Open-Source Artificial Intelligence System Supports Diagnosis of Mendelian Diseases in Acutely Ill Infants
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants
2.2. Dx29: Technology and Data Protection
2.3. File Types Relevant to This Study
- Variant Call Format (VCF) files
- ○
- Following DNA sequencing, the raw sequencing data are compared to a genomic standard to identify the proband’s genetic variants. These variants are compiled into a VCF file. We used trio VCFs (merged data from the proband and two parents) for our genetic analysis in Dx29.
- Pedigree (PED) files
- ○
- A PED file is a structured text document that explains the relation between multiple genetic samples. For each case, we prepared a PED file that listed the sex of each genetic sample and indicated which sample corresponded to the proband. Dx29 requires a PED file when performing trio analyses.
- PDF files
- ○
- Dx29 can analyze and extract a patient’s phenotype from PDFs, text documents, or images of documents. All records uploaded to Dx29 for this study were PDFs, including scanned images saved as PDFs.
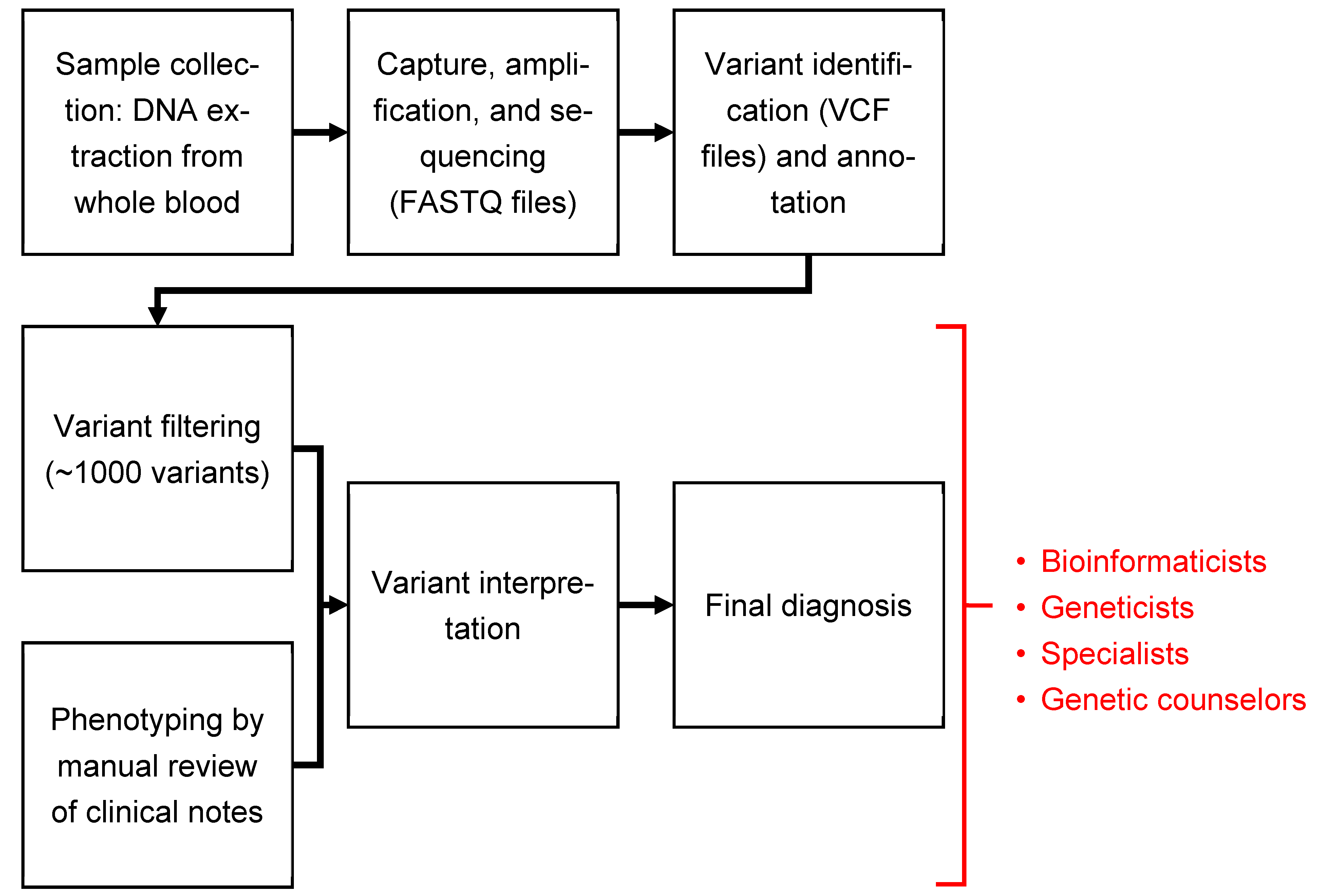
2.4. Dx29 Workflow
- Enter the case ID (required) and demographic information (optional).
