MicroRNA Profiling as a Novel Tool in the Diagnostics of Late-Onset Neonatal Sepsis: A Scoping Review
Highlights
- Many studies show altered expression of miRNAs in septic neonates compared to controls.
- Existing studies are heterogenous, showing either up- or downregulation of specific miRNAs.
- MicroRNAs as novel biomarkers could play an important role in prompt and accurate diagnosis of late-onset sepsis (LOS), offering neonatologists a valuable tool.
- These findings require validation in larger cohorts and/or randomized control trials to determine their clinical utility.
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility Criteria
2.2. Exclusion Criteria
2.3. Selection Process
2.4. Generalizability and Transferability
3. Results
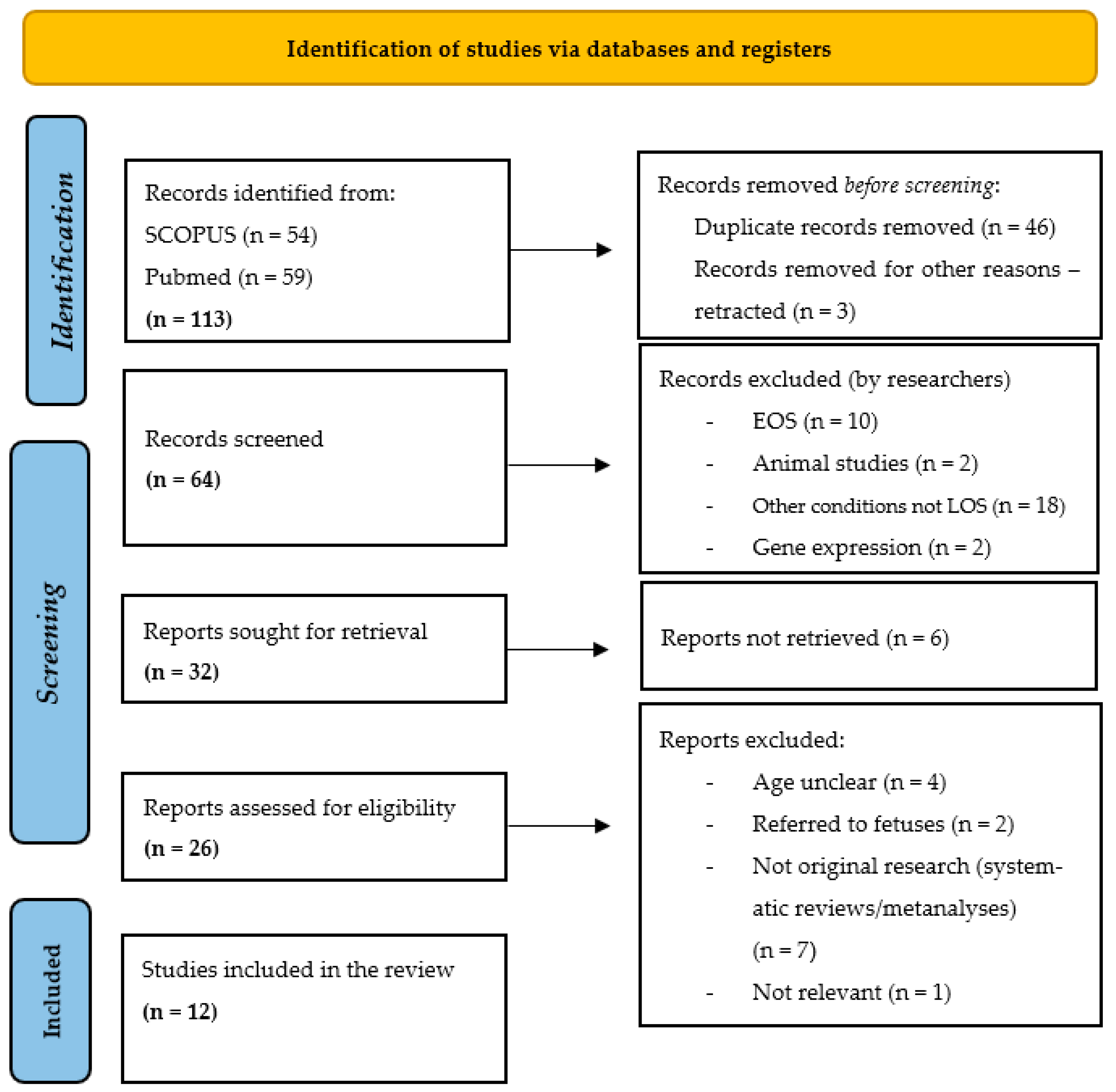
3.1. Study Selection
3.2. MicroRNA Profile
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| LOS | Late-onset sepsis |
| MiRNA | MicroRNA |
| PRISMA | Preferred Reporting Items for Systematic Reviews and Meta-Analyses |
| RISC | RNA-induced silencing complex (RISC) |
| CRP | C-reactive protein |
| PCT | Procalcitonin |
| IL6 | Interleukin 6 |
| IL8 | Interleukin 8 |
| RISC | RNA-induced silencing complex |
| RNA | Ribonucleic acid |
| PICO | Population, intervention, comparison, and outcome |
| SIRS | Systematic inflammatory response syndrome |
| EOS | Early-onset sepsis |
| ROC | Receiver operating characteristic |
| VLBW | Very low birth weight |
| SNAP II | Score for neonatal acute physiology II |
| AUC | Area under the curve |
| LPS | Lipopolysaccharide |
| PCR | Polymerase chain reaction |
| RT-qPCR | Quantitative reverse-transcription polymerase chain reaction |
References
- Liu, L.; Oza, S.; Hogan, D.; Chu, Y.; Perin, J.; Zhu, J.; Lawn, J.E.; Cousens, S.; Mathers, C.; Black, R.E. Global, Regional, and National Causes of under-5 Mortality in 2000–15: An Updated Systematic Analysis with Implications for the Sustainable Development Goals. Lancet 2016, 388, 3027–3035. [Google Scholar] [CrossRef] [PubMed]
