Segmentation and Identification of Vertebrae in CT Scans Using CNN, k-Means Clustering and k-NN
Abstract
1. Introduction
2. Related Work
3. Materials
4. Methods
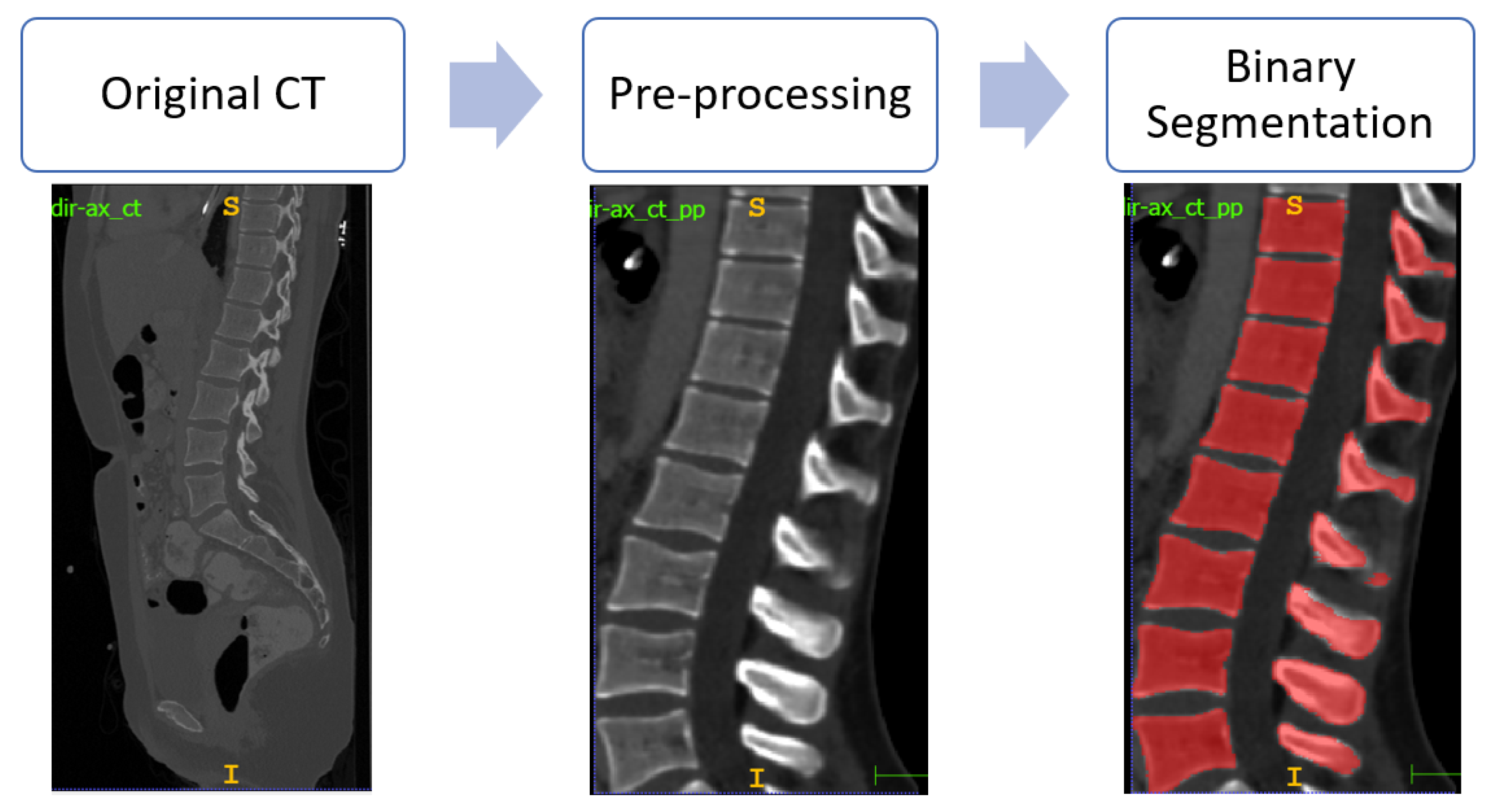
4.1. Spine Segmentation
4.2. Vertebrae Identification
- Vertebrae Number Selection. This step requires an input by the user, which has to insert the number of the vertebrae given the binary segmentation. The user has to provide the name of the first vertebra (from the top to the bottom) in order to perform the correct labeling according to the legends provided by VerSe.
