Bacterial Cultural Media Containing Lipopeptides for Heavy Oil Recovery Enhancement: The Results of Sand-Packed Column Experiment
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolation and Characterization of Microorganisms Capable of Producing Biosurfactants
2.1.1. Strain Screening and Growth Parameters
2.1.2. Molecular Analysis
2.2. Characterization of Biosurfactants
2.2.1. Isolation and Purification of Biosurfactants
2.2.2. Emulsification Test and Surface Tension Measurement
2.2.3. FTIR and TLC
2.3. MEOR Simulation
2.4. Statistical Analysis
3. Results and Discussions
3.1. Characterization of Isolates
3.2. Characterization of Biosurfactants
3.3. MEOR Simulation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Youssef, N.; Simpson, D.R.; Duncan, K.E.; McInerney, M.J.; Folmsbee, M.; Fincher, T.; Knapp, R.M. In Situ Biosurfactant Production by Bacillus Strains Injected into a Limestone Petroleum Reservoir. Appl. Environ. Microbiol. 2007, 73, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, T.; Olugbade, T.O.; Nwankwo, I. Determination of the Effect of Oil Exploration on Galvanized Steel in Niger Delta, Nigeria. J. Sci. Res. Rep. 2016, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Gudiña, E.J.; Pereira, J.F.B.; Costa, R.; Coutinho, J.A.P.; Teixeira, J.A.; Rodrigues, L.R. Biosurfactant-Producing and Oil-Degrading Bacillus Subtilis Strains Enhance Oil Recovery in Laboratory Sand-Pack Columns. J. Hazard. Mater. 2013, 261, 106–113. [Google Scholar] [CrossRef] [PubMed]
- Gudiña, E.J.; Teixeira, J.A. Bacillus Licheniformis: The Unexplored Alternative for the Anaerobic Production of Lipopeptide Biosurfactants? Biotechnol. Adv. 2022, 60, 108013. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Niu, J.; Yu, Y.; Wang, C.; Lu, S.; Zhang, S.; Lv, J.; Peng, B. Production, Characterization and Application of Biosurfactant Produced by Bacillus Licheniformis L20 for Microbial Enhanced Oil Recovery. J. Clean. Prod. 2021, 307, 127193. [Google Scholar] [CrossRef]
- Purwasena, I.A.; Astuti, D.I.; Syukron, M.; Amaniyah, M.; Sugai, Y. Stability Test of Biosurfactant Produced by Bacillus Licheniformis DS1 Using Experimental Design and Its Application for MEOR. J. Pet. Sci. Eng. 2019, 183, 106383. [Google Scholar] [CrossRef]
