Acetone–Butanol–Ethanol Fermentation Phenomenological Models for Process Studies: Parameter Estimation and Multi-Response Model Reduction with Statistical Analysis
Abstract
:1. Introduction
1.1. n-Butanol: Applications and Market
1.2. ABE Fermentation Mathematical Modeling
| Reference | Inhibition Term |
|---|---|
| Mulchandani & Volesky (1986) [3] | |
| Yang and Tsao (1994) [5] | |
| Velázquez-Sánchez and Aguilar-López (2019) [23] | |
| Eom et al. (2015) [24] | |
| Buehler and Mesbah (2016) [19] | |
| Rochón et al. (2017) [6] |
1.3. The Present Work
2. Methods
2.1. ABE Fermentation Model Description
- (1)
- The carbon source is the only limiting substrate;
- (2)
- There is no limitation of nitrogen and nutrients;
- (3)
- There is product inhibition;
- (4)
- Acetic acid and butyric acid are reduced to acetone and butanol, respectively;
- (5)
- Acetone and butanol are also produced directly from carbon substrates;
- (6)
- Ethanol is produced from carbon substrates only;
- (7)
- Fermentation is performed at 37 °C and a pH of 4.5 under anaerobic conditions;
- (8)
- All cells are considered metabolically active and viable.
2.2. Parameter Estimation
- : nth response of the model where n is a response index (n = 1, 2,…, NR);
- NR: Number of model responses; for the ABE fermentation model NR = 7;
- I, k: Experiment indexes (i, k = 1, 2,…, NE);
- NE: Number of experiments; for the ABE fermentation model NE = 27;
- : NRx1 vector of observed response values for experiment ;
- : NRxNE matrix of observed responses for all experiments;
- : Unknown NRx1 vector of correct response values for experiment ;
- : NIx1 vector of independent (input) variables of the model(NI is number of inputs);
- : Known vector for experiment ;
- : Unknown NPx1 vector of correct model parameters;
- ();
- : NPx1 vector of estimated model parameters;
- : NRx1 vector of estimated responses for experiment ;
- :NRxNE matrix of estimated responses for all experiments;
- : Weight matrix for responses of experiment i; this matrix is related to the inverse of the variance–covariance matrix of responses for experiment i;
- : Unknown fundamental variance of the problem;