- Slice Extraction. The algorithm extracts a 2D sagittal slice from the binary segmentation, starting from the middle of the image, since it has a higher probability of showing well-clustered vertebrae. Kindly note that, in some cases, e.g., patients affected by severe scoliosis, this consideration may not hold, resulting in lower segmentation performances. Inside the selected slice, the following sub-steps have been carried out:
- –
- Morphological and Connected Components Analysis. The morphological analysis aims at removing small points which can be wrongly considered as standalone components, whereas the purpose of the connected components analysis is labeling each component with a different value.
- –
- Shape Descriptor and Clustering for Arches and Bodies. It is worth noting that every single component is either a vertebral body or a vertebral arch, so it is important to correctly assign each component to the appropriate category. This stage carries out the above process by considering proper shape descriptors of the individual components.
- –
- Arch/Body Coupling. This step connects each vertebral arch to the nearest vertebral body, by assigning the same label to both.
- –
- Centroids’ Computation and Slice Showing. If the output vertebrae number matches with the input number from the first step, the algorithm goes further with the computation of centroids’ positions for each vertebra; otherwise, the process has to be repeated from another slice.
- Best Slice Selection and Centroids’ Storage. The algorithm repeats the workflow until it reaches a slice without connected components. Then, the user chooses the best slice among the showed ones, and the algorithm stores the centroids’ position.
- 3D Multi-class Segmentation. Centroids are used in a k-nearest neighbors (k-NN) classifier to produce a 3D segmentation map in which each vertebra has its own label.
4.2.1. Morphological and Connected Components Analysis
4.2.2. Shape Descriptor and Clustering
- Area: The number of pixels of the region.
- Centroid: The centroid’s position of the region.
- Extent: Ratio of pixels in the region to pixels in the total bounding box of the region.
- Perimeter: Approximation of the perimeter of the region.
- Eccentricity: Ratio of the focal distance (distance between focal points) over the major axis length.
- Solidity: Ratio of pixels in the region to pixels of the convex hull image (the smallest convex polygon that encloses the region).
4.2.3. Arch/Body Coupling
4.2.4. Multi-Class Segmentation
- The learning phase, which is not mandatory, results in the partitioning of the hyperspace in clusters based on samples’ positions.
- The distance computation phase consists of the computation of all the distances between samples and centroids (the most used distance metric is the Euclidean Distance, as we did in this work, but it is also possible to use Manhattan Distance or other distances).
- The classification phase assigns each sample to the class of the nearest cluster’s centroid.