- Provide phenotype information by performing the following:
- Entering patient’s phenotype manually as HPO terms.
- Uploading patient medical records for automated extraction of phenotypic information.
- Typing a medical description of the patient and then using the automated extraction.
- Phenotype extraction: If medical records were uploaded as PDFs, text documents, or images of documents, Dx29 will then review each record and identify symptoms and translate them to HPO terms. This step is multilingual, supporting 50+ languages. The accuracy of the HPO identification depends on the languages, and user validation is important.
- A user may optionally review the extracted HPO terms within Dx29. Under each term, Dx29 will show, in context, where it was identified in the files provided. Terms deemed inaccurate or irrelevant can be removed from the subsequent analysis.
- Genotype analysis: The trio’s merged VCF file and corresponding pedigree file are uploaded; Dx29 then filters and annotates the variants according to preset parameters. The variants are then ranked by likelihood of causing disease based on the predicted variant pathogenicity and the clinical significance of the affected gene. Those most likely to be disease-causing are prioritized in the ranking of the final differential diagnosis.
- Dx29 allows some exploration of the salient variants, including type of mutation, ClinVar status, in silico pathogenicity scores, and references to relevant literature. At the time of this study, there was no functionality in Dx29 that allowed for the removal of a particular variant from consideration when building the patient’s differential diagnosis.
- Generation of differential diagnosis: Manually provided or automatically extracted phenotypic information is compared to the candidate variants to generate a differential diagnosis.
- The generated differential diagnosis consists of up to 100 diseases that are ranked by how plausibly that diagnosis could explain the patient’s symptoms and findings, while also considering how the patient’s genotype does or does not support that potential diagnosis.
- For each diagnosis on the list, Dx29 will show how the patient’s symptoms overlapped with the expected phenotype for that disease and show the potentially causative variant.
- Because HPO terms are organized hierarchically, when making comparisons between a patient’s phenotype and the expected phenotype of a disease, Dx29 can extrapolate on imperfect matches. For example, if a patient is assigned the HPO term “abnormal aortic valve morphology” (HP:0001646), which is found under “abnormal heart valve morphology” (HP:0001654) in HPO hierarchy, this is considered by Dx29 to be a match between terms.
2.5. Study Design
2.5.1. Phenotype Analysis
- The patient’s medical records were uploaded to Dx29, and HPO terms were extracted automatically from the text and not manually reviewed.
- The patient’s records were uploaded to Dx29 for automatic extraction of HPO terms, and then each term was reviewed to determine if it had been extracted in error (Section 2.4, step 3a). Terms that were incorrectly identified (e.g., “cerebral palsy” being identified in a document that uses the abbreviation “cp” for some other purpose) were then removed from the analysis. Terms were removed strictly based on whether they had been correctly identified by Dx29 from the provided records, and not on judgment of their perceived relevance to a genetic diagnosis.
- The HPO terms used by the RapSeq team in reaching the original diagnosis were uploaded to Dx29 without any of the patient’s medical records; these terms were generated by manual review of the patient’s records by genetic counselors in the RapSeq pipeline.
