 103–113. [Google Scholar] [CrossRef]
- López-Garzón, C.S.; Straathof, A.J.J. Recovery of Carboxylic Acids Produced by Fermentation. Biotechnol. Adv. 2014, 32, 873–904. [Google Scholar] [CrossRef]
- Strazzera, G.; Battista, F.; Garcia, N.H.; Frison, N.; Bolzonella, D. Volatile Fatty Acids Production from Food Wastes for BioreFi Nery Platforms: A Review. J. Environ. Manag. 2018, 226, 278–288. [Google Scholar] [CrossRef]
- Atasoy, M.; Eyice, O.; Schnürer, A.; Cetecioglu, Z. Fatty Acids Production via Mixed Culture Fermentation: Revealing the Link between pH, Inoculum Type and Bacterial Composition. Bioresour. Technol. 2019, 292, 121889. [Google Scholar] [CrossRef]
- Wu, Q.; Bao, X.; Guo, W.; Wang, B.; Li, Y.; Luo, H.; Wang, H.; Ren, N. Medium Chain Carboxylic Acids Production from Waste Biomass: Current Advances and Perspectives. Biotechnol. Adv. 2019, 37, 599–615. [Google Scholar] [CrossRef]
- Kleerebezem, R.; Joosse, B.; Rozendal, R.; Van Loosdrecht, M.C.M. Anaerobic Digestion without Biogas? Rev. Environ. Sci. Biotechnol. 2015, 14, 787–801. [Google Scholar] [CrossRef]
- Li, D.; Yin, F.; Ma, X. Towards Biodegradable Polyhydroxyalkanoate Production from Wood Waste: Using Volatile Fatty Acids as Conversion Medium. Bioresour. Technol. 2020, 299, 122629. [Google Scholar] [CrossRef]
- Gameiro, T.; Sousa, F.; Silva, F.C.; Couras, C.; Lopes, M.; Louros, V.; Nadais, H.; Capela, I. Olive Oil Mill Wastewater to Volatile Fatty Acids: Statistical Study of the Acidogenic Process. Water Air Soil Pollut. 2015, 226, 115. [Google Scholar] [CrossRef]
- Wilks, J.C.; Slonczewski, J.L. pH of the Cytoplasm and Periplasm of Escherichia Coli: Rapid Measurement by Green Fluorescent Protein Fluorimetry. J. Bacteriol. 2007, 189, 5601–5607. [Google Scholar] [CrossRef]
- De Jonge, R.; Takumi, K.; Ritmeester, W.S.; Van Leusden, F.M. The Adaptive Response of Escherichia Coli O157 in an Environment with Changing pH. J. Appl. Microbiol. 2003, 94, 555–560. [Google Scholar] [CrossRef]
- Cao, Q.; Zhang, W.; Lian, T.; Wang, S.; Yin, F.; Zhou, T.; Zhang, H.; Zhu, J.; Dong, H. Roles of Micro-Aeration on Enhancing Volatile Fatty Acids and Lactic Acid Production from Agricultural Wastes. Bioresour. Technol. 2022, 347, 126656. [Google Scholar] [CrossRef]
