Etiology and Epidemiology of Croup before and throughout the COVID-19 Pandemic, 2018–2022, South Korea
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Exclusion Criteria
2.3. Sample Collection and Processing
2.4. Respiratory Virus Identification
2.5. Study Period Division
2.6. Statistical Analyses
2.7. Ethical Consideration
3. Results
3.1. Case Numbers and Demographics
3.2. Viral Pathogens
3.3. Multiple Respiratory Virus Detections
3.4. Viral Identification Rate and Number of Tests Performed
3.5. Epidemiological Association
4. Discussion
Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef] [PubMed]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Twohig, K.A.; Nyberg, T.; Zaidi, A.; Thelwall, S.; Sinnathamby, M.A.; Aliabadi, S.; Seaman, S.R.; Harris, R.J.; Hope, R.; Lopez-Bernal, J.; et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: A cohort study. Lancet Infect. Dis. 2022, 22, 35–42. [Google Scholar] [CrossRef]
- Forrest, C.B.; Burrows, E.K.; Mejias, A.; Razzaghi, H.; Christakis, D.; Jhaveri, R.; Lee, G.M.; Pajor, N.M.; Rao, S.; Thacker, D.; et al. Severity of Acute COVID-19 in Children <18 Years Old March 2020 to December 2021. Pediatrics 2022, 149, e2021055765. [Google Scholar] [CrossRef] [PubMed]
- Karron, R.A.; Hetrich, M.K.; Na, Y.B.; Knoll, M.D.; Schappell, E.; Meece, J.; Hanson, E.; Tong, S.; Lee, J.S.; Veguilla, V.; et al. Assessment of Clinical and Virological Characteristics of SARS-CoV-2 Infection Among Children Aged 0 to 4 Years and Their Household Members. JAMA Netw. Open 2022, 5, e2227348. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, P.; Curtis, N. Why Does the Severity of COVID-19 Differ with Age?: Understanding the Mechanisms Underlying the Age Gradient in Outcome Following SARS-CoV-2 Infection. Pediatr. Infect. Dis. J. 2022, 41, e36–e45. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Abdullah, F.; Myers, J.; Basu, D.; Tintinger, G.; Ueckermann, V.; Mathebula, M.; Ramlall, R.; Spoor, S.; de Villiers, T.; Van der Walt, Z.; et al. Decreased severity of disease during the first global omicron variant covid-19 outbreak in a large hospital in tshwane, south africa. Int. J. Infect. Dis. 2022, 116, 38–42. [Google Scholar] [CrossRef]
- Hui, K.P.Y.; Ho, J.C.W.; Cheung, M.C.; Ng, K.C.; Ching, R.H.H.; Lai, K.L.; Kam, T.T.; Gu, H.; Sit, K.Y.; Hsin, M.K.Y.; et al. SARS-CoV-2 Omicron variant replication in human bronchus and lung ex vivo. Nature 2022, 603, 715–720. [Google Scholar] [CrossRef]
- Adenaiye, O.O.; Lai, J.; Bueno de Mesquita, P.J.; Hong, F.; Youssefi, S.; German, J.; Tai, S.H.S.; Albert, B.; Schanz, M.; Weston, S.; et al. Infectious Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) in Exhaled Aerosols and Efficacy of Masks During Early Mild Infection. Clin. Infect. Dis. 2022, 75, e241–e248. [Google Scholar] [CrossRef]