- Eras-Muñoz, E.; Farré, A.; Sánchez, A.; Font, X.; Gea, T. Microbial Biosurfactants: A Review of Recent Environmental Applications. Bioengineered 2022, 13, 12365–12391. [Google Scholar] [CrossRef] [PubMed]
- Biktasheva, L.; Gordeev, A.; Selivanovskaya, S.; Galitskaya, P. Di-and Mono-Rhamnolipids Produced by the Pseudomonas Putida PP021 Isolate Significantly Enhance the Degree of Recovery of Heavy Oil from the Romashkino Oil Field (Tatarstan, Russia). Processes 2022, 10, 779. [Google Scholar] [CrossRef]
- Zhang, J.; Xue, Q.; Gao, H.; Lai, H.; Wang, P. Production of Lipopeptide Biosurfactants by Bacillus Atrophaeus 5-2a and Their Potential Use in Microbial Enhanced Oil Recovery. Microb. Cell Factories 2016, 15, 168. [Google Scholar] [CrossRef]
- Varjani, S.J.; Upasani, V.N. Carbon Spectrum Utilization by an Indigenous Strain of Pseudomonas Aeruginosa NCIM 5514: Production, Characterization and Surface Active Properties of Biosurfactant. Bioresour. Technol. 2016, 221, 510–516. [Google Scholar] [CrossRef]
- Benincasa, M.; Abalos, A.; Oliveira, I.; Manresa, A. Chemical Structure, Surface Properties and Biological Activities of the Biosurfactant Produced by Pseudomonas Aeruginosa LBI from Soapstock. Antonie Van Leeuwenhoek 2004, 85, 1–8. [Google Scholar] [CrossRef]
- Benincasa, M.; Accorsini, F.R. Pseudomonas Aeruginosa LBI Production as an Integrated Process Using the Wastes from Sunflower-Oil Refining as a Substrate. Bioresour. Technol. 2008, 99, 3843–3849. [Google Scholar] [CrossRef] [PubMed]
- Gong, H.; Li, Y.; Dong, M.; Ma, S.; Liu, W. Effect of Wettability Alteration on Enhanced Heavy Oil Recovery by Alkaline Flooding. Colloids Surf. A Physicochem. Eng. Asp. 2016, 488, 28–35. [Google Scholar] [CrossRef]
- Sarafzadeh, P.; Niazi, A.; Oboodi, V.; Ravanbakhsh, M.; Hezave, A.Z.; Ayatollahi, S.S.; Raeissi, S. Investigating the Efficiency of MEOR Processes Using Enterobacter Cloacae and Bacillus Stearothermophilus SUCPM# 14 (Biosurfactant-Producing Strains) in Carbonated Reservoirs. J. Pet. Sci. Eng. 2014, 113, 46–53. [Google Scholar]
- Aboelkhair, H.; Diaz, P.; Attia, A. Biosurfactant Production Using Egyptian Oil Fields Indigenous Bacteria for Microbial Enhanced Oil Recovery. J. Pet. Sci. Eng. 2022, 208, 109601. [Google Scholar] [CrossRef]