- Coggins, S.A.; Glaser, K. Updates in Late-Onset Sepsis: Risk Assessment, Therapy, and Outcomes. Neoreviews 2022, 23, 738–755. [Google Scholar] [CrossRef] [PubMed]
- Stoll, B.J.; Hansen, N.; Fanaroff, A.A.; Wright, L.L.; Carlo, W.A.; Ehrenkranz, R.A.; Lemons, J.A.; Donovan, E.F.; Stark, A.R.; Tyson, J.E.; et al. Late-Onset Sepsis in Very Low Birth Weight Neonates: The Experience of the NICHD Neonatal Research Network. Pediatrics 2002, 110, 285–291. [Google Scholar] [CrossRef]
- Gkentzi, D.; Dimitriou, G. Antimicrobial Stewardship in the Neonatal Intensive Care Unit: An Update. Curr. Pediatr. Rev. 2019, 15, 47–52. [Google Scholar] [CrossRef]
- Kellogg, J.A.; Ferrentino, F.L.; Goodstein, M.H.; Liss, J.; Shapiro, S.L.; Bankert, D.A. Frequency of Low Level Bacteremia in Infants from Birth to Two Months of Age. Pediatr. Infect. Dis. J. 1997, 16, 381–385. [Google Scholar] [CrossRef]
- Connell, T.G.; Rele, M.; Cowley, D.; Buttery, J.P.; Curtis, N. How Reliable Is a Negative Blood Culture Result? Volume of Blood Submitted for Culture in Routine Practice in a Children’s Hospital. Pediatrics 2007, 119, 891–896. [Google Scholar] [CrossRef]
- Kumar, Y.; Qunibi, M.; Neal, T.J.; Yoxall, C.W. Time to Positivity of Neonatal Blood Cultures. Arch. Dis. Child. Fetal Neonatal Ed. 2001, 85, F182–F186. [Google Scholar] [CrossRef]
- Pammi, M.; Flores, A.; Leeflang, M.; Versalovic, J. Molecular Assays in the Diagnosis of Neonatal Sepsis: A Systematic Review and Meta-Analysis. Pediatrics 2011, 128, e973–e985. [Google Scholar] [CrossRef]
- Sharma, D.; Farahbakhsh, N.; Shastri, S.; Sharma, P. Biomarkers for Diagnosis of Neonatal Sepsis: A Literature Review. J. Matern. Fetal Neonatal Med. 2018, 31, 1646–1659. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Yan, A.; Zhang, L.; Hu, X.; Chen, L.; Cui, J.; Fan, Z.; Li, Y. Comparative Analysis of Inflammatory Biomarkers for the Diagnosis of Neonatal Sepsis: IL-6, IL-8, SAA, CRP, and PCT. Open Life Sci. 2025, 20, 20221005. [Google Scholar] [CrossRef]