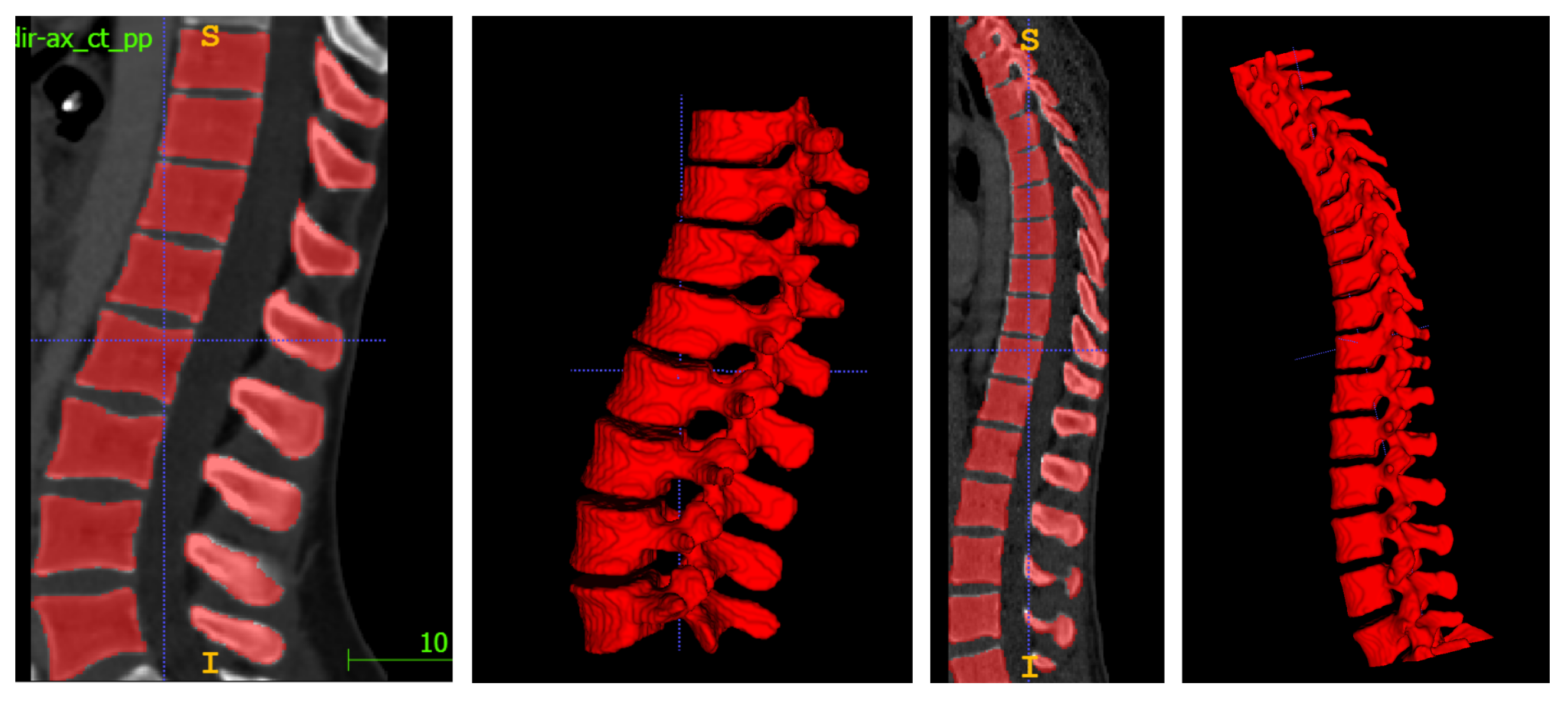
4.3. Visualization Tool
5. Results
5.1. Quality Measures
- measures based on volumetric overlap, such as Dice coefficient (), and . Such metrics allow to compute a similarity degree between the prediction and the ground truth;
- measures based on the concept of surface distance, such as maximum symmetric surface distance () and average symmetric surface distance ().