- Cooper, D.G.; Goldenberg, B.G. Surface-Active Agents from Two Bacillus Species. Appl. Environ. Microbiol. 1987, 53, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Ndlovu, T.; Rautenbach, M.; Khan, S.; Khan, W. Variants of Lipopeptides and Glycolipids Produced by Bacillus Amyloliquefaciens and Pseudomonas Aeruginosa Cultured in Different Carbon Substrates. AMB Express 2017, 7, 109. [Google Scholar] [CrossRef] [PubMed]
- Simpson, D.R.; Natraj, N.R.; McInerney, M.J.; Duncan, K.E. Biosurfactant-Producing Bacillus Are Present in Produced Brines from Oklahoma Oil Reservoirs with a Wide Range of Salinities. Appl. Microbiol. Biotechnol. 2011, 91, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Plaza, G.; Chojniak, J.; Rudnicka, K.; Paraszkiewicz, K.; Bernat, P. Detection of Biosurfactants in Bacillus Species: Genes and Products Identification. J. Appl. Microbiol. 2015, 119, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.; Kong, H.; Buyer, J.S.; Lakshman, D.K.; Lydon, J.; Kim, S.-D.; Roberts, D.P. Isolation and Partial Characterization of Bacillus Subtilis ME488 for Suppression of Soilborne Pathogens of Cucumber and Pepper. Appl. Microbiol. Biotechnol. 2008, 80, 115–123. [Google Scholar] [CrossRef]
- Galitskaya, P.; Karamova, K.; Biktasheva, L.; Galieva, G.; Gordeev, A.; Selivanovskaya, S. Lipopeptides Produced by Bacillus Mojavensis P1709 as an Efficient Tool to Maintain Postharvest Cherry Tomato Quality and Quantity. Agriculture 2022, 12, 609. [Google Scholar] [CrossRef]
- Zhao, F.; Wang, Q.; Zhang, Y.; Lei, L. Anaerobic Biosynthesis of Rhamnolipids by Pseudomonas Aeruginosa: Performance, Mechanism and Its Application Potential for Enhanced Oil Recovery. Microb. Cell Factories 2021, 20, 103. [Google Scholar] [CrossRef] [PubMed]
- Suthar, H.; Hingurao, K.; Desai, A.; Nerurkar, A. Evaluation of Bioemulsifier Mediated Microbial Enhanced Oil Recovery Using Sand Pack Column. J Microbiol Methods 2008, 75, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Gordadze, G.N.; Tikhomirov, V.I. On the Oil Sources in the Northeast of Tatarstan. Pet. Chem. 2007, 47, 389–398. [Google Scholar] [CrossRef]