2.5.2. Patient Medical Records
2.5.3. Preparing for Genotype Analysis
2.6. Outcomes
- The processing time for each case, beginning with creation of a patient case in Dx29, progressing through phenotypic and genotypic analysis, and ending when the ranked differential diagnosis was generated (Section 2.4, steps 1–5);
- How often the patient’s correct diagnosis was identified in the top 5 or top 10 potential diagnoses generated by Dx29.
3. Results
3.1. Analysis with Automated Extraction of HPO Terms
3.2. Analysis with Automated Extraction of HPO Terms Followed by Manual Review
3.3. Analysis Using HPO Terms Utilized in the Standard Diagnostic Pipeline
3.4. Analysis Using Only the Patient’s Phenotype
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kapil, S.; Fishler, K.P.; Euteneuer, J.C.; Brunelli, L. Many newborns in level IV NICUs are eligible for rapid DNA sequencing. Am. J. Med. Genet. Part A 2019, 179, 280–284. [Google Scholar] [CrossRef] [PubMed]
- Blencowe, H.; Moorthie, S.; Petrou, M.; Hamamy, H.; Povey, S.; Bittles, A.H.; Gibbons, S.; Darlison, M.; Modell, B.; Bittles, A.H.; et al. Rare single gene disorders: Estimating baseline prevalence and outcomes worldwide. J. Community Genet. 2018, 9, 397–406. [Google Scholar] [CrossRef]
- Willig, L.K.; Petrikin, J.E.; Smith, L.D.; Saunders, C.J.; Thiffault, I.; Miller, N.A.; Soden, S.E.; Cakici, J.A.; Herd, S.M.; Twist, G.; et al. Whole-genome sequencing for identification of Mendelian disorders in critically ill infants: A retrospective analysis of diagnostic and clinical findings. Lancet Respir. Med. 2015, 3, 377–387. [Google Scholar] [CrossRef]
- French, C.E.; Delon, I.; Dolling, H.; Sanchis-Juan, A.; Shamardina, O.; Mégy, K.; Abbs, S.; Austin, T.; Bowdin, S.; Branco, R.G.; et al. Whole genome sequencing reveals that genetic conditions are frequent in intensively ill children. Intensive Care Med. 2019, 45, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Farnaes, L.; Hildreth, A.; Sweeney, N.M.; Clark, M.M.; Chowdhury, S.; Nahas, S.; Cakici, J.A.; Benson, W.; Kaplan, R.H.; Kronick, R.; et al. Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization. NPJ Genom. Med. 2018, 3, 10. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Lu, Y.; Dong, X.; Lu, G.; Cheng, G.; Qian, Y.; Ni, Q.; Zhang, P.; Yang, L.; Wu, B.; et al. Optimized trio genome sequencing (OTGS) as a first-tier genetic test in critically ill infants: Practice in China. Hum. Genet. 2020, 139, 473–482. [Google Scholar] [CrossRef]
- Brunelli, L.; Jenkins, S.M.; Gudgeon, J.M.; Bleyl, S.B.; Miller, C.E.; Tvrdik, T.; Dames, S.A.; Ostrander, B.; Daboub, J.A.F.; Zielinski, B.A.; et al. Targeted gene panel sequencing for the rapid diagnosis of acutely ill infants. Mol. Genet. Genom. Med. 2019, 7, e00796. [Google Scholar] [CrossRef]
- Brunelli, L.; Mao, R.; Jenkins, S.M.; Bleyl, S.B.; Dames, S.A.; Miller, C.E.; Ostrander, B.; Tvrdik, T.; Andrews, S.; Flores, J.; et al. A rapid gene sequencing panel strategy to facilitate precision neonatal medicine. Am. J. Med. Genet. Part A 2017, 173, 1979–1982. [Google Scholar] [CrossRef]
- Wright, C.F.; Campbell, P.; Eberhardt, R.Y.; Aitken, S.; Perrett, D.; Brent, S.; Danecek, P.; Gardner, E.J.; Chundru, V.K.; Lindsay, S.J.; et al. Genomic Diagnosis of Rare Pediatric Disease in the United Kingdom and Ireland. N. Engl. J. Med. 2023, 388, 1559–1571. [Google Scholar] [CrossRef]