- Aulin, L.B.S.; de Lange, D.W.; Saleh, M.A.A.; van der Graaf, P.H.; Völler, S.; van Hasselt, J.G.C. Biomarker-Guided Individualization of Antibiotic Therapy. Clin. Pharmacol. Ther. 2021, 110, 346–360. [Google Scholar] [CrossRef]
- Kyriazopoulou, E.; Leventogiannis, K.; Tavoulareas, G.; Mainas, E.; Toutouzas, K.; Mathas, C.; Prekates, A.; Sakka, V.; Panagopoulos, P.; Syrigos, K.; et al. Presepsin as a Diagnostic and Prognostic Biomarker of Severe Bacterial Infections and COVID-19. Sci. Rep. 2023, 13, 3814. [Google Scholar] [CrossRef]
- Ruan, L.; Chen, G.Y.; Liu, Z.; Zhao, Y.; Xu, G.Y.; Li, S.F.; Li, C.N.; Chen, L.S.; Tao, Z. The Combination of Procalcitonin and C-Reactive Protein or Presepsin Alone Improves the Accuracy of Diagnosis of Neonatal Sepsis: A Meta-Analysis and Systematic Review. Crit. Care 2018, 22, 316. [Google Scholar] [CrossRef]
- Sodero, G.; Gentili, C.; Mariani, F.; Pulcinelli, V.; Valentini, P.; Buonsenso, D. Procalcitonin and Presepsin as Markers of Infectious Respiratory Diseases in Children: A Scoping Review of the Literature. Children 2024, 11, 350. [Google Scholar] [CrossRef]
- Rodgers, O.; De Beer, A.; Waterfield, T. MicroRNA Biomarkers in Paediatric Infection Diagnostics—Bridging the Gap Between Evidence and Clinical Application: A Scoping Review. Noncoding RNA 2025, 11, 71. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. Elegans Heterochronic Gene Lin-4 Encodes Small RNAs with Antisense Complementarity to Lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional Regulation of the Heterochronic Gene Lin-14 by Lin-4 Mediates Temporal Pattern Formation in C. Elegans. Cell 1993, 75, 855–862. [Google Scholar] [CrossRef] [PubMed]
- Press Release: The Nobel Prize in Chemistry 2024—NobelPrize.Org. Available online: https://www.nobelprize.org/prizes/chemistry/2024/press-release/ (accessed on 23 June 2025).
- MiRBase. Available online: https://www.mirbase.org/ (accessed on 18 March 2025).