- : NRxNR variance–covariance matrix of responses of experiment i;
- : NRxNP Jacobian matrix of model responses to parameters for experiment i.
- (1)
- The experiments are independent; i.e., data from experiment i are not influenced by other experiments;
- (2)
- Experimental responses are independent and each one obeys a normal distribution around the correct response value with the variance given by ;
- (3)
- The model is correct; i.e., with the correct parameter vector (), the model generates the correct response vector () for experiment i with inputs .
2.3. Statistical Analysis of Estimated Parameters
2.4. Analysis of Sensitivity to Input Variables
3. Results and Discussion
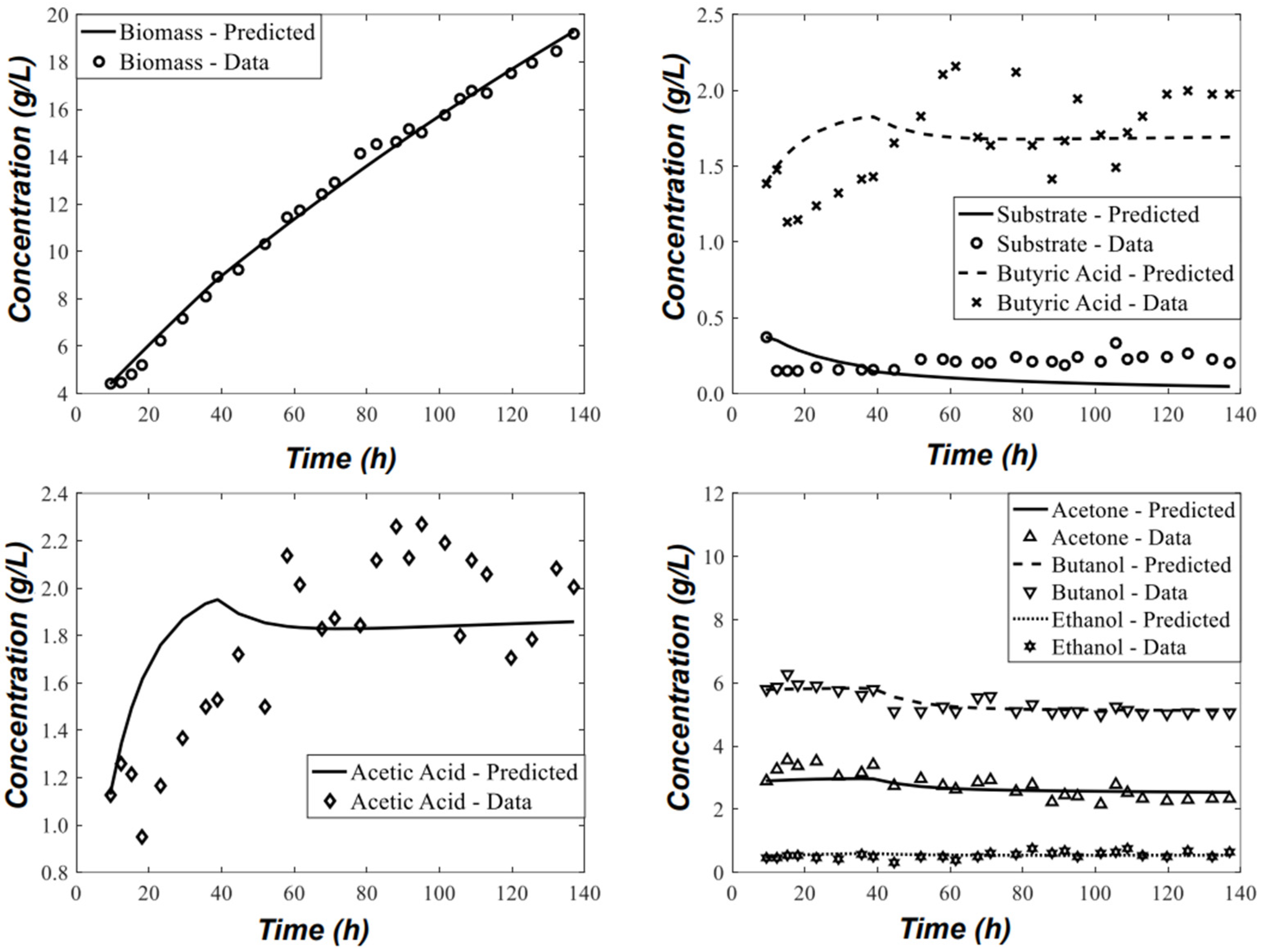
3.1. Parameter Estimation and Statistical Analysis
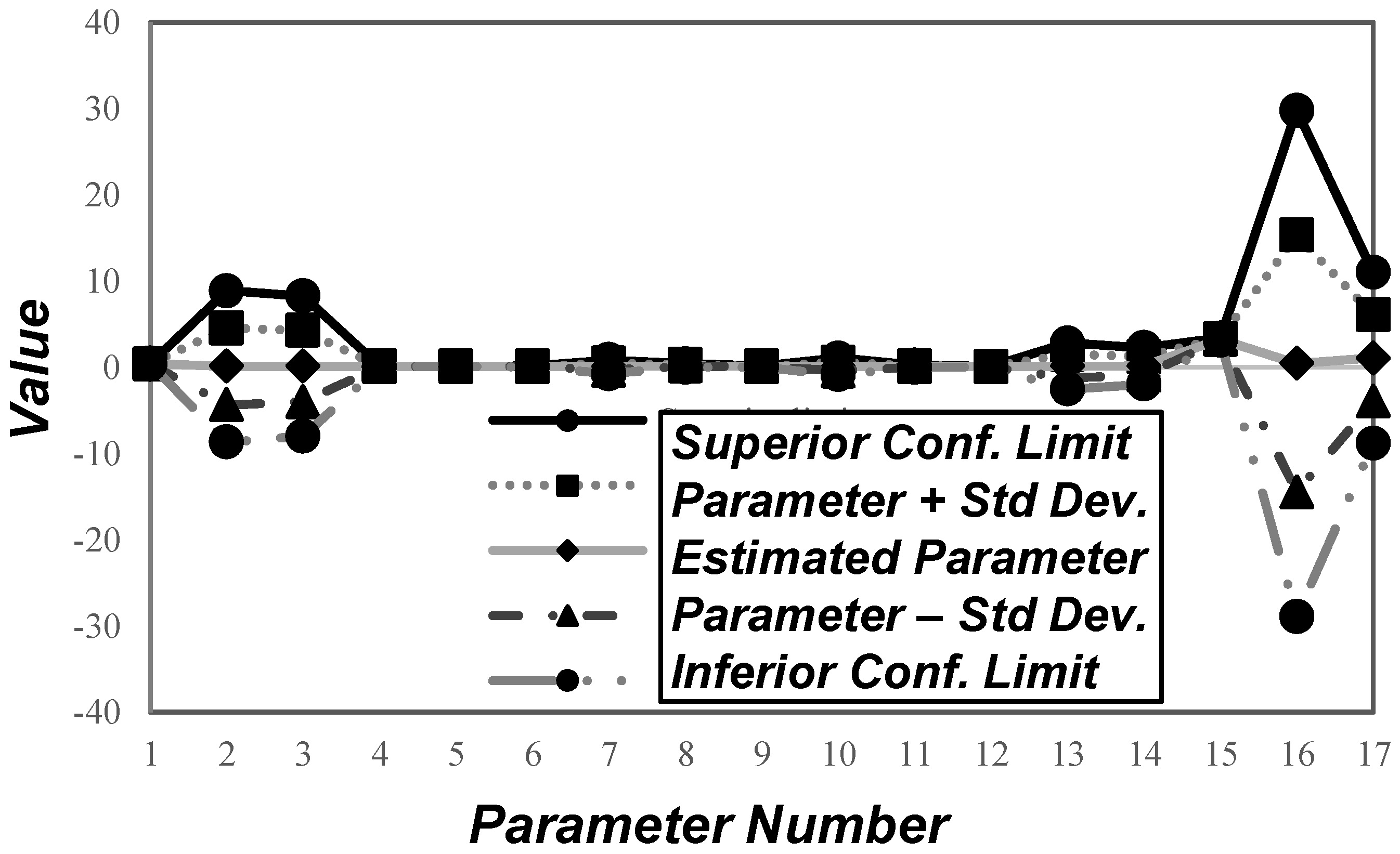
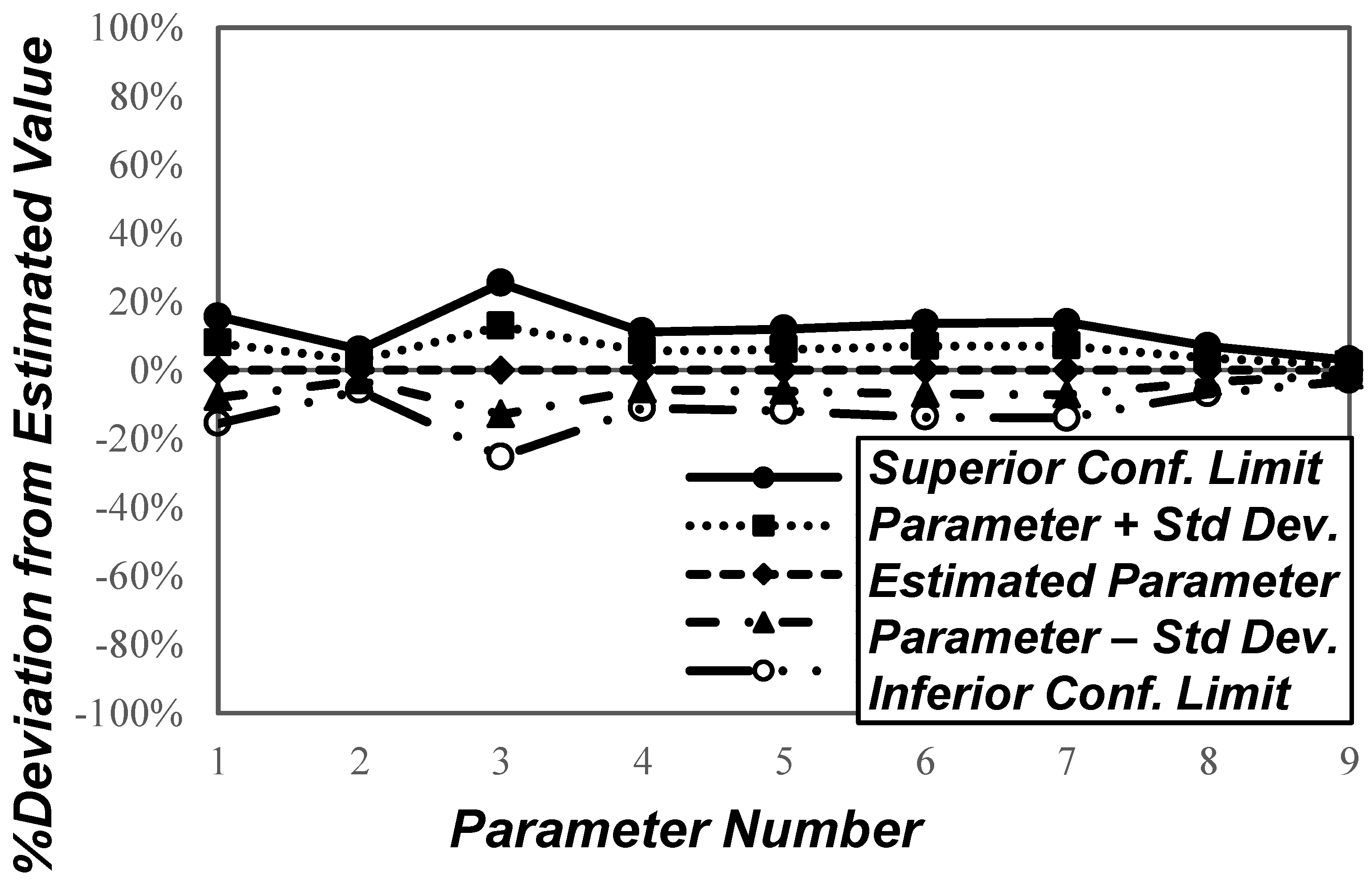
3.2. Input Variables Sensitivity Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABE | Acetone–Butanol–Ethanol |
| MV-Model | Mulchandani–Volesky |
| ABE | fermentation model |
| probability density function | |
| Nomenclature | |
| A | Acetone concentration (g/L) |
| AA | Acetic acid concentration (g/L) |
| B | Butanol concentration (g/L) |
| BA | Butyric acid concentration (g/L) |
| D | Dilution rate (h−1) |
| E | Ethanol concentration (g/L) |
| Substrate affinity constant (g/L) | |
| Cell maintenance coefficient (g/g.h) | |
| NE | Number of experiments |
| NP | Number of parameters |
| NR | Number of responses |
| S | Substrate concentration (g/L) |
| S0 | Substrate feeding concentration (g/L) |
| X | Biomass concentration (g/L) |
| Acetone yield coefficient (g/g) | |
| Acetic acid yield coefficient (g/g) | |
| Butanol yield coefficient (g/g) | |
| Butyric acid yield coefficient (g/g) | |
| Ethanol yield coefficient (g/g) | |
| Biomass yield coefficient (g/g) | |
| Maximum cellular growth rate (h−1) | |
References
- Ndaba, B.; Chiyanzu, I.; Marx, S. n-Butanol derived from biochemical and chemical routes: A review. Biotechnol. Rep. 2015, 8, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Weizmann, C. Production of Acetone and Alcohol by Bacteriological Processes. US Patent 1315585A, 9 September 1919. [Google Scholar]
