An Integrated Approach Using HLAMatchmaker and Pirche II for Epitopic Matching in Pediatric Kidney Transplant—A Romanian Single-Center Study
Abstract
:1. Introduction
2. Patients and Methods
2.1. Patient Population
2.2. PIRCHE Analysis
2.3. HLAMatchmaker Analysis
2.4. Statistical Analysis
3. Results
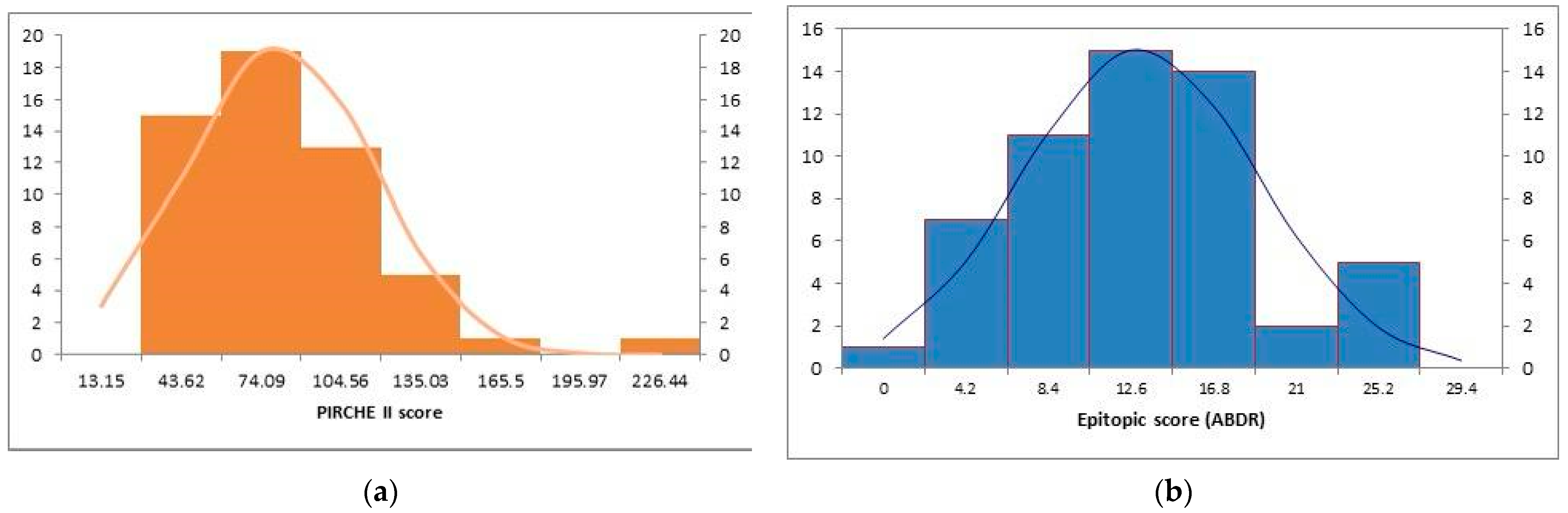
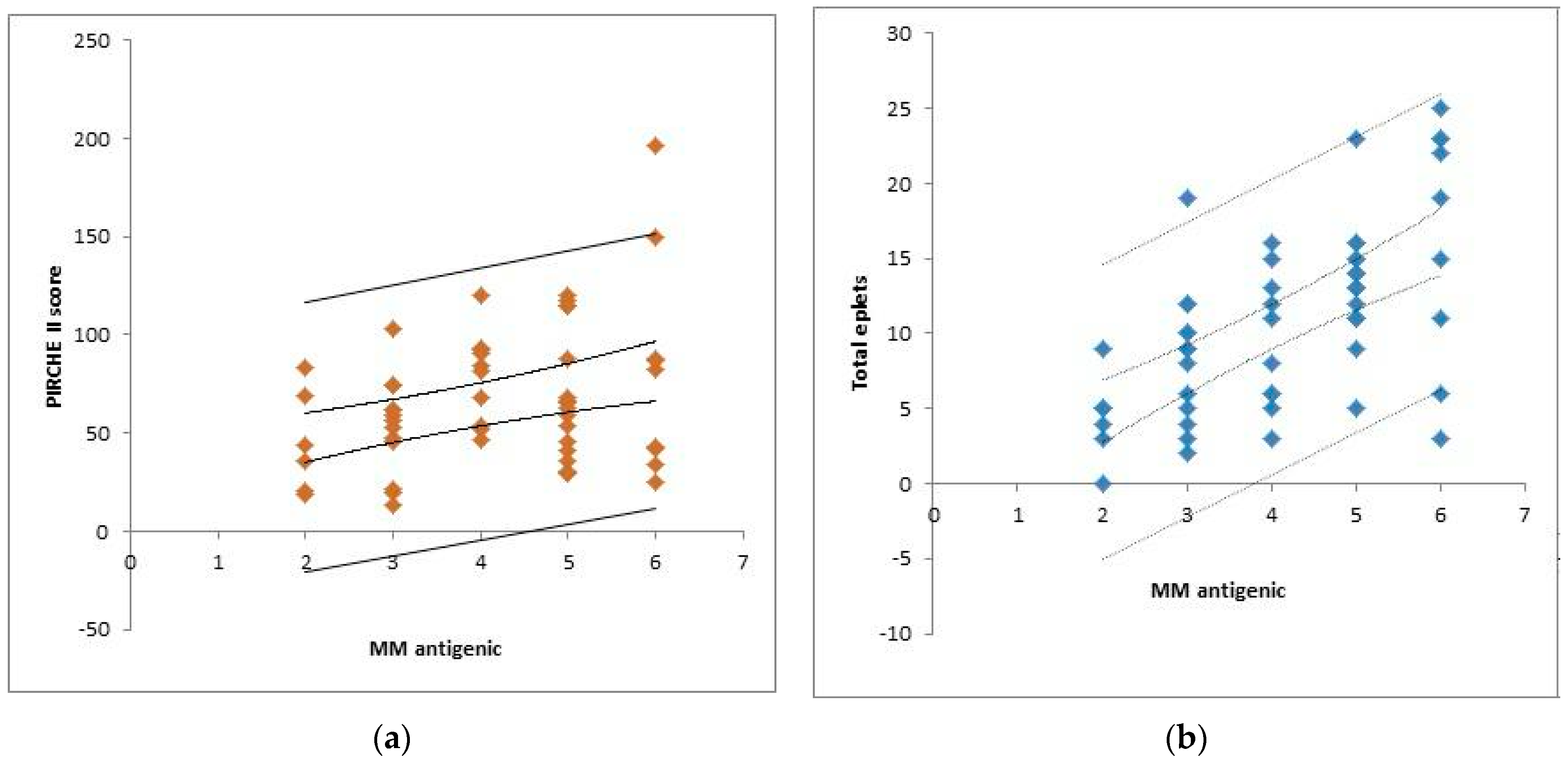
3.1. Descriptive Analysis of the Cohort (n = 55)
3.2. Defining Low-Risk and High-Risk Patients
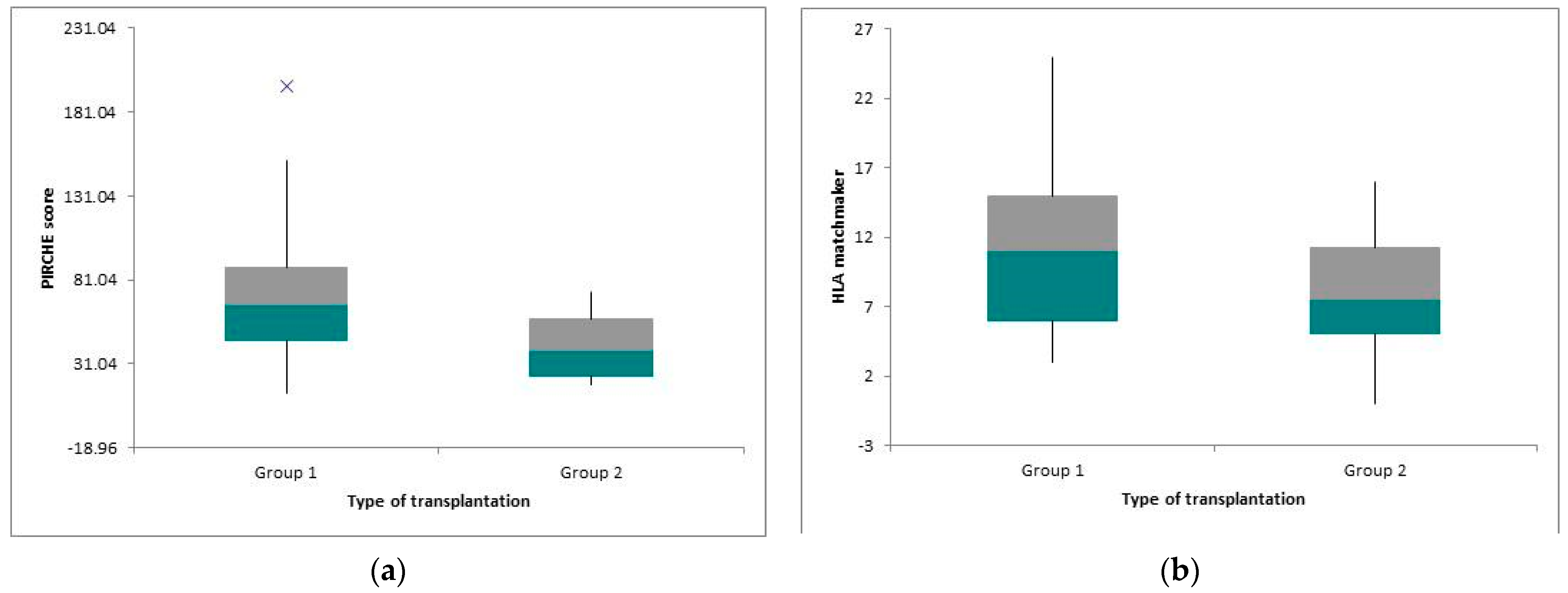
3.3. Statistical Analysis Depending on Other Parameters (Type of Donor and Transplantation)
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shapiro, R.; Sarwal, M.M. Pediatric kidney transplantation. Pediatr. Clin. N. Am. 2010, 57, 393–400. [Google Scholar] [CrossRef]
- Charnaya, O.; Levy Erez, D.; Amaral, S.; Monos, D.S. Pediatric kidney transplantation—Can we do better? The promise and limitations of Epitope/Eplet matching. Front. Pediatr. 2022, 10, 893002. [Google Scholar] [CrossRef]
- Wiebe, C.; Gibson, I.W.; Blydt-Hansen, T.D.; Karpinski, M.; Ho, J.; Storsley, L.J.; Goldberg, A.; Birk, P.E.; Rush, D.N.; Nickerson, P.W. Evolution and clinical pathologic correlations of de novo donor-specific HLA antibody post kidney transplant: Clinical pathologic correlations of DE Novo DSA. Am. J. Transplant. 2012, 12, 1157–1167. [Google Scholar] [CrossRef]
- Tran, J.N.; Günther, O.P.; Sherwood, K.R.; Fenninger, F.; Allan, L.L.; Lan, J.; Sapir-Pichhadze, R.; Duquesnoy, R.; Claas, F.; Marsh, S.G.; et al. High-throughput sequencing defines donor and recipient HLA B-cell epitope frequencies for prospective matching in transplantation. Commun. Biol. 2021, 4, 583. [Google Scholar] [CrossRef]