- Schwarze, K.; Buchanan, J.; Taylor, J.C.; Wordsworth, S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet. Med. 2018, 20, 1122–1130. [Google Scholar] [CrossRef]
- Franck, L.S.; Kriz, R.M.; Rego, S.; Garman, K.; Hobbs, C.; Dimmock, D. Implementing Rapid Whole-Genome Sequencing in Critical Care: A Qualitative Study of Facilitators and Barriers to New Technology Adoption. J. Pediatr. 2021, 237, 237–243.e232. [Google Scholar] [CrossRef]
- Elliott, A.M.; du Souich, C.; Lehman, A.; Guella, I.; Evans, D.M.; Candido, T.; Tooman, L.; Armstrong, L.; Clarke, L.; Gibson, W.; et al. RAPIDOMICS: Rapid genome-wide sequencing in a neonatal intensive care unit—Successes and challenges. Eur. J. Pediatr. 2019, 178, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Mestek-Boukhibar, L.; Clement, E.; Jones, W.D.; Drury, S.; Ocaka, L.; Gagunashvili, A.; Le Quesne Stabej, P.; Bacchelli, C.; Jani, N.; Rahman, S.; et al. Rapid Paediatric Sequencing (RaPS): Comprehensive real-life workflow for rapid diagnosis of critically ill children. J. Med. Genet. 2018, 55, 721–728. [Google Scholar] [CrossRef]
- Sanford, E.F.; Clark, M.M.; Farnaes, L.; Williams, M.R.; Perry, J.C.; Ingulli, E.G.; Sweeney, N.M.; Doshi, A.; Gold, J.J.; Briggs, B.; et al. Rapid Whole Genome Sequencing Has Clinical Utility in Children in the PICU. Pediatr. Crit. Care Med. 2019, 20, 1007–1020. [Google Scholar] [CrossRef] [PubMed]
- Corominas, J.; Smeekens, S.P.; Nelen, M.R.; Yntema, H.G.; Kamsteeg, E.-J.; Pfundt, R.; Gilissen, C. Clinical exome sequencing—Mistakes and caveats. Hum. Mutat. 2022, 43, 1041–1055. [Google Scholar] [CrossRef]
- Fast Facts on U.S. Hospitals. 2022. AHA. Available online: https://www.aha.org/statistics/fast-facts-us-hospitals (accessed on 16 May 2023).
- NICU Directory–Neonatology Solutions. Available online: https://neonatologysolutions.com/nicu-directory/ (accessed on 16 May 2023).
- Clark, M.M.; Hildreth, A.; Batalov, S.; Ding, Y.; Chowdhury, S.; Watkins, K.; Ellsworth, K.; Camp, B.; Kint, C.I.; Yacoubian, C.; et al. Diagnosis of genetic diseases in seriously ill children by rapid whole-genome sequencing and automated phenotyping and interpretation. Sci. Transl. Med. 2019, 11, eaat6177. [Google Scholar] [CrossRef] [PubMed]
- Peterson, B.; Hernandez, E.J.; Hobbs, C.; Malone Jenkins, S.; Moore, B.; Rosales, E.; Zoucha, S.; Sanford, E.; Bainbridge, M.N.; Frise, E.; et al. Automated prioritization of sick newborns for whole genome sequencing using clinical natural language processing and machine learning. Genome Med. 2023, 15, 18. [Google Scholar] [CrossRef]
- De La Vega, F.M.; Chowdhury, S.; Moore, B.; Frise, E.; McCarthy, J.; Hernandez, E.J.; Wong, T.; James, K.; Guidugli, L.; Agrawal, P.B.; et al. Artificial intelligence enables comprehensive genome interpretation and nomination of candidate diagnoses for rare genetic diseases. Genome Med. 2021, 13, 153. [Google Scholar] [CrossRef]
- James, K.N.; Clark, M.M.; Camp, B.; Kint, C.; Schols, P.; Batalov, S.; Briggs, B.; Veeraraghavan, N.; Chowdhury, S.; Kingsmore, S.F. Partially automated whole-genome sequencing reanalysis of previously undiagnosed pediatric patients can efficiently yield new diagnoses. Npj Genom. Med. 2020, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Dias, R.; Torkamani, A. Artificial intelligence in clinical and genomic diagnostics. Genome Med. 2019, 11, 70. [Google Scholar] [CrossRef]
- Dx29-Foundation29. Available online: https://www.foundation29.org/our-work/dx29/ (accessed on 2 August 2021).