- Mulchandani, A.; Volesky, B. Modelling of the acetone-butanol fermentation with cell retention. Can. J. Chem. Eng. 1986, 64, 625–631. [Google Scholar] [CrossRef]
- Tao, L.; He, X.; Tan, E.C.D.; Zhang, M.; Aden, A. Comparative Techno-Economic Analysis and Reviews of n-Butanol Production from Corn Grain and Corn Stover. Biofuels Bioprod. Biorefining 2014, 8, 342–361. [Google Scholar] [CrossRef]
- Yang, X.; Tsao, G.T. Mathematical modeling of inhibition kinetics in acetone-butanol fermentation by Clostridium acetobutylicum. Biotechnol. Prog. 1994, 10, 532–538. [Google Scholar] [CrossRef]
- Rochón, E.; Ferrari, M.D.; Lareo, C. Integrated ABE fermentation-gas stripping process for enhanced butanol production from sugarcane-sweet sorghum juices. Biomass Bioenergy 2017, 98, 153–160. [Google Scholar] [CrossRef]
- Jiang, Y.; Xu, C.; Dong, F.; Yang, Y.; Jiang, W.; Yang, S. Disruption of the acetoacetate decarboxylase gene in solvent-producing Clostridium acetobutylicum increases the butanol ratio. Metab. Eng. 2009, 11, 284–291. [Google Scholar] [CrossRef] [PubMed]
- Jiménez-Bonilla, P.; Zhang, J.; Wang, Y.; Blersch, D.; de-Bashan, L.-E.; Guo, L.; Wang, Y. Enhancing the tolerance of Clostridium saccharoperbutylacetonicum to lignocellulosic-biomass-derived inhibitors for efficient biobutanol production by overexpressing efflux pumps genes from Pseudomonas putida. Bioresour. Technol. 2020, 312, 123532. [Google Scholar] [CrossRef]
- Iyyappan, J.; Bharathiraja, B.; Varjani, S.; PraveenKumar, R.; Muthu Kumar, S. Anaerobic biobutanol production from black strap molasses using Clostridium acetobutylicum MTCC11274: Media engineering and kinetic analysis. Bioresour. Technol. 2021, 346, 126405. [Google Scholar] [CrossRef] [PubMed]
- López-Linares, J.C.; García-Cubero, M.T.; Coca, M.; Lucas, S. Efficient biobutanol production by acetone-butanol-ethanol fermentation from spent coffee grounds with microwave assisted dilute sulfuric acid pretreatment. Bioresour. Technol. 2021, 320, 124348. [Google Scholar] [CrossRef]