5.2. Experimental Results
6. Conclusions and Future Works
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sekuboyina, A.; Bayat, A.; Husseini, M.E.; Löffler, M.; Li, H.; Tetteh, G.; Kukačka, J.; Payer, C.; Štern, D.; Urschler, M.; et al. VerSe: A Vertebrae Labelling and Segmentation Benchmark for Multi-detector CT Images. arXiv 2020, arXiv:2001.09193. [Google Scholar]
- Williams, A.L.; Al-Busaidi, A.; Sparrow, P.J.; Adams, J.E.; Whitehouse, R.W. Under-reporting of osteoporotic vertebral fractures on computed tomography. Eur. J. Radiol. 2009, 69, 179–183. [Google Scholar] [CrossRef] [PubMed]
- Vania, M.; Mureja, D.; Lee, D. Automatic spine segmentation from CT images using Convolutional Neural Network via redundant generation of class labels. J. Comput. Des. Eng. 2019, 6, 224–232. [Google Scholar] [CrossRef]
- Korez, R.; Ibragimov, B.; Likar, B.; Pernuš, F.; Vrtovec, T. A Framework for Automated Spine and Vertebrae Interpolation-Based Detection and Model-Based Segmentation. IEEE Trans. Med. Imaging 2015, 34, 1649–1662. [Google Scholar] [CrossRef]
- Yao, J.; Burns, J.E.; Forsberg, D.; Seitel, A.; Rasoulian, A.; Abolmaesumi, P.; Hammernik, K.; Urschler, M.; Ibragimov, B.; Korez, R.; et al. A multi-center milestone study of clinical vertebral CT segmentation. Comput. Med. Imaging Graph. 2016, 49, 16–28. [Google Scholar] [CrossRef]
- Löffler, M.T.; Sekuboyina, A.; Jacob, A.; Grau, A.L.; Scharr, A.; El Husseini, M.; Kallweit, M.; Zimmer, C.; Baum, T.; Kirschke, J.S. A Vertebral Segmentation Dataset with Fracture Grading. Radiol. Artif. Intell. 2020, 2, e190138. [Google Scholar] [CrossRef] [PubMed]
- Sekuboyina, A.; Rempfler, M.; Valentinitsch, A.; Menze, B.H.; Kirschke, J.S. Labeling Vertebrae with Two-dimensional Reformations of Multidetector CT Images: An Adversarial Approach for Incorporating Prior Knowledge of Spine Anatomy. Radiol. Artif. Intell. 2020, 2, e190074. [Google Scholar] [CrossRef]
- Yao, J.; Burns, J.E.; Munoz, H.; Summers, R.M. Detection of Vertebral Body Fractures Based on Cortical Shell Unwrapping; Springer: Berlin/Heidelberg, Germany, 2012; pp. 509–516. [Google Scholar]
- Lessmann, N.; van Ginneken, B.; de Jong, P.A.; Išgum, I. Iterative fully convolutional neural networks for automatic vertebra segmentation and identification. Med. Image Anal. 2019, 53, 142–155. [Google Scholar] [CrossRef]
- Kim, Y.J.; Ganbold, B.; Kim, K.G. Web-based spine segmentation using deep learning in computed tomography images. Healthc. Inform. Res. 2020, 26, 61–67. [Google Scholar] [CrossRef]
- Furqan Qadri, S.; Ai, D.; Hu, G.; Ahmad, M.; Huang, Y.; Wang, Y.; Yang, J. Automatic Deep Feature Learning via Patch-Based Deep Belief Network for Vertebrae Segmentation in CT Images. Appl. Sci. 2019, 9, 69. [Google Scholar] [CrossRef]
- Zareie, M.; Parsaei, H.; Amiri, S.; Awan, M.S.; Ghofrani, M. Automatic segmentation of vertebrae in 3D CT images using adaptive fast 3D pulse coupled neural networks. Australas. Phys. Eng. Sci. Med. 2018, 41, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. Lect. Notes Comput. Sci. 2015, 9351, 234–241. [Google Scholar] [CrossRef]