- MacFarlane, L.-A.; Murphy, P.R. MicroRNA: Biogenesis, Function and Role in Cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA Genes Are Transcribed by RNA Polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of MicroRNA Biogenesis and Its Crosstalk with Other Cellular Pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Kim, H.; Lee, Y.-Y.; Kim, V.N. The Biogenesis and Regulation of Animal MicroRNAs. Nat. Rev. Mol. Cell Biol. 2025, 26, 276–296. [Google Scholar] [CrossRef]
- Kosaka, N.; Iguchi, H.; Ochiya, T. Circulating MicroRNA in Body Fluid: A New Potential Biomarker for Cancer Diagnosis and Prognosis. Cancer Sci. 2010, 101, 2087–2092. [Google Scholar] [CrossRef]
- Vickers, K.C.; Remaley, A.T. Lipid-Based Carriers of MicroRNAs and Intercellular Communication. Curr. Opin. Lipidol. 2012, 23, 91–97. [Google Scholar] [CrossRef]
- Vasudevan, S.; Tong, Y.; Steitz, J.A. Switching from Repression to Activation: MicroRNAs Can up-Regulate Translation. Science 2007, 318, 1931–1934. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 Regulate Insulin Sensitivity. Nature 2011, 474, 649–653. [Google Scholar] [CrossRef]
- Roderburg, C.; Benz, F.; Koch, A.; Loosen, S.H.; Spehlmann, M.; Luedde, M.; Wree, A.; Vucur, M.; Trautwein, C.; Tacke, F.; et al. A Combined Score of Circulating MiRNAs Allows Outcome Prediction in Critically Ill Patients. J. Clin. Med. 2019, 8, 1644. [Google Scholar] [CrossRef]
- Searles, C.D. MicroRNAs and Cardiovascular Disease Risk. Curr. Cardiol. Rep. 2014, 26, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Lv, B.; He, S.; Li, P.; Jiang, S.; Li, D.; Lin, J.; Feinberg, M.W. MicroRNA-181 in Cardiovascular Disease: Emerging Biomarkers and Therapeutic Targets. FASEB J. 2024, 38, e23635. [Google Scholar] [CrossRef] [PubMed]
- Canale, P.; Borghini, A. Mitochondrial MicroRNAs: New Emerging Players in Vascular Senescence and Atherosclerotic Cardiovascular Disease. Int. J. Mol. Sci. 2024, 25, 6620. [Google Scholar] [CrossRef]
- Hochreuter, M.Y.; Dall, M.; Treebak, J.T.; Barrès, R. MicroRNAs in Non-Alcoholic Fatty Liver Disease: Progress and Perspectives. Mol. Metab. 2022, 65, 101581. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Y.; MacKowiak, B.; Gao, B. MicroRNAs as Regulators, Biomarkers and Therapeutic Targets in Liver Diseases. Gut 2021, 70, 784–795. [Google Scholar] [CrossRef] [PubMed]
- Siniscalco, D.; Galderisi, U.; Peluso, G.; Finicelli, M. Circulating MicroRNAs in Cancer: A 5-Year Update with a Focus on Breast and Lung Cancers. Int. J. Mol. Sci. 2024, 25, 3140. [Google Scholar] [CrossRef] [PubMed]
- Abdel Rhman, M.; PMO, O. Potential Therapeutic Applications of MicroRNAs in Cancer Diagnosis and Treatment: Sharpening a Double-Edged Sword? Eur. J. Pharmacol. 2022, 932, 175210. [Google Scholar] [CrossRef] [PubMed]
- Syed, S.N.; Brüne, B. Exosomal and Non-Exosomal MicroRNAs: New Kids on the Block for Cancer Therapy. Int. J. Mol. Sci. 2022, 23, 4493. [Google Scholar] [CrossRef]
- Orefice, N.S.; Petrillo, G.; Pignataro, C.; Mascolo, M.; De Luca, G.; Verde, S.; Pentimalli, F.; Condorelli, G.; Quintavalle, C. Extracellular Vesicles and MicroRNAs in Cancer Progression. In Advances in Clinical Chemistry; Academic Press Inc.: Cambridge, MA, USA, 2025. [Google Scholar]