- Martin, B.; DeWitt, P.E.; Russell, S.; Sanchez-Pinto, L.N.; Haendel, M.A.; Moffitt, R.; Bennett, T.D. Acute Upper Airway Disease in Children with the Omicron (B.1.1.529) Variant of SARS-CoV-2-A Report from the US National COVID Cohort Collaborative. JAMA Pediatr. 2022, 176, 819–821. [Google Scholar] [CrossRef] [PubMed]
- Hanna, J.; Brauer, P.R.; Morse, E.; Berson, E.; Mehra, S. Epidemiological analysis of croup in the emergency department using two national datasets. Int. J. Pediatr. Otorhinolaryngol. 2019, 126, 109641. [Google Scholar] [CrossRef] [PubMed]
- Thompson, M.; Vodicka, T.A.; Blair, P.S.; Buckley, D.I.; Heneghan, C.; Hay, A.D.; Team, T.P. Duration of symptoms of respiratory tract infections in children: Systematic review. BMJ 2013, 347, f7027. [Google Scholar] [CrossRef] [PubMed]
- Brewster, R.C.; Parsons, C.; Laird-Gion, J.; Hilker, S.; Irwin, M.; Sommerschield, A.; Michaelis, K.A.; Lam, M.; Parsons, A.; Mansbach, J.M. COVID-19-Associated Croup in Children. Pediatrics 2022, 149, e2022056492. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Tomari, K.; Matsuoka, T. Children with Croup and SARS-CoV-2 Infection During the Large Outbreak of Omicron. Pediatr. Infect. Dis. J. 2022, 41, e249. [Google Scholar] [CrossRef]
- Lefchak, B.; Nickel, A.; Lammers, S.; Watson, D.; Hester, G.Z.; Bergmann, K.R. Analysis of COVID-19-Related Croup and SARS-CoV-2 Variant Predominance in the US. JAMA Netw. Open 2022, 5, e2220060. [Google Scholar] [CrossRef]
- Sharma, S.; Agha, B.; Delgado, C.; Walson, K.; Woods, C.; Gonzalez, M.D.; Jerris, R.; Sysyn, G.; Beiter, J.; Kamidani, S.; et al. Croup Associated With SARS-CoV-2: Pediatric Laryngotracheitis During the Omicron Surge. J. Pediatr. Infect. Dis. Soc. 2022, 11, 371–374. [Google Scholar] [CrossRef]
- Counihan, M.E.; Shay, D.K.; Holman, R.C.; Lowther, S.A.; Anderson, L.J. Human parainfluenza virus-associated hospitalizations among children less than five years of age in the United States. Pediatr. Infect. Dis. J. 2001, 20, 646–653. [Google Scholar] [CrossRef]
- Van der Hoek, L.; Sure, K.; Ihorst, G.; Stang, A.; Pyrc, K.; Jebbink, M.F.; Petersen, G.; Forster, J.; Berkhout, B.; Uberla, K. Croup is associated with the novel coronavirus NL63. PLoS Med. 2005, 2, e240. [Google Scholar] [CrossRef]
- Sung, J.Y.; Lee, H.J.; Eun, B.W.; Kim, S.H.; Lee, S.Y.; Lee, J.Y.; Park, K.U.; Choi, E.H. Role of human coronavirus NL63 in hospitalized children with croup. Pediatr. Infect. Dis. J. 2010, 29, 822–826. [Google Scholar] [CrossRef]
- Xie, X.; Xue, Q.; Zhou, Y.; Zhu, K.; Liu, Q.; Zhang, J.; Song, R. Mental Health Status Among Children in Home Confinement During the Coronavirus Disease 2019 Outbreak in Hubei Province, China. JAMA Pediatr. 2020, 174, 898–900. [Google Scholar] [CrossRef] [PubMed]