- Al-Sakkaf, M.K.; Onaizi, S.A. Rheology, Characteristics, Stability, and pH-Responsiveness of Biosurfactant-Stabilized Crude Oil/Water Nanoemulsions. Fuel 2022, 307, 121845. [Google Scholar] [CrossRef]
- Salehizadeh, H.; Mohammadizad, S. Microbial Enhanced Oil Recovery Using Biosurfactant Produced by Alcaligenes Faecalis. Iran. J. Biotechnol. 2009, 7, 216–223. [Google Scholar]
- Mora, I.; Cabrefiga, J.; Montesinos, E. Antimicrobial Peptide Genes in Bacillus Strains from Plant Environments. Int. Microbiol 2011, 14, 213–223. [Google Scholar] [PubMed]
- Batista, R.M.; Rufino, R.D.; Luna, J.M.; de Souza, J.E.G.; Sarubbo, L.A. Effect of Medium Components on the Production of a Biosurfactant from Candida Tropicalis Applied to the Removal of Hydrophobic Contaminants in Soil. Water Environ. Res. 2010, 82, 418–425. [Google Scholar] [CrossRef]
- Joshi, S.J.; Al-Wahaibi, Y.M.; Al-Bahry, S.N.; Elshafie, A.E.; Al-Bemani, A.S.; Al-Bahri, A.; Al-Mandhari, M.S. Production, Characterization, and Application of Bacillus Licheniformis W16 Biosurfactant in Enhancing Oil Recovery. Front. Microbiol. 2016, 7, 1853. [Google Scholar] [CrossRef]
- Kiran, G.S.; Priyadharsini, S.; Sajayan, A.; Priyadharsini, G.B.; Poulose, N.; Selvin, J. Production of Lipopeptide Biosurfactant by a Marine Nesterenkonia Sp. and Its Application in Food Industry. Front. Microbiol. 2017, 8, 1138. [Google Scholar] [CrossRef]
- Pathak, K.V.; Keharia, H. Application of Extracellular Lipopeptide Biosurfactant Produced by Endophytic Bacillus Subtilis K1 Isolated from Aerial Roots of Banyan (Ficus Benghalensis) in Microbially Enhanced Oil Recovery (MEOR). 3 Biotech 2014, 4, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Gittins, P.; Iglauer, S.; Pentland, C.H.; Al-Mansoori, S.; Al-Sayari, S.; Bijeljic, B.; Blunt, M.J. Nonwetting phase residual saturation in sand packs. J. Porous Media 2010, 13, 591–599. [Google Scholar] [CrossRef]
- Kögler, F.; Mahler, E.; Dopffel, N.; Schulze-Makuch, D.; Borovina, A.; Visser, F.; Herold, A.; Alkan, H. The Microbial Enhanced Oil Recovery (MEOR) Potential of Halanaerobiales under Dynamic Conditions in Different Porous Media. J. Pet. Sci. Eng. 2021, 196, 107578. [Google Scholar] [CrossRef]