- Roderburg, C.; Luedde, M.; Vargas Cardenas, D.; Vucur, M.; Scholten, D.; Frey, N.; Koch, A.; Trautwein, C.; Tacke, F.; Luedde, T. Circulating MicroRNA-150 Serum Levels Predict Survival in Patients with Critical Illness and Sepsis. PLoS ONE 2013, 8, e54612. [Google Scholar] [CrossRef]
- Hermann, S.; Brandes, F.; Kirchner, B.; Buschmann, D.; Borrmann, M.; Klein, M.; Kotschote, S.; Bonin, M.; Reithmair, M.; Kaufmann, I.; et al. Diagnostic Potential of Circulating Cell-Free MicroRNAs for Community-Acquired Pneumonia and Pneumonia-Related Sepsis. J. Cell Mol. Med. 2020, 24, 12054–12064. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, K.; Jin, C. Clinical Value of MicroRNA-378a-3p in Sepsis and Its Role in Sepsis-Induced Inflammation and Cardiac Dysfunction. Bioengineered 2021, 12, 8496–8504. [Google Scholar] [CrossRef]
- Yao, J.; Lui, K.Y.; Hu, X.; Liu, E.; Zhang, T.; Tong, L.; Xu, J.; Huang, F.; Zhu, Y.; Lu, M.; et al. Circulating MicroRNAs as Novel Diagnostic Biomarkers and Prognostic Predictors for Septic Patients. Infect. Genet. Evol. 2021, 95, 105082. [Google Scholar] [CrossRef]
- Wang, S.; Yang, Y.; Suen, A.; Zhu, J.; Williams, B.; Hu, J.; Chen, F.; Kozar, R.; Shen, S.; Li, Z.; et al. Role of Extracellular MicroRNA-146a-5p in Host Innate Immunity and Bacterial Sepsis. iScience 2021, 24, 103441. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef]
- Zhang, C.; Li, X.; Liu, N.; Feng, Z.; Zhang, C. MicroRNA-96 is Downregulated in Sepsis Neonates and Attenuates LPSInduced Inflammatory Response by Inhibiting IL-16 in Monocytes. Comb. Chem. High Throughput Screen. 2022, 25, 90–96. [Google Scholar] [CrossRef] [PubMed]
- Sankar, S.; Maruthai, K.; Bobby, Z.; Adhisivam, B. MicroRNA Expression in Neonates with Late-onset Sepsis—A Cross-sectional Comparative Study. Immunol. Investig. 2022, 51, 1647–1659. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xu, X.; Wang, M. Clinical significance of serum miR-101-3p expression in patients with neonatal sepsis. Pers. Med. 2021, 18, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Wang, H.; Zhang, X.; Chen, R. Identification of Potential Biomarkers in Neonatal Sepsis by Establishing a Competitive Endogenous RNA Network. Comb. Chem. High Throughput Screen. 2020, 23, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Jiang, S.; Cao, Y.; Yang, Y. Altered miRNAs expression profiles and modulation of immune response genes and proteins during neonatal sepsis. J. Clin. Immunol. 2014, 34, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.C.; Yu, H.R.; Huang, L.T.; Huang, H.C.; Chen, R.F.; Lin, I.C.; Ou, C.Y.; Hsu, T.Y.; Yang, K.D. miRNA-125b regulates TNF-α production in CD14+ neonatal monocytes via post-transcriptional regulation. J. Leukoc. Biol. 2012, 92, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, X.; Liu, X.; Xu, J.; Hou, S.; Zhang, X.; Ding, Y. MiR-15a/16 Are Upreuglated in the Serum of Neonatal Sepsis Patients and Inhibit the LPS-Induced Inflammatory Pathway. Int. J. Clin. Exp. Med. 2015, 8, 5683. [Google Scholar] [PubMed]
- Wang, D.; Han, L. Downregulation of MiR-1184 Serves as a Diagnostic Biomarker in Neonatal Sepsis and Regulates LPS-Induced Inflammatory Response by Inhibiting IL-16 in Monocytes. Exp. Ther. Med. 2021, 21, 350. [Google Scholar] [CrossRef]