- What Is the Text Analytics for Health in Azure Cognitive Service for Language?-Azure Cognitive Services. Microsoft Learn. Available online: https://learn.microsoft.com/en-us/azure/cognitive-services/language-service/text-analytics-for-health/overview?tabs=ner (accessed on 25 February 2023).
- Human Phenotype Ontology. Available online: https://hpo.jax.org/app/ (accessed on 17 February 2021).
- Robinson, P.N.; Köhler, S.; Oellrich, A.; Genetics, S.M.; Wang, K.; Mungall, C.J.; Lewis, S.E.; Washington, N.; Bauer, S.; Seelow, D.; et al. Improved exome prioritization of disease genes through cross-species phenotype comparison. Genome Res. 2014, 24, 340–348. [Google Scholar] [CrossRef]
- Cipriani, V.; Pontikos, N.; Arno, G.; Sergouniotis, P.I.; Lenassi, E.; Thawong, P.; Danis, D.; Michaelides, M.; Webster, A.R.; Moore, A.T.; et al. An improved phenotype-driven tool for rare mendelian variant prioritization: Benchmarking exomiser on real patient whole-exome data. Genes 2020, 11, 460. [Google Scholar] [CrossRef] [PubMed]
- Orphanet-Home. Available online: https://www.orpha.net/consor/cgi-bin/index.php (accessed on 27 July 2021).
- Home-OMIM. Available online: https://www.omim.org/ (accessed on 25 February 2023).
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; De Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
| ID | Gene | Diagnosis/OMIM Diseases | Automatic | Automatic with Review | RapSeq Terms | Phenotype Only |
|---|---|---|---|---|---|---|
| 001 | CHAT | Congenital presynaptic myasthenic syndrome 6 | 31 | 31 | 30 | 20 |
| 004 | FNLA | X-linked periventricular nodular heterotopia | 14 | 6 | 4 | unranked |
| 005 | FANCB | X-linked VACTERL with hydrocephalus syndrome | 2 | 2 | 2 | 42 |
| 007 | KMTD2 | Kabuki syndrome 1 | 1 | 1 | 2 | unranked |
| 009 | CHD7 | CHARGE syndrome | 1 | 1 | 1 | 2 |
| 013 | ASXL1 | Bohring–Opitz syndrome | 1 | 1 | 1 | 8 |
| 014 | FBN1 | Neonatal Marfan | 1 | 1 | 1 | 3 |
| 023 | PAX3 | Craniofacial–deafness–hand syndrome, Waardenburg syndrome, type 1 and type 3 | 1 | 1 | 1 | unranked |
| 026 | CACNA1A | Developmental and epileptic encephalopathy 42 | 5 | 4 | 1 | unranked |
| 027 | KCNQ2 | Early infantile epileptic encephalopathy 7 | 1 | 1 | 1 | 44 |
| 028 | HDAC8 | Cornelia de Lange syndrome 5 | 1 | 1 | 1 | 1 |
| 029 | AHCY | Hypermethioninemia with deficiency of S-adenosylhomocysteine hydrolase | 1 | 1 | 1 | 60 |
| 035 | ACAD9 | Mitochondrial complex I deficiency, nuclear type 20 | 1 | 1 | 3 | 43 |
| 036 | CDAN1 | Dyserythropoietic anemia, congenital, type Ia | 3 | 3 | 1 | 60 |
| 037 | AMER1 | Osteopathia striata with cranial sclerosis | 1 | 1 | 2 | unranked |
| 041 | TCIRG1 | Osteopetrosis 1 | 1 | 1 | 1 | 9 |