- Tambur, A.R.; Claas, F.H.J. HLA epitopes as viewed by antibodies: What is it all about?: HLA epitopes as viewed by antibodies. Am. J. Transplant. 2015, 15, 1148–1154. [Google Scholar] [CrossRef]
- Sypek, M.P.; Hughes, P. HLA Eplet mismatches in kidney transplantation: More than just adding things up. KI Rep. 2021, 6, 1500–1502. [Google Scholar] [CrossRef]
- Engen, R.M.; Jedraszko, A.M.; Conciatori, M.A.; Tambur, A.R. Substituting imputation of HLA antigens for high-resolution HLA typing: Evaluation of a multiethnic population and implications for clinical decision making in Transplantation. Am. J. Transplant. 2021, 21, 344–352. [Google Scholar] [CrossRef]
- Duquesnoy, R.J. Antibody-reactive epitope determination with HLAMatchmaker and its clinical applications. Tissue Antigens 2011, 77, 525–534. [Google Scholar] [CrossRef]
- Otten, H.G.; Calis, J.J.; Keşmir, C.; van Zuilen, A.D.; Spierings, E. Predicted indirectly recognisable HLA epitopes presented by HLA-DR correlate with the de novo development of donor-specific HLA IgG antibodies after kidney transplantation. Hum. Immunol. 2013, 74, 290–296. [Google Scholar] [CrossRef]
- Tambur, A.R.; Campbell, P.; Claas, F.H.; Feng, S.; Gebel, H.M.; Jackson, A.M.; Mannon, R.B.; Reed, E.F.; Tinckam, K.; Askar, M.; et al. Sensitisation in transplantation: Assessment of risk (STAR) 2019 Working Group Meeting Report. Am. J. Transplant. 2020, 20, 2652–2668. [Google Scholar] [CrossRef]
- Geneugelijk, K.; Thus, K.A.; Spierings, E. Predicting alloreactivity in transplantation. JIR 2014, 2014, 159479. [Google Scholar] [CrossRef] [PubMed]