- Ahmed, I.A.; Elfadle, A.A.; Abdelmotaleb, G.S. Value of Serum MiRNA 146a as Biomarker for Early Diagnosis of Neonatal Sepsis. Bull. Egypt. Soc. Physiol. Sci. 2017, 37, 59–68. [Google Scholar] [CrossRef]
- Salim, R.F.; Sobeih, A.A.; Abd El Kareem, H.M. Evaluation of the Clinical Value of Circulating MiR-101, MiR-187 and MiR-21 in Neonatal Sepsis Diagnosis and Prognosis. Egypt. J. Med. Hum. Genet. 2020, 21, 12. [Google Scholar] [CrossRef]
- Liu, G.; Liu, W.; Guo, J. Clinical Significance of MiR-181a in Patients with Neonatal Sepsis and Its Regulatory Role in the Lipopolysaccharide-Induced Inflammatory Response. Exp. Ther. Med. 2020, 19, 1977–1983. [Google Scholar] [CrossRef]
- Fatmi, A.; Rebiahi, S.A.; Chabni, N.; Zerrouki, H.; Azzaoui, H.; Elhabiri, Y.; Benmassour, S.; Ibáñez-Cabellos, J.S.; Smahi, M.C.E.; Aribi, M.; et al. MiRNA-23b as a Biomarker of Culture-Positive Neonatal Sepsis. Mol. Med. 2020, 26, 94. [Google Scholar] [CrossRef]
- Li, M.; Huang, X.; Zhuo, Q.; Zhang, J.; Ju, X. Clinical Significance of MiR-129-5p in Patients with Neonatal Sepsis and Its Regulatory Role in the Lipopolysaccharide-Induced Inflammatory Response. Bosn. J. Basic. Med. Sci. 2022, 22, 185–190. [Google Scholar] [CrossRef]
- Mao, Y.Y.; Su, C.; Fang, C.C.; Fan, X.P.; Wang, L.P.; Zhu, S.S.; Yao, H.M. Clinical Significance of the Serum MiR-455-5p Expression in Patients with Neonatal Sepsis. Bioengineered 2021, 12, 4174–4182. [Google Scholar] [CrossRef] [PubMed]
- Serna, E.; Parra-Llorca, A.; Panadero, J.; Vento, M.; Cernada, M. MiRNomic Signature in Very Low Birth-Weight Neonates Discriminates Late-Onset Gram-Positive Sepsis from Controls. Diagnostics 2021, 11, 1389. [Google Scholar] [CrossRef] [PubMed]
- Abdelaleem, O.O.; Mohammed, S.R.; El Sayed, H.S.; Hussein, S.K.; Ali, D.Y.; Abdelwahed, M.Y.; Gaber, S.N.; Hemeda, N.F.; Abd El-Hmid, R.G. Serum MiR-34a-5p and MiR-199a-3p as New Biomarkers of Neonatal Sepsis. PLoS ONE 2022, 17, e262339. [Google Scholar] [CrossRef]
- Ali, M.A.; Hussein, S.K.; Ali Mohamed, E.; Ezzat, M.A.; Abdelmoktader, A.; Habib, M.A.; Kamal, M.; Ahmed, F.A.; Ali, D.Y. Diagnostic and Prognostic Values of MiR181b-5p and MiR21-5p for Neonatal Sepsis Risk and Their Link to SNAP II Score and Disease Mortality. Noncoding RNA Res. 2023, 8, 115–125. [Google Scholar] [CrossRef]
- Katta, D.; Sridharan, K.S.; Balakrishnan, U.M.; Amboiram, P.; Kumar, K. Sensitivity of MiRNA-181a, MiRNA-23b and MiRNA-16 in the Late-Onset Neonatal Sepsis: A Diagnostic Study. Matern. Fetal Med. 2024, 6, 243–248. [Google Scholar] [CrossRef] [PubMed]
- Lawrie, C.H.; Gal, S.; Dunlop, H.M.; Pushkaran, B.; Liggins, A.P.; Pulford, K.; Banham, A.H.; Pezzella, F.; Boultwood, J.; Wainscoat, J.S.; et al. Detection of Elevated Levels of Tumour-Associated MicroRNAs in Serum of Patients with Diffuse Large B-Cell Lymphoma. Br. J. Haematol. 2008, 141, 672–675. [Google Scholar] [CrossRef]
- Clinical Evaluation of Circulating MicroRNA-25 Level Change in Sepsis and Its Potential Relationship with Oxidative Stress. Request PDF. Available online: https://www.researchgate.net/publication/281513272_Clinical_evaluation_of_circulating_microRNA-25_level_change_in_sepsis_and_its_potential_relationship_with_oxidative_stress (accessed on 11 March 2025).