- Nerurkar, A.S.; Suthar, H.G.; Desai, A.J. Biosystem Development for Microbial Enhanced Oil Recovery (MEOR). In Microorganisms in Sustainable Agriculture and Biotechnology; Satyanarayana, T., Johri, B.N., Eds.; Springer: Dordrecht, The Netherlands, 2012; pp. 711–737. ISBN 978-94-007-2214-9. [Google Scholar]
- Gandomkar, A.; Rahimpour, M.R. Investigation of Low-Salinity Waterflooding in Secondary and Tertiary Enhanced Oil Recovery in Limestone Reservoirs. Energy Fuels 2015, 29, 7781–7792. [Google Scholar] [CrossRef]
- Nazar, M.F.; Shah, S.S.; Khosa, M.A. Microemulsions in Enhanced Oil Recovery: A Review. Pet. Sci. Technol. 2011, 29, 1353–1365. [Google Scholar] [CrossRef]
- Hamzah, A.F.; Al-Mossawy, M.I.; Al-Tamimi, W.H.; Al-Najm, F.M.; Hameed, Z.M. Enhancing the Spontaneous Imbibition Process Using Biosurfactants Produced from Bacteria Isolated from Al-Rafidiya Oil Field for Improved Oil Recovery. J. Pet. Explor. Prod. Technol. 2020, 10, 3767–3777. [Google Scholar] [CrossRef]
- Bera, A.; Kumar, T.; Ojha, K.; Mandal, A. Adsorption of Surfactants on Sand Surface in Enhanced Oil Recovery: Isotherms, Kinetics and Thermodynamic Studies. Appl. Surf. Sci. 2013, 284, 87–99. [Google Scholar] [CrossRef]
- Saravanan, A.; Kumar, P.S.; Vardhan, K.H.; Jeevanantham, S.; Karishma, S.B.; Yaashikaa, P.R.; Vellaichamy, P. A Review on Systematic Approach for Microbial Enhanced Oil Recovery Technologies: Opportunities and Challenges. J. Clean. Prod. 2020, 258, 120777. [Google Scholar] [CrossRef]
- Hamzah, A.F.; Al-Tamimi, W.H. Enhanced Oil Recovery by Sand Packed Column Supplemented with Biosurfactants Produced by Local Oil Fields Bacteria. Marsh Bull. 2021, 16, 135–143. [Google Scholar]
- Makkar, R.S.; Cameotra, S.S. Production of Biosurfactant at Mesophilic and Thermophilic Conditions by a Strain of Bacillus Subtilis. J. Ind. Microbiol. Biotechnol. 1998, 20, 48–52. [Google Scholar] [CrossRef]
- Wang, H.; Xiu, J.; Huang, L.; Yu, L.; Wu, B. Study on the Application Potential of Lipopeptide Fermentation Broth in Oil Recovery. Energy Sci. Eng. 2022, 10, 2065–2075. [Google Scholar] [CrossRef]