- Tun, E.M.; Koid Jia Shin, C.; Usoro, E.; Thomas-Smith, S.E.; Trehan, I.; Migita, R.T.; Keilman, A.E. Croup during the Coronavirus Disease 2019 Omicron Variant Surge. J. Pediatr. 2022, 247, 147–149. [Google Scholar] [CrossRef] [PubMed]
- Madewell, Z.J.; Yang, Y.; Longini, I.M., Jr.; Halloran, M.E.; Dean, N.E. Household Secondary Attack Rates of SARS-CoV-2 by Variant and Vaccination Status: An Updated Systematic Review and Meta-analysis. JAMA Netw. Open 2022, 5, e229317. [Google Scholar] [CrossRef] [PubMed]
- Laws, R.L.; Chancey, R.J.; Rabold, E.M.; Chu, V.T.; Lewis, N.M.; Fajans, M.; Reses, H.E.; Duca, L.M.; Dawson, P.; Conners, E.E.; et al. Symptoms and Transmission of SARS-CoV-2 Among Children—Utah and Wisconsin, March-May 2020. Pediatrics 2021, 147, e2020027268. [Google Scholar] [CrossRef]
- Jorgensen, S.B.; Nygard, K.; Kacelnik, O.; Telle, K. Secondary Attack Rates for Omicron and Delta Variants of SARS-CoV-2 in Norwegian Households. JAMA 2022, 327, 1610–1611. [Google Scholar] [CrossRef]
- Van der Hoek, L.; Pyrc, K.; Jebbink, M.F.; Vermeulen-Oost, W.; Berkhout, R.J.; Wolthers, K.C.; Wertheim-van Dillen, P.M.; Kaandorp, J.; Spaargaren, J.; Berkhout, B. Identification of a new human coronavirus. Nat. Med. 2004, 10, 368–373. [Google Scholar] [CrossRef]
- Kuypers, J.; Martin, E.T.; Heugel, J.; Wright, N.; Morrow, R.; Englund, J.A. Clinical disease in children associated with newly described coronavirus subtypes. Pediatrics 2007, 119, e70–e76. [Google Scholar] [CrossRef]
- Chu, H.Y.; Englund, J.A.; Strelitz, B.; Lacombe, K.; Jones, C.; Follmer, K.; Martin, E.K.; Bradford, M.; Qin, X.; Kuypers, J.; et al. Rhinovirus Disease in Children Seeking Care in a Tertiary Pediatric Emergency Department. J. Pediatr. Infect. Dis. Soc. 2016, 5, 29–38. [Google Scholar] [CrossRef]
- Winther, B.; Gwaltney, J.M., Jr.; Mygind, N.; Hendley, J.O. Viral-induced rhinitis. Am. J. Rhinol. 1998, 12, 17–20. [Google Scholar] [CrossRef]
- Peltola, V.; Waris, M.; Osterback, R.; Susi, P.; Ruuskanen, O.; Hyypia, T. Rhinovirus transmission within families with children: Incidence of symptomatic and asymptomatic infections. J. Infect. Dis. 2008, 197, 382–389. [Google Scholar] [CrossRef]
- Cherry, J.D. Clinical practice. Croup. N. Engl. J. Med. 2008, 358, 384–391. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.M.; Kang, J.H. Effects of nasopharyngeal microbiota in respiratory infections and allergies. Clin. Exp. Pediatr. 2021, 64, 543–551. [Google Scholar] [CrossRef] [PubMed]
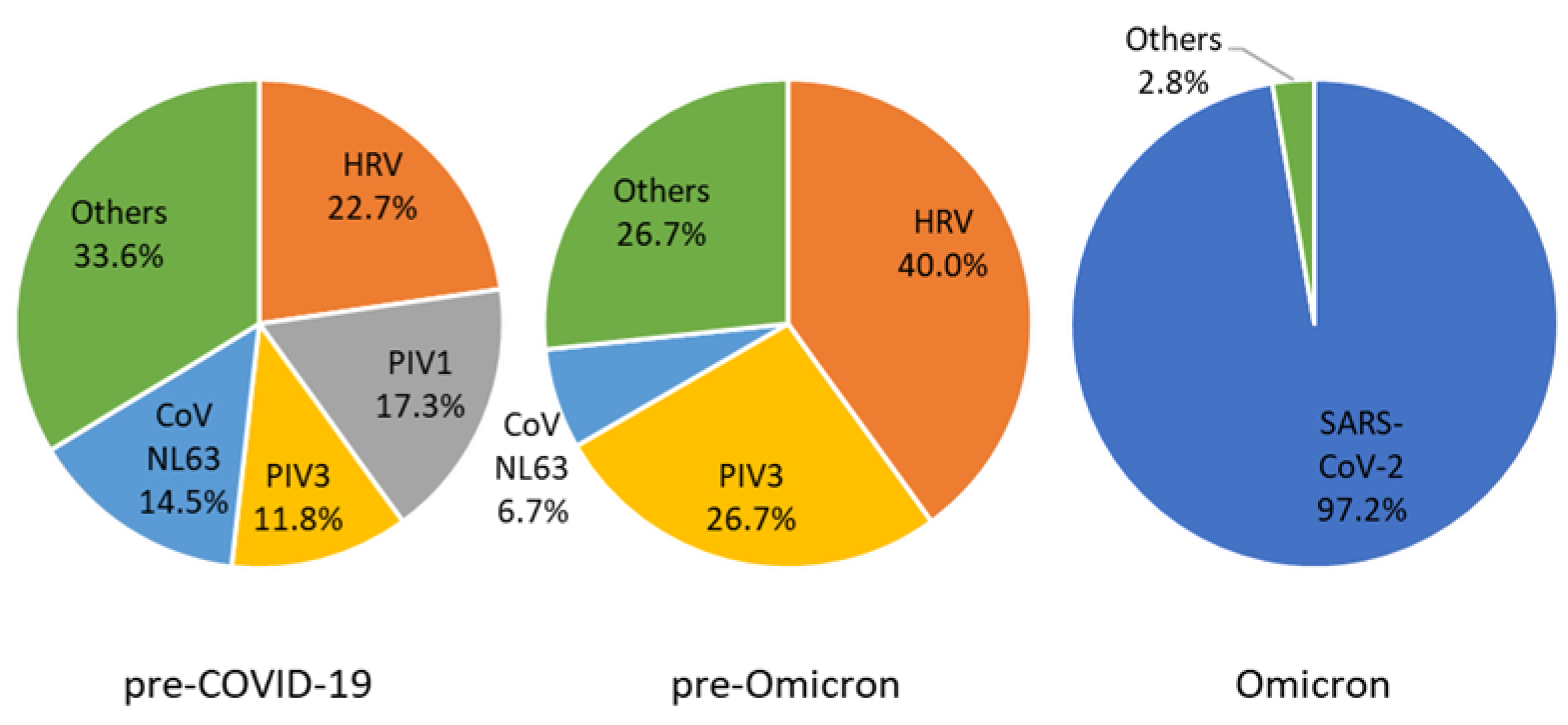

| Characteristic | Total | Pre-COVID-19 (Jan 2018–3rd Week 2020) | Pre-Omicron (4th Week 2020–3rd Week 2022) | Omicron (4th Week 2022–Mar 2022) |
|---|---|---|---|---|
| Total weeks | 220 | 107 | 104 | 9 |
| Case number | 879 | 670 | 145 | 64 |