- Hinton, G.E.; Osindero, S.; Teh, Y.W. A fast learning algorithm for deep belief nets. Neural Comput. 2006, 18, 1527–1554. [Google Scholar] [CrossRef] [PubMed]
- Chang, Q.; Shi, J.; Xiao, Z. A New 3D Segmentation Algorithm Based on 3D PCNN for Lung CT Slices. In Proceedings of the 2009 2nd International Conference on Biomedical Engineering and Informatics, Tianjin, China, 17–19 October 2009; pp. 1–5. [Google Scholar] [CrossRef]
- Bae, H.J.; Hyun, H.; Byeon, Y.; Shin, K.; Cho, Y.; Song, Y.J.; Yi, S.; Kuh, S.U.; Yeom, J.S.; Kim, N. Fully automated 3D segmentation and separation of multiple cervical vertebrae in CT images using a 2D convolutional neural network. Comput. Methods Programs Biomed. 2020, 184. [Google Scholar] [CrossRef] [PubMed]
- Payer, C.; Štern, D.; Bischof, H.; Urschler, M. Coarse to fine vertebrae localization and segmentation with spatialconfiguration-Net and U-Net. VISIGRAPP 2020, 5, 124–133. [Google Scholar] [CrossRef]
- Sekuboyina, A.; Rempfler, M.; Kukačka, J.; Tetteh, G.; Valentinitsch, A.; Kirschke, J.S.; Menze, B.H. Btrfly Net: Vertebrae Labelling with Energy-Based Adversarial Learning of Local Spine Prior. Lect. Notes Comput. Sci. 2018, 11073 LNCS, 649–657. [Google Scholar] [CrossRef]
- Glocker, B.; Zikic, D.; Konukoglu, E.; Haynor, D.R.; Criminisi, A. Vertebrae Localization in Pathological Spine CT via Dense Classification from Sparse Annotations; Springer: Berlin/Heidelberg, Germany, 2013; pp. 262–270. [Google Scholar]
- Glocker, B.; Feulner, J.; Criminisi, A.; Haynor, D.R.; Konukoglu, E. Automatic Localization and Identification of Vertebrae in Arbitrary Field-of-View CT Scans; Springer: Berlin/Heidelberg, Germany, 2012; pp. 590–598. [Google Scholar]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully Convolutional Networks for Semantic Segmentation. arXiv 2014, arXiv:1411.4038. [Google Scholar]
- Çiçek, O.; Abdulkadir, A.; Lienkamp, S.S.; Brox, T.; Ronneberger, O. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation. Lect. Notes Comput. Sci. 2016, 9901 LNCS, 424–432. [Google Scholar] [CrossRef]
- Milletari, F.; Navab, N.; Ahmadi, S.A.A. V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. In Proceedings of the 2016 Fourth International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 565–571. [Google Scholar] [CrossRef]
- Liu, L.; Cheng, J.; Quan, Q.; Wu, F.X.; Wang, Y.P.; Wang, J. A survey on U-shaped networks in medical image segmentations. Neurocomputing 2020, 409, 244–258. [Google Scholar] [CrossRef]
- Lateef, F.; Ruichek, Y. Survey on semantic segmentation using deep learning techniques. Neurocomputing 2019, 338, 321–348. [Google Scholar] [CrossRef]
- Altini, N.; Cascarano, G.D.; Brunetti, A.; Marino, F.; Rocchetti, M.T.; Matino, S.; Venere, U.; Rossini, M.; Pesce, F.; Gesualdo, L.; et al. Semantic Segmentation Framework for Glomeruli Detection and Classification in Kidney Histological Sections. Electronics 2020, 9, 503. [Google Scholar] [CrossRef]