- Saadatinavaz, F.; Karimi, K.; Denayer, J.F.M. Hydrothermal pretreatment: An efficient process for improvement of biobutanol, biohydrogen, and biogas production from orange waste via a biorefinery approach. Bioresour. Technol. 2021, 341, 125834. [Google Scholar] [CrossRef]
- Setlhaku, M.; Heitmann, S.; Górak, A.; Wichmann, R. Investigation of gas stripping and pervaporation for improved feasibility of two-stage butanol production process. Bioresour. Technol. 2013, 136, 102–108. [Google Scholar] [CrossRef]
- Valles, A.; Álvarez-Hornos, J.; Capilla, M.; San-Valero, P.; Gabaldón, C. Fed-batch simultaneous saccharification and fermentation including in-situ recovery for enhanced butanol production from rice straw. Bioresour. Technol. 2021, 342, 126020. [Google Scholar] [CrossRef]
- Mascal, M. Chemicals from biobutanol: Technologies and markets. Biofuels Bioprod. Biorefining 2012, 6, 483–493. [Google Scholar] [CrossRef]
- Da Silva Trindade, W.R.; dos Santos, R.G. Review on the characteristics of butanol, its production and use as fuel in internal combustion engines. Renew. Sustain. Energy Rev. 2017, 69, 642–651. [Google Scholar] [CrossRef]
- Bharathiraja, B.; Jayamuthunagai, J.; Sudharsanaa, T.; Bharghavi, A.; Praveenkumar, R.; Chakravarthy, M.; Yuvaraj, D. Biobutanol—An impending biofuel for future: A review on upstream and downstream processing tecniques. Renew. Sustain. Energy Rev. 2017, 68, 788–807. [Google Scholar] [CrossRef]
- Papoutsakis, E.T. Equations and calculations for fermentations of butyric acid bacteria. Biotechnol. Bioeng. 1984, 26, 174–187. [Google Scholar] [CrossRef]
- Shinto, H.; Tashiro, Y.; Yamashita, M.; Kobayashi, G.; Sekiguchi, T.; Hanai, T.; Kuriya, Y.; Okamoto, M.; Sonomoto, K. Kinetic modeling and sensitivity analysis of acetone–butanol–ethanol production. J. Biotechnol. 2007, 131, 45–56. [Google Scholar] [CrossRef]
- Buehler, E.A.; Mesbah, A. Kinetic Study of Acetone-Butanol-Ethanol Fermentation in Continuous Culture. PLoS ONE 2016, 11, e0158243. [Google Scholar] [CrossRef]
- Lian, T.; Zhang, W.; Cao, Q.; Wang, S.; Yin, F.; Chen, Y.; Zhou, T.; Dong, H. Optimization of lactate production from co-fermentation of swine manure with apple waste and dynamics of microbial communities. Bioresour. Technol. 2021, 336, 125307. [Google Scholar] [CrossRef]