- Tacke, F.; Roderburg, C.; Benz, F.; Cardenas, D.V.; Luedde, M.; Hippe, H.J.; Frey, N.; Vucur, M.; Gautheron, J.; Koch, A.; et al. Levels of Circulating MiR-133a Are Elevated in Sepsis and Predict Mortality in Critically Ill Patients. Crit. Care Med. 2014, 42, 1096–1104. [Google Scholar] [CrossRef]
- Wang, J.F.; Yu, M.L.; Yu, G.; Bian, J.J.; Deng, X.M.; Wan, X.J.; Zhu, K.M. Serum MiR-146a and MiR-223 as Potential New Biomarkers for Sepsis. Biochem. Biophys. Res. Commun. 2010, 394, 184–188. [Google Scholar] [CrossRef]
- Wang, L.; Wang, H.C.; Chen, C.; Zeng, J.; Wang, Q.; Zheng, L.; Yu, H. Du Differential Expression of Plasma MiR-146a in Sepsis Patients Compared with Non-Sepsis-SIRS Patients. Exp. Ther. Med. 2013, 5, 1101. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhang, P.; Chen, W.; Feng, D.; Jia, Y.; Xie, L. Serum MicroRNA Signatures Identified by Solexa Sequencing Predict Sepsis Patients’ Mortality: A Prospective Observational Study. PLoS ONE 2012, 7, e38885. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.; Zhang, J.; Huang, Y.; Tong, J.; Zhang, L.; Zhang, Z.; Yu, W.; Qiu, Y. Accuracy of Circulating MicroRNAs in Diagnosis of Sepsis: A Systematic Review and Meta-Analysis. J. Intensive Care 2020, 8. [Google Scholar] [CrossRef]
- Han, W.; Li, S.; Wang, N.; Chen, X.; Ma, J.; Liang, J.; Hao, L.; Ren, C. MiRNAs as Biomarkers for Diagnosis of Neonatal Sepsis: A Systematic Review and Meta-Analysis. J. Matern.-Fetal Neonatal Med. 2023, 36, 2217317. [Google Scholar] [CrossRef] [PubMed]
- Bindayna, K. MicroRNA as Sepsis Biomarkers: A Comprehensive Review. Int. J. Mol. Sci. 2024, 25, 6476. [Google Scholar] [CrossRef]
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. MiRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276. [Google Scholar] [CrossRef]
| Study Title and Author | Year | Origin | Population | Used for miRNA Extraction | MiRNA Profile/Result Upregulated (U) Downregulated (D) |
|---|---|---|---|---|---|
| miRNA-15a/16 are upregulated in the serum of neonatal sepsis patients and inhibit the LPS-induced inflammatory pathway, Wang et al. [50] | 2015 | China | 46 neonates with sepsis and 41 neonates with respiratory infection/pneumonia | Serum | miRNA-15a/16 (U) |
| Downregulation of miRNA-1184 serves as a diagnostic biomarker in neonatal sepsis and regulates LPS-induced inflammatory response by inhibiting IL-16 in monocytes, Wang et al. [51] | 2021 | China | 72 neonates with neonatal sepsis (including 6 cases of EOS and 66 cases of LOS) and 56 neonates with respiratory infection or pneumonia | Serum | miRNA-1184 (D) |
| Value of Serum miRNA 146a as Biomarker for Early Diagnosis of Neonatal Sepsis. Ahmed et al. [52] | 2017 | Egypt | 24 neonates with LOS, 22 neonates at high risk of sepsis and 17 healthy controls | Serum | miRNA-146a (U) |
| Evaluation of the clinical value of circulating miRNA-101, miRNA-187 and miRNA-21 in neonatal sepsis diagnosis and prognosis, Salim et al. [53] | 2020 | Egypt | 100 neonates (50 with LOS with positive blood culture, 30 with SIRS with negative blood culture and 20 healthy neonates) | Serum | miRNA-101 (U), miRNA-187 (U), miRNA-21 (U) |
| Clinical significance of miRNA-181a, in patients with neonatal sepsis and its regulatory role in the lipopolysaccharide-induced inflammatory response, Liu et al. [54] | 2020 | China | 102 cases with LOS and 50 healthy controls | Serum | miRNA-181a (D) |