- Spitznagel, T.; Matter, L.S.; Kaufmann, Y.L.; Nilsson, J.; von Moos, S.; Schachtner, T. PIRCHE-II scores prove useful as a predictive biomarker among kidney transplant recipients with rejection: An analysis of indication and follow-up biopsies. Front. Immunol. 2022, 13, 949933. [Google Scholar] [CrossRef] [PubMed]
- Geneugelijk, K.; Niemann, M.; Drylewicz, J.; van Zuilen, A.D.; Joosten, I.; Allebes, W.A.; van der Meer, A.; Hilbrands, L.B.; Baas, M.C.; Hack, C.E.; et al. Pirche-II is related to graft failure after kidney transplantation. Front. Immunol. 2018, 9, 321. [Google Scholar] [CrossRef] [PubMed]
- Unterrainer, C.; Döhler, B.; Niemann, M.; Lachmann, N.; Süsal, C. Can Pirche-II Matching outmatch traditional HLA matching? Front. Immunol. 2021, 12, 631246. [Google Scholar] [CrossRef] [PubMed]
- Rãchişan, A.L.; Dubois, V.; Ranchin, B.; Sellier-Leclerc, A.; Thomas, A.B.; Cochat, P.; Bacchetta, J. Eplet incompatibility in pediatric renal transplantation. Pediatr. Transplant. 2020, 24, e13721. [Google Scholar] [CrossRef]
- Branco, F.; Almeida, F.; Cavadas, V.; Ribeiro, S.; Osório, L.; Rocha, A.; Ramos, M.; Martins, L.; Dias, L.; Castro-Henriques, A.; et al. Pediatric kidney transplantation: A single center experience with 134 procedures. Transpl. Proc. 2013, 45, 1057–1059. [Google Scholar] [CrossRef]
- Kausman, J.Y.; Walker, A.M.; Cantwell, L.S.; Quinlan, C.; Sypek, M.P.; Ierino, F.L. Application of an epitope-based allocation system in pediatric kidney transplantation. Pediatr. Transplant. 2016, 20, 931–938. [Google Scholar] [CrossRef]
- Bryan, C.F.; Chadha, V.; Warady, B.A. Donor selection in pediatric kidney transplantation using DR and DQ eplet mismatching: A new histocompatibility paradigm. Pediatr. Transplant. 2016, 20, 926–930. [Google Scholar] [CrossRef]