- Martinez-Burgos, W.J.; Sydney, E.B.; de Paula, D.R.; Medeiros, A.B.P.; de Carvalho, J.C.; Molina, D.; Soccol, C.R. Hydrogen production by dark fermentation using a new low-cost culture medium composed of corn steep liquor and cassava processing water: Process optimization and scale-up. Bioresour. Technol. 2021, 320, 124370. [Google Scholar] [CrossRef]
- Mota, T.R.; Oliveira, D.M.; Simister, R.; Whitehead, C.; Lanot, A.; dos Santos, W.D.; Rezende, C.A.; McQueen-Mason, S.J.; Gomez, L.D. Design of experiments driven optimization of alkaline pretreatment and saccharification for sugarcane bagasse. Bioresour. Technol. 2021, 321, 124499. [Google Scholar] [CrossRef] [PubMed]
- Velázquez-Sánchez, H.I.; Aguilar-López, R. Multi-Objective Optimization of an ABE Fermentation System for Butanol Production as Biofuel. Int. J. Chem. React. Eng. 2019, 17, 20180214. [Google Scholar] [CrossRef]
- Eom, M.-H.; Kim, B.; Jang, H.; Lee, S.-H.; Kim, W.; Shin, Y.-A.; Lee, J.H. Dynamic Modeling of a Fermentation Process with Ex situ Butanol Recovery (ESBR) for Continuous Biobutanol Production. Energy Fuels 2015, 29, 7254–7265. [Google Scholar] [CrossRef]
- Gattermayr, F.; Herwig, C.; Leitner, V. Effect of changes in continuous carboxylate feeding on the specific production rate of butanol using Clostridium saccharoperbutylacetonicum. Bioresour. Technol. 2021, 332, 125057. [Google Scholar] [CrossRef] [PubMed]
- Petre, E.; Selişteanu, D.; Roman, M. Advanced nonlinear control strategies for a fermentation bioreactor used for ethanol production. Bioresour. Technol. 2021, 328, 124836. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, N.; d’Ippolito, G.; Dipasquale, L.; Esposito, G.; Panico, A.; Lens, P.N.L.; Fontana, A. Kinetic modeling of hydrogen and L-lactic acid production by Thermotoga neapolitana via capnophilic lactic fermentation of starch. Bioresour. Technol. 2021, 332, 125127. [Google Scholar] [CrossRef] [PubMed]
- Monod, J. The Growth of Bacterial Cultures. Annu. Rev. Microbiol. 1949, 3, 371–394. [Google Scholar] [CrossRef] [Green Version]