- Bordoloi, N.K.; Konwar, B.K. Microbial Surfactant-Enhanced Mineral Oil Recovery under Laboratory Conditions. Colloids Surf. B Biointerfaces 2008, 63, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Waghmode, S.; Sharma, D.; Joshi, S.J.; Banpurkar, A.; Satpute, S. Prospective Use of a Cell-Free Supernatant from the Biosurfactant-Producing Marine Planococcus Maritimus SAMP MCC 3013 for Oil Removal and Enhanced Oil Recovery. Environ. Qual. Manag. 2023. [Google Scholar] [CrossRef]
- Zargar, A.N.; Patil, N.; Kumar, M.; Srivastava, P. Enhanced Oil Recovery Using a Combination of Biosurfactants. J. Vis. Exp. 2022, 184, e63207. [Google Scholar] [CrossRef]
- Kuyukina, M.S.; Ivshina, I.B.; Philp, J.C.; Christofi, N.; Dunbar, S.A.; Ritchkova, M.I. Recovery of Rhodococcus Biosurfactants Using Methyl Tertiary-Butyl Ether Extraction. J. Microbiol. Methods 2001, 46, 149–156. [Google Scholar] [CrossRef] [PubMed]
- Jimoh, A.A.; Lin, J. Biosurfactant: A New Frontier for Greener Technology and Environmental Sustainability. Ecotoxicol. Environ. Saf. 2019, 184, 109607. [Google Scholar] [CrossRef] [PubMed]
- Pereira, J.F.B.; Gudiña, E.J.; Costa, R.; Vitorino, R.; Teixeira, J.A.; Coutinho, J.A.P.; Rodrigues, L.R. Optimization and Characterization of Biosurfactant Production by Bacillus Subtilis Isolates towards Microbial Enhanced Oil Recovery Applications. Fuel 2013, 111, 259–268. [Google Scholar] [CrossRef]
- de Castilho, L.V.A.; Pasqualino, I.P.; Duarte, A.M.; Waldow, V.D.A.; de Sousa, M.P.; Seldin, L.; Freire, D.M.G. Biosurfactant Versus Commercial Surfactant: Study on Effectiveness for Application in EOR; American Society of Mechanical Engineers Digital Collection: Pittsburgh, PA, USA, 2018. [Google Scholar]
- Wu, B.; Xiu, J.; Yu, L.; Huang, L.; Yi, L.; Ma, Y. Biosurfactant Production by Bacillus Subtilis SL and Its Potential for Enhanced Oil Recovery in Low Permeability Reservoirs. Sci. Rep. 2022, 12, 7785. [Google Scholar] [CrossRef]
- Aparna, A.; Srinikethan, G.; Smitha, H. Production and Characterization of Biosurfactant Produced by a Novel Pseudomonas Sp. 2B. Colloids Surf. B Biointerfaces 2012, 95, 23–29. [Google Scholar] [CrossRef]
- Aguirre-Ramírez, M.; Silva-Jiménez, H.; Banat, I.M.; Díaz De Rienzo, M.A. Surfactants: Physicochemical Interactions with Biological Macromolecules. Biotechnol. Lett. 2021, 43, 523–535. [Google Scholar] [CrossRef]