| MiRNA-23b as a biomarker of culture-positive neonatal sepsis, Fatmi et al. [55] | 2020 | Algeria | 48 neonates with sepsis (27 with EOS, 21 with LOS) | Serum | miRNA-23b (D) |
| Clinical significance of miRNA-129-5p in patients with neonatal sepsis and its regulatory role in the lipopolysaccharide-induced inflammatory response, Li et al. [56] | 2020 | China | 75 newborns with neonatal sepsis and 84 newborns with respiratory infection or pneumonia but without a picture of sepsis | Serum | miRNA-129-5p (D) |
| Clinical significance of the serum miRNA-455-5p expression in patients with neonatal sepsis, Mao et al. [57] | 2021 | China | 99 cases with LOS and 88 healthy controls | Serum | miRNA-455-5p (U) |
| MiRNomic signature in very low birth-weight neonates discriminates late-onset Gram-positive sepsis from controls, Serna et al. [58] | 2021 | Spain | 11 very-low birth-weight neonates with late-onset Gram-positive sepsis and 16 controls | Not clear | miRNA-31-5p, miRNA-1271-5p, miRNA-326, miRNA-146b-5p, miRNA-140-5p, miRNA-409-5p, miRNA-668-3p, miRNA-27b-3p, miRNA-28-5p, miRNA-152-3p, miRNA-431-5p, miRNA-106b-5p, miRNA-151a-3p, miRNA-15a-5p, miRNA-339-5p, miRNA-30b-5p, miRNA-146a-5p, miRNA-20a-5p, miRNA-20b-5p, miRNA-532-5p, miRNA-30c-5p, miRNA-106a-5p, miRNA-17-5p, miRNA-23b-3p, miRNA-93-5p, miRNA-425-5p, miRNA-1298-5p, miRNA-107, miRNA-103a-3p, miRNA-372-3p, miRNA-760, miRNA-6088 |
| Serum miRNA-34a-5p and miRNA-199a-3p as new biomarkers of neonatal sepsis, Abdelaleem et al. [59] | 2022 | Egypt | 60 cases with LOS and 60 healthy controls | Serum | miRNA-34a-5p (D), miRNA-199a-3p (D) |
| Diagnostic and prognostic values of miRNA181b-5p and miRNA21-5p for neonatal sepsis risk and their link to SNAP II score and disease mortality, Ali et al. [60] | 2023 | Egypt | 60 cases with LOS and 60 healthy controls | Serum | miRNA181b-5p (D), miRNA21-5p (D) |
| Sensitivity of miRNA-181a, miRNA-23b and miRNA-16 in the Late-Onset Neonatal Sepsis: A Diagnostic Study, Katta et al. [61] | 2024 | India | 50 cases of culture-proven LOS and 50 healthy controls | Serum | miRNA-181a (D), miRNA-23b (D), miRNA-16 (U) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Papachatzi, E.; Gkouti, E.; Kouvela, A.; Benou, S.; Dimitriou, G.; Fouzas, S.; Stamatopoulou, V.; Gkentzi, D. MicroRNA Profiling as a Novel Tool in the Diagnostics of Late-Onset Neonatal Sepsis: A Scoping Review. Children 2025, 12, 1573. https://doi.org/10.3390/children12111573
Papachatzi E, Gkouti E, Kouvela A, Benou S, Dimitriou G, Fouzas S, Stamatopoulou V, Gkentzi D. MicroRNA Profiling as a Novel Tool in the Diagnostics of Late-Onset Neonatal Sepsis: A Scoping Review. Children. 2025; 12(11):1573. https://doi.org/10.3390/children12111573
Chicago/Turabian StylePapachatzi, Eleni, Eleni Gkouti, Adamantia Kouvela, Sofia Benou, Gabriel Dimitriou, Sotiris Fouzas, Vassiliki Stamatopoulou, and Despoina Gkentzi. 2025. "MicroRNA Profiling as a Novel Tool in the Diagnostics of Late-Onset Neonatal Sepsis: A Scoping Review" Children 12, no. 11: 1573. https://doi.org/10.3390/children12111573
APA StylePapachatzi, E., Gkouti, E., Kouvela, A., Benou, S., Dimitriou, G., Fouzas, S., Stamatopoulou, V., & Gkentzi, D. (2025). MicroRNA Profiling as a Novel Tool in the Diagnostics of Late-Onset Neonatal Sepsis: A Scoping Review. Children, 12(11), 1573. https://doi.org/10.3390/children12111573