- Pirt, S.J. Principles of Microbe and Cell Cultivation; John Wiley & Sons: New York, NY, USA, 1975. [Google Scholar]
- De Medeiros, J.L.; Barbosa, L.C.; Araújo, O.Q.F. Equilibrium Approach for CO2 and H2S Absorption with Aqueous Solutions of Alkanolamines: Theory and Parameter Estimation. Ind. Eng. Chem. Res. 2013, 52, 9203–9226. [Google Scholar] [CrossRef]

| Code | Type of Disturbance | Substrate Feed Concentration (g/L) | Feed Flowrate (m3/h) |
|---|---|---|---|
| 0 | Undisturbed Base-Case | 35 | 35.6 |
| +1/0 | +43% Feed Concentration Disturbance | 50 | 35.6 |
| −1/0 | −43% Feed Concentration Disturbance | 20 | 35.6 |
| 0/+1 | +85% Feed Flowrate Disturbance | 35 | 66 |
| 0/−1 | −89% Feed Flowrate Disturbance | 35 | 4 |
| Parameter Number | Parameter | Estimated Value | Unit | Confidence Interval (95% Probability) |
|---|---|---|---|---|
| 1 | 0.3488 | h−1 | ||
| 2 | 0.0588 | gsubst/gcel.h | ||
| 3 | 0.0972 | gsubst/gcel.h | ||
| 4 | 0.0564 | gcel/gsubst | ||
| 5 | 0.0575 | gsubst/gcel.h | ||
| 6 | 0.0573 | gBA/gsubst | ||
| 7 | 0.0298 | gBA/gcel.h | ||
| 8 | 0.1613 | gB/gsubst | ||
| 9 | 0.0650 | gAA/gsubst | ||
| 10 | 0.0487 | gAA/gcel.h | ||
| 11 | 0.0728 | gA/gsubst | ||
| 12 | 0.0177 | gE/gsubst | ||
| 13 | 0.0967 | gB/gcel.h | ||
| 14 | 0.1143 | gA/gcel.h | ||
| 15 | 3.2054 | g/L | ||
| 16 | 0.3975 | g/L | ||
| 17 | 1.0405 | g/L |
| Parameter Number | Parameter | Test Value | Fisher Test Abscissa φ1−α | Result |
|---|---|---|---|---|
| 1 | 110.3981 | 3.8961 | Significant | |
| 2 | 1.743 × 10−4 | 3.8961 | Nonsignificant | |
| 3 | 5.576 × 10−4 | 3.8961 | Nonsignificant | |
| 4 | 10.1117 | 3.8961 | Significant | |
| 5 | 44.4632 | 3.8961 | Significant | |
| 6 | 4.3046 | 3.8961 | Significant | |
| 7 | 0.0054 | 3.8961 | Nonsignificant | |
| 8 | 2.4946 | 3.8961 | Probably Nonsignificant | |
| 9 | 4.5595 | 3.8961 | Significant | |
| 10 | 0.0096 | 3.8961 | Nonsignificant | |
| 11 | 1.6276 | 3.8961 | Probably Nonsignificant | |
| 12 | 17.2535 | 3.8961 | Significant | |
| 13 | 0.0051 | 3.8961 | Nonsignificant | |
| 14 | 0.0113 | 3.8961 | Nonsignificant | |
| 15 | 1931.0 | 3.8961 | Significant | |