- Atasoy, M.; Cetecioglu, Z. Butyric Acid Dominant Volatile Fatty Acids Production: Bio-Augmentation of Mixed Culture Fermentation by Clostridium Butyricum. J. Environ. Chem. Eng. 2020, 8, 104496. [Google Scholar] [CrossRef]
- Liang, J.; Fang, W.; Chang, J.; Zhang, G.; Ma, W.; Nabi, M.; Zubair, M.; Zhang, R.; Chen, L.; Huang, J.; et al. Long-Term Rumen Microorganism Fermentation of Corn Stover in Vitro for Volatile Fatty Acid Production. Bioresour. Technol. 2022, 358, 127447. [Google Scholar] [CrossRef]
- Djemiel, C.; Maron, P.A.; Terrat, S.; Dequiedt, S.; Cottin, A.; Ranjard, L. Inferring Microbiota Functions from Taxonomic Genes: A Review. Gigascience 2022, 11, 1–30. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Langille, M.G.I. A Primer and Discussion on DNA-Based Microbiome Data and Related Bioinformatics Analyses. Peer Community J. 2021, 1. [Google Scholar] [CrossRef]
- Shao, Q.; Zhang, Q.; Fang, S.; Huang, W.; Li, Z.; Fang, X. Upgrading Volatile Fatty Acids Production from Anaerobic Co-Fermentation of Orange Peel Waste and Sewage Sludge: Critical Roles of Limonene on Functional Consortia and Microbial Metabolic Traits. Bioresour. Technol. 2022, 362, 127773. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Fang, W.; Wang, Q.; Zubair, M.; Zhang, G.; Ma, W.; Cai, Y.; Zhang, P. Metagenomic Analysis of Community, Enzymes and Metabolic Pathways during Corn Straw Fermentation with Rumen Microorganisms for Volatile Fatty Acid Production. Bioresour. Technol. 2021, 342, 126004. [Google Scholar] [CrossRef]
- Zhou, M.; Yan, B.; Wong, J.W.C.C.; Zhang, Y. Enhanced Volatile Fatty Acids Production from Anaerobic Fermentation of Food Waste: A Mini-Review Focusing on Acidogenic Metabolic Pathways. Bioresour. Technol. 2018, 248, 68–78. [Google Scholar] [CrossRef]
- Ware, A.; Power, N. Modelling Methane Production Kinetics of Complex Poultry Slaughterhouse Wastes Using Sigmoidal Growth Functions. Renew. Energy 2017, 104, 50–59. [Google Scholar] [CrossRef]
- Hartmann, F.S.F.; Udugama, I.A.; Seibold, G.M.; Sugiyama, H.; Gernaey, K.V. Digital Models in Biotechnology: Towards Multi-Scale Integration and Implementation. Biotechnol. Adv. 2022, 60, 108015. [Google Scholar] [CrossRef]