- Lachmann, N.; Niemann, M.; Reinke, P.; Budde, K.; Schmidt, D.; Halleck, F.; Pruß, A.; Schönemann, C.; Spierings, E.; Staeck, O. Donor–Recipient Matching Based on Predicted Indirectly Recognizable HLA Epitopes Independently Predicts the Incidence of De Novo Donor-Specific HLA Antibodies Following Renal Transplantation. Am. J. Transplant. 2017, 17, 3076–3086. [Google Scholar] [CrossRef]
- Geneugelijk, K.; Wissing, J.; Koppenaal, D.; Niemann, M.; Spierings, E. Computational Approaches to Facilitate Epitope-Based HLA Matching in Solid Organ Transplantation. J. Immunol. Res. 2017, 2017, 9130879. [Google Scholar] [CrossRef]
- Marlais, M.; Hudson, A.; Pankhurst, L.; Fuggle, S.V.; Marks, S.D. Living Donation Has a Greater Impact on Renal Allograft Survival Than HLA Matching in Pediatric Renal Transplant Recipients. Transplantation 2016, 100, 2717–2722. [Google Scholar] [CrossRef]
- Opelz, G.; Döhler, B.; Middleton, D.; Süsal, C.; Report, A.C.T.S. HLA Matching in Pediatric Kidney Transplantation: HLA Poorly Matched Living Donor Transplants versus HLA Well-Matched Deceased Donor Transplants. Transplantation 2017, 101, 2789–2792. [Google Scholar] [CrossRef]
- Rana Magar, R.; Knight, S.; Stojanovic, J.; Marks, S.D.; Lafranca, J.A.; Turner, S.; Dor, F.; Pengel, L.H.M. Is Preemptive Kidney Transplantation Associated with Improved Outcomes when Compared to Non-preemptive Kidney Transplantation in Children? A Systematic Review and Meta-Analysis. Transpl. Int. 2022, 35, 10315. [Google Scholar] [CrossRef]
- Xiang, F.F.; Zhu, J.M.; Cao, X.S.; Shen, B.; Zou, J.Z.; Liu, Z.H.; Zhang, H.; Teng, J.; Liu, H.; Ding, X.Q. Lymphocyte depletion and subset alteration correlate to renal function in chronic kidney disease patients. Ren. Fail 2016, 38, 7–14. [Google Scholar] [CrossRef]