- Kosaric, N.; Sukan, F.V. Biosurfactants: Production and Utilization-Processes, Technologies, and Economics; CRC Press: Boca Raton, FL, USA, 2014; ISBN 978-1-4665-9670-2. [Google Scholar]
- Invally, K.; Sancheti, A.; Ju, L.-K. A New Approach for Downstream Purification of Rhamnolipid Biosurfactants. Food Bioprod. Process. 2019, 114, 122–131. [Google Scholar] [CrossRef]
- Johnson, P.; Trybala, A.; Starov, V.; Pinfield, V.J. Effect of Synthetic Surfactants on the Environment and the Potential for Substitution by Biosurfactants. Adv. Colloid Interface Sci. 2021, 288, 102340. [Google Scholar] [CrossRef] [PubMed]
- Rezaei Gomari, S.; Elyasi Gomari, K.; Islam, M.; Hughes, D. New Insight into the Influence of Rhamnolipid Bio-Surfactant on the Carbonate Rock/Water/Oil Interaction at Elevated Temperature. Resources 2018, 7, 75. [Google Scholar] [CrossRef]
- Jahanbani Veshareh, M.; Ganji Azad, E.; Deihimi, T.; Niazi, A.; Ayatollahi, S. Isolation and Screening of Bacillus Subtilis MJ01 for MEOR Application: Biosurfactant Characterization, Production Optimization and Wetting Effect on Carbonate Surfaces. J. Pet. Explor. Prod. Technol. 2019, 9, 233–245. [Google Scholar] [CrossRef]
- Vecino Bello, X.; Devesa-Rey, R.; Cruz, J.M.; Moldes, A.B. Study of the Synergistic Effects of Salinity, pH, and Temperature on the Surface-Active Properties of Biosurfactants Produced by Lactobacillus Pentosus. J. Agric. Food Chem. 2012, 60, 1258–1265. [Google Scholar] [CrossRef] [PubMed]
- Uzoigwe, C.; Burgess, J.G.; Ennis, C.J.; Rahman, P.K.S.M. Bioemulsifiers Are Not Biosurfactants and Require Different Screening Approaches. Front. Microbiol. 2015, 6, 245. [Google Scholar] [CrossRef]

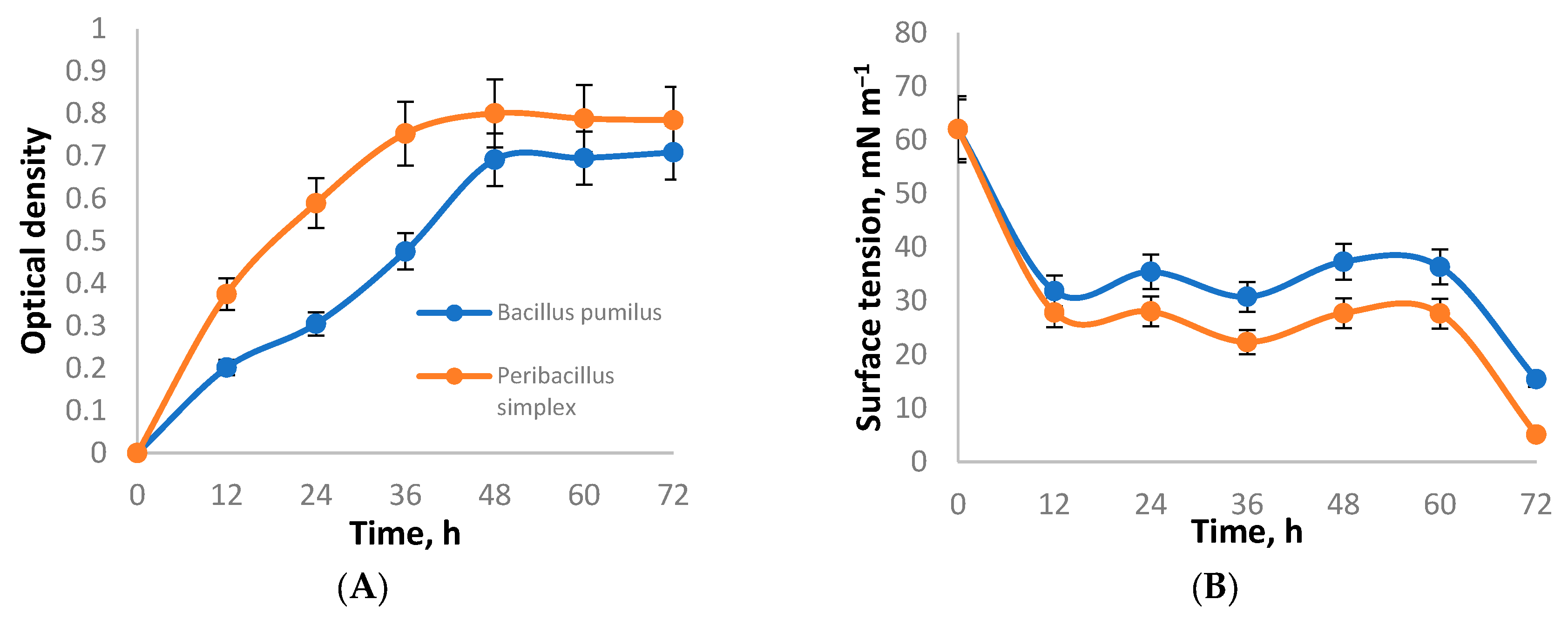


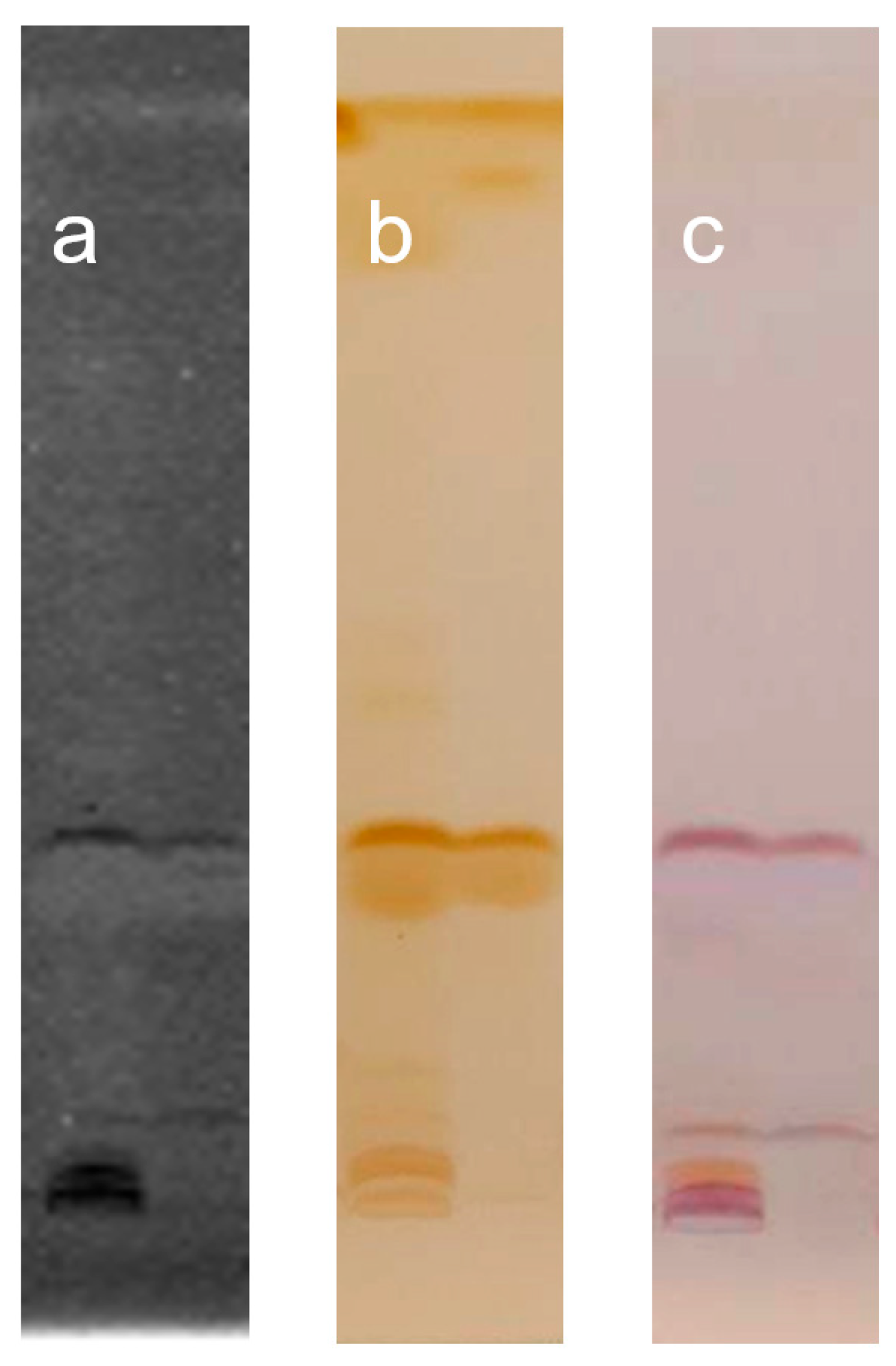
| Strains | Emulsification Index, E24, % | Surface Tension, mN m−1 |
|---|---|---|
| 2A | 50 ± 5 | 29.42 ± 0.09 |
| 2B | 70 ± 2 | 25.17 ± 0.04 |
| 8A | 5 ± 0 | 63.43 ± 0.53 |
| 8B | 10 ± 1 | 64.91 ± 0.31 |
| 14A | 30 ± 2 | 59.78 ± 0.22 |
| 1A | 50 ± 1 | 48.61 ± 0.18 |
| 10A | 30 ± 2 | 29.92 ± 0.07 |
| 3A | 10 ± 1 | 51.50 ± 0.17 |
| 3B1 | 10 ± 0 | 62.45 ± 0.32 |
| 3B2 | 20 ± 3 | 47.90 ± 0.19 |
| 3B3 | 50 ± 4 | 29.15 ± 0.04 |
| 4B | 20 ± 5 | 55.33 ± 0.15 |