- Altini, N.; Cascarano, G.D.; Brunetti, A.; De Feudis, D.I.; Buongiorno, D.; Rossini, M.; Pesce, F.; Gesualdo, L.; Bevilacqua, V. A Deep Learning Instance Segmentation Approach for Global Glomerulosclerosis Assessment in Donor Kidney Biopsies. Electronics 2020, 9, 1768. [Google Scholar] [CrossRef]
- Bevilacqua, V.; Brunetti, A.; Cascarano, G.D.; Guerriero, A.; Pesce, F.; Moschetta, M.; Gesualdo, L. A comparison between two semantic deep learning frameworks for the autosomal dominant polycystic kidney disease segmentation based on magnetic resonance images. BMC Med. Inform. Decis. Mak. 2019, 19, 244. [Google Scholar] [CrossRef]
- Altini, N.; Prencipe, B.; Brunetti, A.; Brunetti, G.; Triggiani, V.; Carnimeo, L.; Marino, F.; Guerriero, A.; Villani, L.; Scardapane, A.; et al. A Tversky Loss-Based Convolutional Neural Network for Liver Vessels Segmentation; Springer: Cham, Switzerland, 2020; Volume 12463 LNCS. [Google Scholar] [CrossRef]
- Adams, J.E.; Mughal, Z.; Damilakis, J.; Offiah, A.C. Radiology. Biol. Dis. 2012. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Delving deep into rectifiers: Surpassing human-level performance on imagenet classification. In Proceedings of the 2015 IEEE International Conference on Computer Vision (ICCV), Santiago, Chile, 7–13 December 2015; pp. 1026–1034. [Google Scholar] [CrossRef]
- Shen, C.; Milletari, F.; Roth, H.R.; Oda, H.; Oda, M.; Hayashi, Y.; Misawa, K.; Mori, K. Improving V-Nets for multi-class abdominal organ segmentation. In Medical Imaging 2019: Image Processing; Angelini, E.D., Landman, B.A., Eds.; International Society for Optics and Photonics, SPIE: Bellingham, DC, USA, 2019; Volume 10949, pp. 76–82. [Google Scholar] [CrossRef]
- Pérez-García, F.; Sparks, R.; Ourselin, S. TorchIO: A Python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning. arXiv 2020, arXiv:2003.04696. [Google Scholar]
- Sonka, M.; Hlavac, V.; Boyle, R. Image Processing, Analysis, and Machine Vision; Cengage Learning: Boston, MA, USA, 2014. [Google Scholar]
- Shakhnarovich, G.; Darrell, T.; Indyk, P. Nearest-neighbor methods in learning and vision. In Neural Information Processing Series; MIT Press: Cambrdige, MA, USA, 2005. [Google Scholar]
- McCormick, M.; Liu, X.; Ibanez, L.; Jomier, J.; Marion, C. ITK: Enabling reproducible research and open science. Front. Neuroinform. 2014, 8, 13. [Google Scholar] [CrossRef]
- Schroeder, W.; Martin, K.L.B. The Visualization Toolkit, 4th ed.; Kitware: Clifton Park, NY, USA, 2006. [Google Scholar]
- Bradski, G. The OpenCV Library. Dr. Dobb’s J. Softw. Tools 2000, 25, 120–125. [Google Scholar]
- Heimann, T.; van Ginneken, B.; Styner, M.M.A.; Arzhaeva, Y.; Aurich, V.; Bauer, C.; Beck, A.; Becker, C.; Beichel, R.; Bekes, G.; et al. Comparison and Evaluation of Methods for Liver Segmentation From CT Datasets. IEEE Trans. Med. Imaging 2009, 28, 1251–1265. [Google Scholar] [CrossRef]
- Bilic, P.; Christ, P.F.; Vorontsov, E.; Chlebus, G.; Chen, H.; Dou, Q.; Fu, C.W.; Han, X.; Heng, P.A.; Hesser, J.; et al. The Liver Tumor Segmentation Benchmark (LiTS). arXiv 2019, arXiv:1901.04056. [Google Scholar]