| Cases/week | 3.99 | 6.26 | 1.39 | 7.11 |
| Sex (male) | 569 (64.7) | 433 (64.6) | 97 (66.9) | 39 (60.9) |
| Median age in month (range) | 22 (2–59) | 22 (2–59) | 24 (2–58) | 16.5 (3–54) |
| mRT-PCR-tested | 106 (12.1) | 94 (14.0) | 11 (7.6) | 1 (1.5) |
| SARS-CoV-2 PCR-tested | 112 (12.7) | NA | 58 (40.0) | 54 (84.4) |
| Viral identification | 130 (14.8) | 83 (12.4) | 11 (7.6) | 36 (56.3) |
| Codetection | 26 (20.0) | 23 (27.7) | 3 (27.3) | 0 (0) |
| Virus | Total | Pre-COVID-19 (Jan 2018–3rd Week 2020) | Pre-Omicron (4th Week 2020–3rd Week 2022) | Omicron (4th Week 2022–Mar 2022) |
|---|---|---|---|---|
| SARS-CoV-2 | 35 (26.9) | NA | 0 (0) | 35 (97.2) |
| HRV | 31 (23.8) | 25 (30.1) | 6 (54.5) | |
| PIV1 | 19 (14.6) | 19 (22.9) | ||
| PIV3 | 17 (13.1) | 13 (15.7) | 4 (36.4) | |
| CoV NL63 | 17 (13.1) | 16 (19.3) | 1 (9.1) | |
| AdV | 8 (6.2) | 8 (9.6) | ||
| HBoV | 8 (6.2) | 5 (6.0) | 3 (27.3) | |
| CoV OC43 | 5 (3.8) | 5 (6.0) | ||
| RSV A | 5 (3.8) | 5 (6.0) | ||
| RSV B | 5 (3.8) | 3 (3.6) | 1 (9.1) | 1 (2.8) |
| Flu B | 3 (2.3) | 3 (3.6) | ||
| PIV2 | 3 (2.3) | 3 (3.6) | ||
| MPV | 3 (2.3) | 3 (3.6) | ||
| Flu A | 1 (0.8) | 1 (1.2) | ||
| HEV | 1 (0.8) | 1 (1.2) | ||
| Total | 161 (100) | 110 (100) | 15 (100) | 36 (100) |
| Characteristic | Total | Dual Detection | Triple Detection |
|---|---|---|---|
| HRV | 17 (29.8) | 13 (31.0) | 4 (26.7) |
| PIV1 | 8 (14.0) | 7 (16.7) | 1 (6.7) |
| AdV | 7 (12.3) | 4 (9.5) | 3 (20.0) |
| CoV NL63 | 6 (10.5) | 5 (11.9) | 1 (6.7) |
| HBoV | 5 (8.8) | 2 (4.8) | 3 (20.0) |
| PIV3 | 4 (7.0) | 3 (7.1) | 1 (6.7) |
| RSV A | 3 (5.3) | 2 (4.8) | 1 (6.7) |
| MPV | 2 (3.5) | 2 (4.8) | |
| CoV OC43 | 2 (3.5) | 1 (2.4) | 1 (6.7) |
| Flu A | 1 (1.8) | 1 (2.4) | |
| HEV | 1 (1.8) | 1 (2.4) | |
| RSV B | 1 (1.8) | 1 (2.4) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, J.K.; Song, S.H.; Ahn, B.; Yun, K.W.; Choi, E.H. Etiology and Epidemiology of Croup before and throughout the COVID-19 Pandemic, 2018–2022, South Korea. Children 2022, 9, 1542. https://doi.org/10.3390/children9101542
Lee JK, Song SH, Ahn B, Yun KW, Choi EH. Etiology and Epidemiology of Croup before and throughout the COVID-19 Pandemic, 2018–2022, South Korea. Children. 2022; 9(10):1542. https://doi.org/10.3390/children9101542
Chicago/Turabian StyleLee, Joon Kee, Seung Ha Song, Bin Ahn, Ki Wook Yun, and Eun Hwa Choi. 2022. "Etiology and Epidemiology of Croup before and throughout the COVID-19 Pandemic, 2018–2022, South Korea" Children 9, no. 10: 1542. https://doi.org/10.3390/children9101542
APA StyleLee, J. K., Song, S. H., Ahn, B., Yun, K. W., & Choi, E. H. (2022). Etiology and Epidemiology of Croup before and throughout the COVID-19 Pandemic, 2018–2022, South Korea. Children, 9(10), 1542. https://doi.org/10.3390/children9101542