| 5A | 20 ± 2 | 57.70 ± 0.66 |
| 5B | 15 ± 0 | 67.66 ± 0.51 |
| 5C | 10 ± 2 | 50.11 ± 0.45 |
| Primer | Biosurfactant | Primer Sequence (5′-3′) | T | References |
|---|---|---|---|---|
| bamC | Bacillomycin | TGCAGGAGGAGAGAGCAGAT AGGTTGTCCGATGTTGCTTC | 60 °C | [17] |
| srfAA | Surfactin | TCGGGACAGGAAGACATCAT CCACTCAAACGGATAATCCTGA | 60 °C | [18] |
| ituC | Iturin | GGCTGCTGCAGATGCTTTAT TCGCAGATAATCGCAGTGAG | 58 °C | [19] |
| fenD | Fengycin | GGCCCGTTCTCTAAATCCAT GTCATGCTGACGAGAGCAAA | 58 °C | [20] |
| Sample | PV, mL | OOIP, mL | Soi, % | Sorbf, mL | Sorsf, mL | The Effectiveness of Biosurfactants in Relation to Chemical Surfactants |
|---|---|---|---|---|---|---|
| Atren | 62.50 ± 0.25 | 21.25 ± 0.40 | 33.90 ± 0.48 | 41.25 ± 0.40 | 2.75 ± 0.14 | - |
| B. pumilus | 55.00 ± 0.40 | 14.00 ± 0.25 | 25.50 ± 0.12 | 41.00 ± 0.75 | 2.50 ± 0.20 | 34% higher |
| P. simplex | 59.00 ± 0.20 | 22.25 ± 0.25 | 37.70 ± 0.40 | 36.25 ± 0.10 | 2.50 ± 0.10 | 16% lower |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galitskaya, P.; Gordeev, A.; Ezhkin, N.; Biktasheva, L.; Kuryntseva, P.; Selivanovskaya, S. Bacterial Cultural Media Containing Lipopeptides for Heavy Oil Recovery Enhancement: The Results of Sand-Packed Column Experiment. Processes 2023, 11, 3203. https://doi.org/10.3390/pr11113203
Galitskaya P, Gordeev A, Ezhkin N, Biktasheva L, Kuryntseva P, Selivanovskaya S. Bacterial Cultural Media Containing Lipopeptides for Heavy Oil Recovery Enhancement: The Results of Sand-Packed Column Experiment. Processes. 2023; 11(11):3203. https://doi.org/10.3390/pr11113203
Chicago/Turabian StyleGalitskaya, Polina, Alexander Gordeev, Nikita Ezhkin, Liliya Biktasheva, Polina Kuryntseva, and Svetlana Selivanovskaya. 2023. "Bacterial Cultural Media Containing Lipopeptides for Heavy Oil Recovery Enhancement: The Results of Sand-Packed Column Experiment" Processes 11, no. 11: 3203. https://doi.org/10.3390/pr11113203
APA StyleGalitskaya, P., Gordeev, A., Ezhkin, N., Biktasheva, L., Kuryntseva, P., & Selivanovskaya, S. (2023). Bacterial Cultural Media Containing Lipopeptides for Heavy Oil Recovery Enhancement: The Results of Sand-Packed Column Experiment. Processes, 11(11), 3203. https://doi.org/10.3390/pr11113203