- Cordero-Lanzac, T.; Ramirez, A.; Navajas, A.; Gevers, L.; Brunialti, S.; Gandía, L.M.; Aguayo, A.T.; Mani Sarathy, S.; Gascon, J. A Techno-Economic and Life Cycle Assessment for the Production of Green Methanol from CO2: Catalyst and Process Bottlenecks. J. Energy Chem. 2022, 68, 255–266. [Google Scholar] [CrossRef]
- Thomassen, G.; Van Dael, M.; Van Passel, S.; You, F. How to Assess the Potential of Emerging Green Technologies? Towards a Prospective Environmental and Techno-Economic Assessment Framework. Green Chem. 2019, 21, 4868–4886. [Google Scholar] [CrossRef]
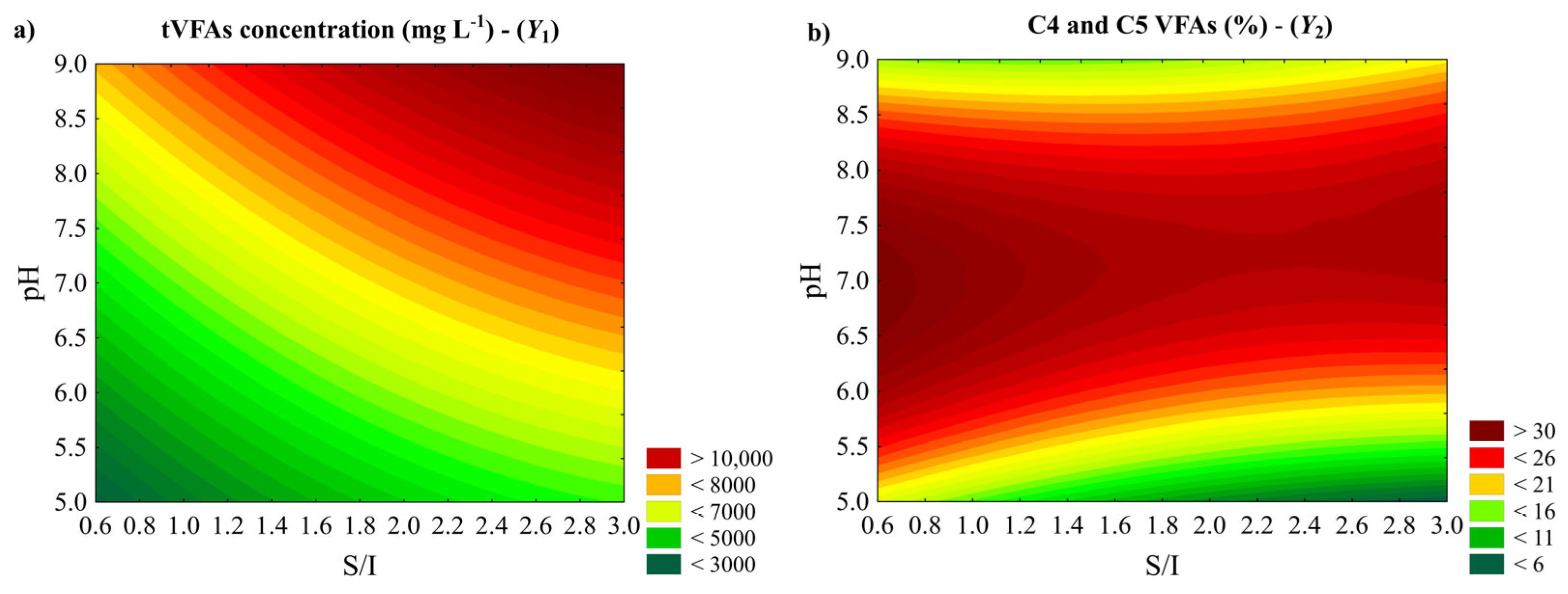

| Experiment Number | Independent Variable (Coded) | Independent Variable (Uncoded) | Dependent Variable | |||
|---|---|---|---|---|---|---|
| S/I | pH | S/I | pH | tVFAs (mg L−1) (Y1) | C4 and C5 VFAs (%) (Y2) | |
| 1 | 1 | 0 | 3 | 7 | 8543.5 ± 531.0 | 27.2 ± 2.7 |
| 2 | 0.5 | 0.866 | 2.4 | 9 | 11302.3 ± 168.1 | 19.0 ± 1.5 |
| 3 | −1 | 0 | 0.5 | 7 | 5228.6 ± 636.6 | 36.4 ± 3.2 |
| 4 | −0.5 | −0.866 | 1.13 | 5 | 3623.6 ± 262.0 | 11.9 ± 1.2 |
| 5 | 0.5 | −0.866 | 2.4 | 5 | 5527.9 ± 168.1 | 8.8 ± 0.2 |
| 6 | −0.5 | 0.866 | 1.13 | 9 | 8816.4 ± 945.2 | 13.4 ± 4.2 |
| 7 | 0 | 0 | 1.75 | 7 | 7605.3 ± 182.6 | 30.3 ± 1.5 |
| 8 | 0 | 0 | 1.76 | 7 | 6517.7 ± 431.8 | 27.9 ±2.9 |
| 9 | 0 | 0 | 1.77 | 7 | 7880.9 ± 319.5 | 30.2 ± 1.1 |
| Kinetic Model | Model Equation |
|---|---|
| First-order | |
| Second-order | |
| Fitzhugh | |
| Monomolecular | |
| Modified Gompertz | |
| Logistic | |
| Transference | |
| Richards |
| tVFAs Concentration (mg L−1) (Y1) | C4 and C5 VFAs (%) (Y2) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Sum of Squares | Degrees of Freedom | Mean Square | F | p | Sum of Squares | Degrees of Freedom | Mean Squares | F | p | |
| S/I | 10,120,033 | 1 | 10,120,033 | 19.479 | 0.048 | 20.718 | 1 | 20.718 | 12.267 | 0.073 |
| (S/I)² | 241,472 | 1 | 241,472 | 0.465 | 0.566 | 6.487 | 1 | 6.487 | 3.841 | 0.189 |
| pH | 30,069,869 | 1 | 30,069,869 | 57.880 | 0.017 | 34.357 | 1 | 34.357 | 20.344 | 0.046 |
| pH² | 19,279 | 1 | 19,279 | 0.037 | 0.865 | 603.865 | 1 | 603.865 | 357.559 | 0.003 |
| S/I x pH | 84,565 | 1 | 84,565 | 0.163 | 0.726 | 19.018 | 1 | 19.018 | 11.261 | 0.079 |
| Lack of fit | 192,712 | 1 | 192,712 | 0.371 | 0.605 | 22.950 | 1 | 22.950 | 13.589 | 0.066 |
| Pure error | 1,039,048 | 2 | 519,524 | 3.378 | 2 | 1.689 | ||||