| 16 | 7.153 × 10−4 | 3.8961 | Nonsignificant | |
| 17 | 0.0431 | 3.8961 | Nonsignificant |
| Parameter Number | Parameter | Estimated Value | Unit | Confidence Interval (95% Probability) |
|---|---|---|---|---|
| 1 | 0.3244 | h−1 | ||
| 2 | 0.0538 | gcel/gsubst | ||
| 3 | 0.0521 | gsubst/gcel.h | ||
| 4 | 0.0541 | gBA/gsubst | ||
| 5 | 0.1688 | gB/gsubst | ||
| 6 | 0.0582 | gAA/gsubst | ||
| 7 | 0.0854 | gA/gsubst | ||
| 8 | 0.0173 | gE/gsubst | ||
| 9 | 2.8923 | g/L |
| Parameter Number | Parameter | Test Value | Fisher Test Abscissa | Result |
|---|---|---|---|---|
| 1 | 160.776 | 3.8936 | Significant | |
| 2 | 1119.0 | 3.8936 | Significant | |
| 3 | 60.746 | 3.8936 | Significant | |
| 4 | 313.904 | 3.8936 | Significant | |
| 5 | 271.948 | 3.8936 | Significant | |
| 6 | 207.651 | 3.8936 | Significant | |
| 7 | 198.535 | 3.8936 | Significant | |
| 8 | 805.819 | 3.8936 | Significant | |
| 9 | 4430.0 | 3.8936 | Significant |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Moura, F.R.; de Medeiros, J.L.; Araújo, O.d.Q.F. Acetone–Butanol–Ethanol Fermentation Phenomenological Models for Process Studies: Parameter Estimation and Multi-Response Model Reduction with Statistical Analysis. Processes 2022, 10, 1978. https://doi.org/10.3390/pr10101978
Moura FR, de Medeiros JL, Araújo OdQF. Acetone–Butanol–Ethanol Fermentation Phenomenological Models for Process Studies: Parameter Estimation and Multi-Response Model Reduction with Statistical Analysis. Processes. 2022; 10(10):1978. https://doi.org/10.3390/pr10101978
Chicago/Turabian StyleMoura, Felipe Ramalho, José Luiz de Medeiros, and Ofélia de Queiroz F. Araújo. 2022. "Acetone–Butanol–Ethanol Fermentation Phenomenological Models for Process Studies: Parameter Estimation and Multi-Response Model Reduction with Statistical Analysis" Processes 10, no. 10: 1978. https://doi.org/10.3390/pr10101978
APA StyleMoura, F. R., de Medeiros, J. L., & Araújo, O. d. Q. F. (2022). Acetone–Butanol–Ethanol Fermentation Phenomenological Models for Process Studies: Parameter Estimation and Multi-Response Model Reduction with Statistical Analysis. Processes, 10(10), 1978. https://doi.org/10.3390/pr10101978