- Wallis, C.B.; Samy, K.P.; Roth, A.E.; Rees, M.A. Kidney paired donation. Nephrol. Dial. Transpl. 2011, 26, 2091–2099. [Google Scholar] [CrossRef] [PubMed]
- Niemann, M.; Lachmann, N.; Geneugelijk, K.; Spierings, E. Computational Eurotransplant kidney allocation simulations demonstrate the feasibility and benefit of T-cell epitope matching. PLoS Comput. Biol. 2021, 17, e1009248. [Google Scholar] [CrossRef] [PubMed]
- Sapir-Pichhadze, R.; Tinckam, K.; Quach, K.; Logan, A.G.; Laupacis, A.; John, R.; Beyene, J.; Kim, S.J. HLA-DR and -DQ Eplet Mismatches and Transplant Glomerulopathy: A Nested Case–Control Study. Am. J. Transplant. 2015, 15, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, S.; Iwasaki, K.; Tomosugi, T.; Niemann, M.; Spierings, E.; Miwa, Y.; Horimi, K.; Takeda, A.; Goto, N.; Narumi, S.; et al. Analysis of T and B Cell Epitopes to Predict the Risk of de novo Donor-Specific Antibody (DSA) Production After Kidney Transplantation: A Two-Center Retrospective Cohort Study. Front. Immunol. 2020, 11, 2000. [Google Scholar] [CrossRef]
- Ming, Y.; Peng, B.; Guo, X.; Luo, W.; Shao, M.; Cheng, K.; Luo, Q.; Zou, Y. Posttransplant-Alloantibodies Against MICA Antigens Associated With Decreased Long-Term Allograft Survival of Kidney Transplant Recipients. Transpl. Proc. 2022, 54, 1801–1808. [Google Scholar] [CrossRef]
- Lemy, A.; Andrien, M.; Lionet, A.; Labalette, M.; Noel, C.; Hiesse, C.; Delahousse, M.; Suberbielle-Boissel, C.; De Meyer, M.; Latinne, D.; et al. Posttransplant major histocompatibility complex class I chain-related gene A antibodies and long-term graft outcomes in a multicenter cohort of 779 kidney transplant recipients. Transplantation 2012, 93, 1258–1264. [Google Scholar] [CrossRef]