| Total SS | 41,806,271 | 8 | 762.282 | 8 | ||||||
| R² | R²a | pregression | R² | R²a | pregression | |||||
| 0.970 | 0.921 | 0.0167 | 0.965 | 0.908 | 0.066 | |||||
| Kinetic Model | Parameters | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| kCA | n | λ | µ | ν | R² | R²a | RMSE | NRMSE | AIC | |
| (d−1) | (d) | (g L−1 d−1) | (%) | |||||||
| First-order | 0.44 | 0.81 | 0.80 | 2.69 | 11.10 | 41.56 | ||||
| Second-order | 0.04 | 0.89 | 0.89 | 2.03 | 8.38 | 30.31 | ||||
| Fitzhugh | 1.07 | 0.41 | 0.81 | 0.79 | 2.69 | 11.10 | 43.56 | |||
| Monomolecular | 0.44 | 0.00 | 0.81 | 0.79 | 2.69 | 11.10 | 43.56 | |||
| Modified Gompertz | 0.00 | 6.24 | 0.77 | 0.75 | 2.93 | 12.08 | 46.94 | |||
| Logistic | 0.00 | 5.85 | 0.78 | 0.75 | 2.91 | 12.03 | 46.77 | |||
| Transference | 0.00 | 9.92 | 0.81 | 0.79 | 2.69 | 11.10 | 43.56 | |||
| Richards | 0.00 | 2.22 | 0.17 | 0.77 | 0.73 | 2.93 | 12.08 | 48.94 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
da Fonseca, Y.A.; de Camargos, A.B.; Gomes, G.S.M.; Fregulia, P.; Silva, S.Q.; Gurgel, L.V.A.; Baêta, B.E.L. Enhancing Biobased Volatile Fatty Acids Production from Olive Mill Solid Waste by Optimization of pH and Substrate to Inoculum Ratio. Processes 2023, 11, 338. https://doi.org/10.3390/pr11020338
da Fonseca YA, de Camargos AB, Gomes GSM, Fregulia P, Silva SQ, Gurgel LVA, Baêta BEL. Enhancing Biobased Volatile Fatty Acids Production from Olive Mill Solid Waste by Optimization of pH and Substrate to Inoculum Ratio. Processes. 2023; 11(2):338. https://doi.org/10.3390/pr11020338
Chicago/Turabian Styleda Fonseca, Yasmim A., Adonai B. de Camargos, Gustavo S. M. Gomes, P. Fregulia, Silvana Q. Silva, Leandro V. A. Gurgel, and Bruno E. L. Baêta. 2023. "Enhancing Biobased Volatile Fatty Acids Production from Olive Mill Solid Waste by Optimization of pH and Substrate to Inoculum Ratio" Processes 11, no. 2: 338. https://doi.org/10.3390/pr11020338
APA Styleda Fonseca, Y. A., de Camargos, A. B., Gomes, G. S. M., Fregulia, P., Silva, S. Q., Gurgel, L. V. A., & Baêta, B. E. L. (2023). Enhancing Biobased Volatile Fatty Acids Production from Olive Mill Solid Waste by Optimization of pH and Substrate to Inoculum Ratio. Processes, 11(2), 338. https://doi.org/10.3390/pr11020338