| 042 | RYR1 | Autosomal recessive and autosomal dominant congenital neuromuscular disease with uniform type 1 fiber and with central core disease | 3 | 1 | 1 | unranked |
| 044 | RYR1 | Autosomal recessive and autosomal dominant congenital neuromuscular disease with uniform type 1 fiber and with central core disease | 5 | 1 | 1 | unranked |
| 045 | GUSB | Autosomal recessive mucopolysaccharidosis VII | 1 | 2 | 2 | unranked |
| 047 | ASNS | Asparagine synthetase deficiency | 7 | 6 | 2 | 57 |
| 050 | CHD7 | CHARGE syndrome | 1 | 1 | 1 | unranked |
| 054 | ACTA1 | Unspecified myopathy | 1 | 1 | 8 | unranked |
| 058 | KCNQ3 | Early infantile epileptic encephalopathy 7 | 1 | 1 | 1 | unranked |
| 068 | ASXL1 | Bohring–Opitz syndrome | 1 | 1 | 2 | 7 |
| 069 | SLC35A2 | Congenital disorder of glycosylation, type IIm | 1 | 2 | 11 | unranked |
| Automatic Extraction of Phenotype | Automatic Extraction + Manual Review | RapSeq Phenotypic Terms | Phenotype Only | |
|---|---|---|---|---|
| Correct diagnosis is in top 5 suggested diseases | 88% | 88% | 88% | 12% |
| Correct diagnosis is in top 10 suggested diseases | 92% | 96% | 92% | 24% |
| Median rank of correct diagnosis | 1 | 1 | 1 | |
| Mean rank of correct diagnosis | 3.48 | 2.92 | 3.28 | |
| Minimum rank of correct diagnosis | 1 | 1 | 1 | |
| Maximum rank of correct diagnosis | 31 | 31 | 30 | |
| Average time elapsed (mean ± SD) in hours | 0.32 ± 0.21 | 1.96 ± 0.85 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Reiley, J.; Botas, P.; Miller, C.E.; Zhao, J.; Malone Jenkins, S.; Best, H.; Grubb, P.H.; Mao, R.; Isla, J.; Brunelli, L. Open-Source Artificial Intelligence System Supports Diagnosis of Mendelian Diseases in Acutely Ill Infants. Children 2023, 10, 991. https://doi.org/10.3390/children10060991
Reiley J, Botas P, Miller CE, Zhao J, Malone Jenkins S, Best H, Grubb PH, Mao R, Isla J, Brunelli L. Open-Source Artificial Intelligence System Supports Diagnosis of Mendelian Diseases in Acutely Ill Infants. Children. 2023; 10(6):991. https://doi.org/10.3390/children10060991
Chicago/Turabian StyleReiley, Joseph, Pablo Botas, Christine E. Miller, Jian Zhao, Sabrina Malone Jenkins, Hunter Best, Peter H. Grubb, Rong Mao, Julián Isla, and Luca Brunelli. 2023. "Open-Source Artificial Intelligence System Supports Diagnosis of Mendelian Diseases in Acutely Ill Infants" Children 10, no. 6: 991. https://doi.org/10.3390/children10060991
APA StyleReiley, J., Botas, P., Miller, C. E., Zhao, J., Malone Jenkins, S., Best, H., Grubb, P. H., Mao, R., Isla, J., & Brunelli, L. (2023). Open-Source Artificial Intelligence System Supports Diagnosis of Mendelian Diseases in Acutely Ill Infants. Children, 10(6), 991. https://doi.org/10.3390/children10060991