- Carapito, R.; Aouadi, I.; Verniquet, M.; Untrau, M.; Pichot, A.; Beaudrey, T.; Bassand, X.; Meyer, S.; Faucher, L.; Posson, J.; et al. The MHC class I MICA gene is a histocompatibility antigen in kidney transplantation. Nat. Med. 2022, 28, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Stastny, P.; Süsal, C.; Döhler, B.; Opelz, G. Antibodies against MICA antigens and kidney-transplant rejection. N. Engl. J. Med. 2007, 357, 1293–1300. [Google Scholar] [CrossRef] [PubMed]

| Sex ratio F/M | 30/25 |
| Age at the transplantation (years) | 12.07 ± 3.44 |
| Preemptive transplantation | 10 (18.1%) |
| Type of dialysis (DP/HD) | 15/30 |
| Living donor | 22 (12.1%) |
| Split HLA mismatches (ABDR) | 4 (2–6) |
| PIRCHE II score | 66.31 ± 35.66 |
| Split epitopes mismatches (ABDR) (HLA matchmaker) | 10.94 ± 5.98 |
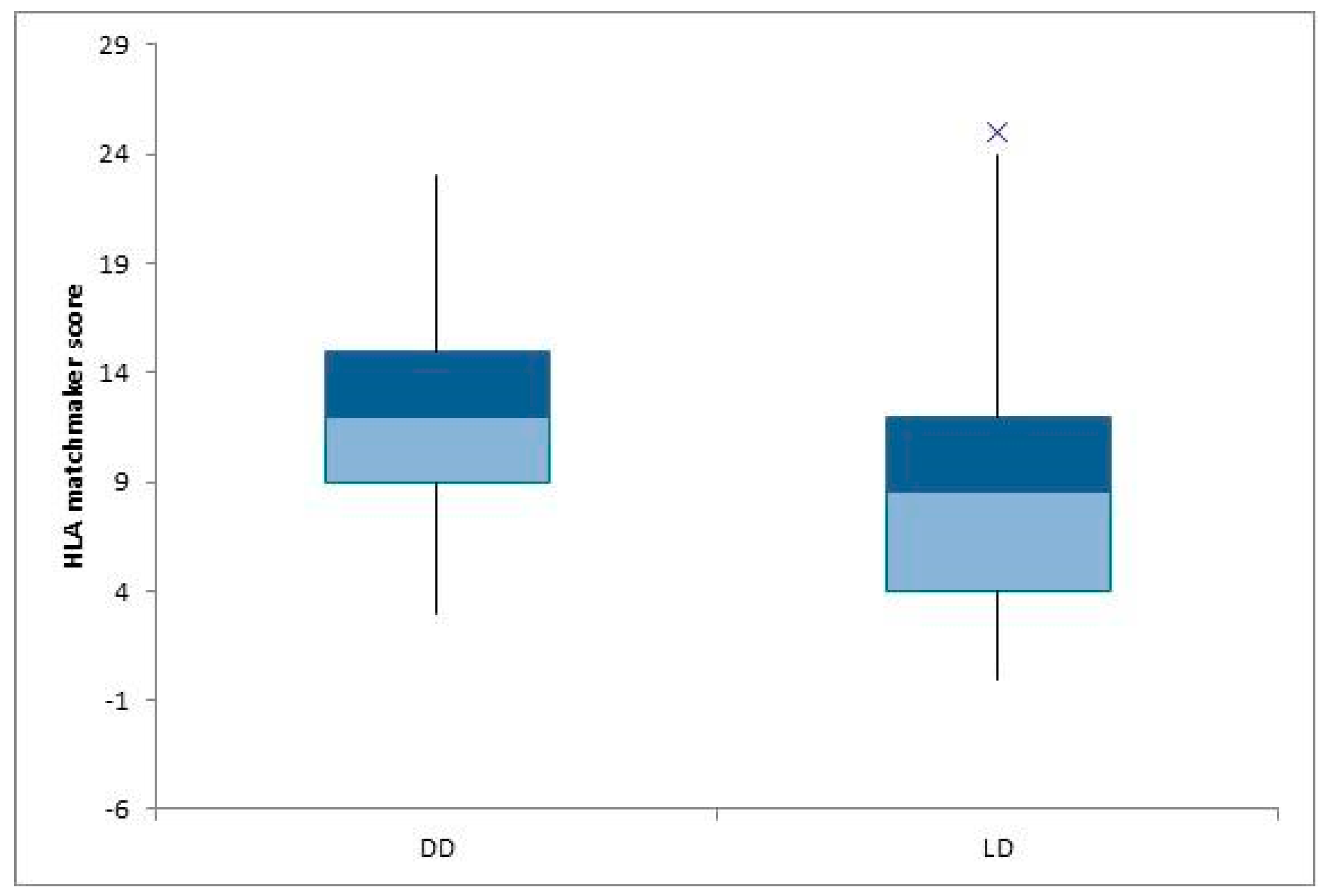
| Deceased Donors (DD) n = 33 | Living Donors (LD) n = 22 | p Value | |
|---|---|---|---|
| Sex ratio F/M | 18/15 | 12/10 | - |
| Age at the transplantation (years) | 11.93 ± 3.85 | 12.29 ± 2.93 | - |
| Preemptive transplantation | 4 (12.12%) | 6 (27.27%) | - |
| Split HLA mismatches (ABDR) | 4.81 | 3.23 | |
| PIRCHE II score | 71.84 ± 37.56 | 57.43 ± 30.6 | NS |
| Split epitopes mismatches (ABDR) (HLA matchmaker) | 12.33 ± 5.41 | 8.86 ± 6.33 | 0.04 |
| References | Age at Transplantation | Number of Patients, Sex Ratio | Mean Period of Follow-Up | % dnDSA+ | Antigenic Incompatibility | Epitopic Incompatibility | Type of Donor (DD/LD) |
|---|---|---|---|---|---|---|---|
| Răchișan et al., 2020 [15] | 11.2 ± 3.9 | 70 (44M/26F) | 3.5 years | 10% | 4.98 ± 1.43 | 15.5 | 60/10 |
| Branco et al., 2013 [16] | 13 | 124 (70M/54F) | 10.16 years | N/A | N/A | N/A | 111/13 |