- Prencipe, B.; Altini, N.; Cascarano, G.D.; Guerriero, A.; Brunetti, A. A Novel Approach Based on Region Growing Algorithm for Liver and Spleen Segmentation from CT Scans. In Intelligent Computing Theories and Application; Huang, D.S., Bevilacqua, V., Hussain, A., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 398–410. [Google Scholar]








| Reference | Method | Test Sample | [%] |
|---|---|---|---|
| Proposed | 3D V-Net | 50 CT scans | |
| Kim et al. [10] | U-Net | 14 CT scans | |
| Vania et al. [3] | CNN | 32 CT scans | |
| Qadri et al. [11] | PaDBN | 3 CT scans | |
| Lessmann et al. [9] | 3D U-Net | 25 CT scans | |
| Zareie et al. [12] | PCNN | 17 CT scans | |
| APCNN | 17 CT scans | ||
| MLPNN | 17 CT scans | ||
| MLPNN1F | 17 CT scans | ||
| APCNN (noise 3%) | 17 CT scans | ||
| MLPNN (noise 3%) | 17 CT scans |
| Dataset | Spine Tract | Sample Size | Modality | Annotations |
|---|---|---|---|---|
| xVertSeg [4] | Lumbar | n = 25 | CT scans | C |
| CSI-Seg 2014 [5,8] | Thoraco-lumbar | n = 20 | CT scans | M |
| CSI-Label 2014 [19,20] | Whole spine | n = 302 | CT scans | C |
| Verse’19 [1,6,7] | Whole spine | n = 160 | CT scans | C + M |
| Verse’20 [1,6,7] | Whole spine | n = 300 | CT scans | C + M |
| Medica Sud s.r.l. | Whole spine | n = 12 | CT scans | S |
| Epochs | [%] | [%] | [%] | [mm] | [mm] |
|---|---|---|---|---|---|
| 100 | 85.07 ± 3.02 | 94.25 ± 3.79 | 77.65 ± 3.78 | 2.46 ± 0.81 | 63.16 ± 23.27 |
| 200 | 88.20 ± 2.66 | 93.46 ± 4.03 | 83.61 ± 2.65 | 1.89 ± 0.61 | 60.87 ± 23.46 |
| 300 | 88.44 ± 2.69 | 93.85 ± 4.49 | 83.78 ± 2.81 | 1.91 ± 0.56 | 64.03 ± 28.96 |
| 400 | 88.34 ± 2.35 | 94.51 ± 3.31 | 83.02 ± 2.81 | 1.85 ± 0.63 | 62.77 ± 27.67 |
| 500 | 89.17 ± 3.63 | 93.60 ± 6.27 | 85.43 ± 2.75 | 1.43 ± 0.63 | 56.69 ± 18.07 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Altini, N.; De Giosa, G.; Fragasso, N.; Coscia, C.; Sibilano, E.; Prencipe, B.; Hussain, S.M.; Brunetti, A.; Buongiorno, D.; Guerriero, A.; et al. Segmentation and Identification of Vertebrae in CT Scans Using CNN, k-Means Clustering and k-NN. Informatics 2021, 8, 40. https://doi.org/10.3390/informatics8020040
Altini N, De Giosa G, Fragasso N, Coscia C, Sibilano E, Prencipe B, Hussain SM, Brunetti A, Buongiorno D, Guerriero A, et al. Segmentation and Identification of Vertebrae in CT Scans Using CNN, k-Means Clustering and k-NN. Informatics. 2021; 8(2):40. https://doi.org/10.3390/informatics8020040
Chicago/Turabian StyleAltini, Nicola, Giuseppe De Giosa, Nicola Fragasso, Claudia Coscia, Elena Sibilano, Berardino Prencipe, Sardar Mehboob Hussain, Antonio Brunetti, Domenico Buongiorno, Andrea Guerriero, and et al. 2021. "Segmentation and Identification of Vertebrae in CT Scans Using CNN, k-Means Clustering and k-NN" Informatics 8, no. 2: 40. https://doi.org/10.3390/informatics8020040
APA StyleAltini, N., De Giosa, G., Fragasso, N., Coscia, C., Sibilano, E., Prencipe, B., Hussain, S. M., Brunetti, A., Buongiorno, D., Guerriero, A., Tatò, I. S., Brunetti, G., Triggiani, V., & Bevilacqua, V. (2021). Segmentation and Identification of Vertebrae in CT Scans Using CNN, k-Means Clustering and k-NN. Informatics, 8(2), 40. https://doi.org/10.3390/informatics8020040