| Kausman et al., 2016 [17] | 13.02 ± 1.25 | 35 (19M/16F) | 1 years | N/A | 4 | 50 | 27/8 |
| Bryan et al., 2016 [18] | 14.1 ± 6.8 | 16 (10M/6F) | N/A | N/A | N/A | 10 DR 17 DQ | 15/1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aldea, P.L.; Santionean, M.D.; Elec, A.; Munteanu, A.; Antal, O.; Loga, L.; Moisoiu, T.; Elec, F.I.; Delean, D.; Bulata, B.; et al. An Integrated Approach Using HLAMatchmaker and Pirche II for Epitopic Matching in Pediatric Kidney Transplant—A Romanian Single-Center Study. Children 2023, 10, 1756. https://doi.org/10.3390/children10111756
Aldea PL, Santionean MD, Elec A, Munteanu A, Antal O, Loga L, Moisoiu T, Elec FI, Delean D, Bulata B, et al. An Integrated Approach Using HLAMatchmaker and Pirche II for Epitopic Matching in Pediatric Kidney Transplant—A Romanian Single-Center Study. Children. 2023; 10(11):1756. https://doi.org/10.3390/children10111756
Chicago/Turabian StyleAldea, Paul Luchian, Maria Diana Santionean, Alina Elec, Adriana Munteanu, Oana Antal, Luminita Loga, Tudor Moisoiu, Florin Ioan Elec, Dan Delean, Bogdan Bulata, and et al. 2023. "An Integrated Approach Using HLAMatchmaker and Pirche II for Epitopic Matching in Pediatric Kidney Transplant—A Romanian Single-Center Study" Children 10, no. 11: 1756. https://doi.org/10.3390/children10111756
APA StyleAldea, P. L., Santionean, M. D., Elec, A., Munteanu, A., Antal, O., Loga, L., Moisoiu, T., Elec, F. I., Delean, D., Bulata, B., & Rachisan, A. L. (2023). An Integrated Approach Using HLAMatchmaker and Pirche II for Epitopic Matching in Pediatric Kidney Transplant—A Romanian Single-Center Study. Children, 10(11), 1756. https://doi.org/10.3390/children10111756






