Platelet miRNA Biosignature Discriminates between Dementia with Lewy Bodies and Alzheimer’s Disease
Abstract
:1. Introduction
2. Materials and Methods
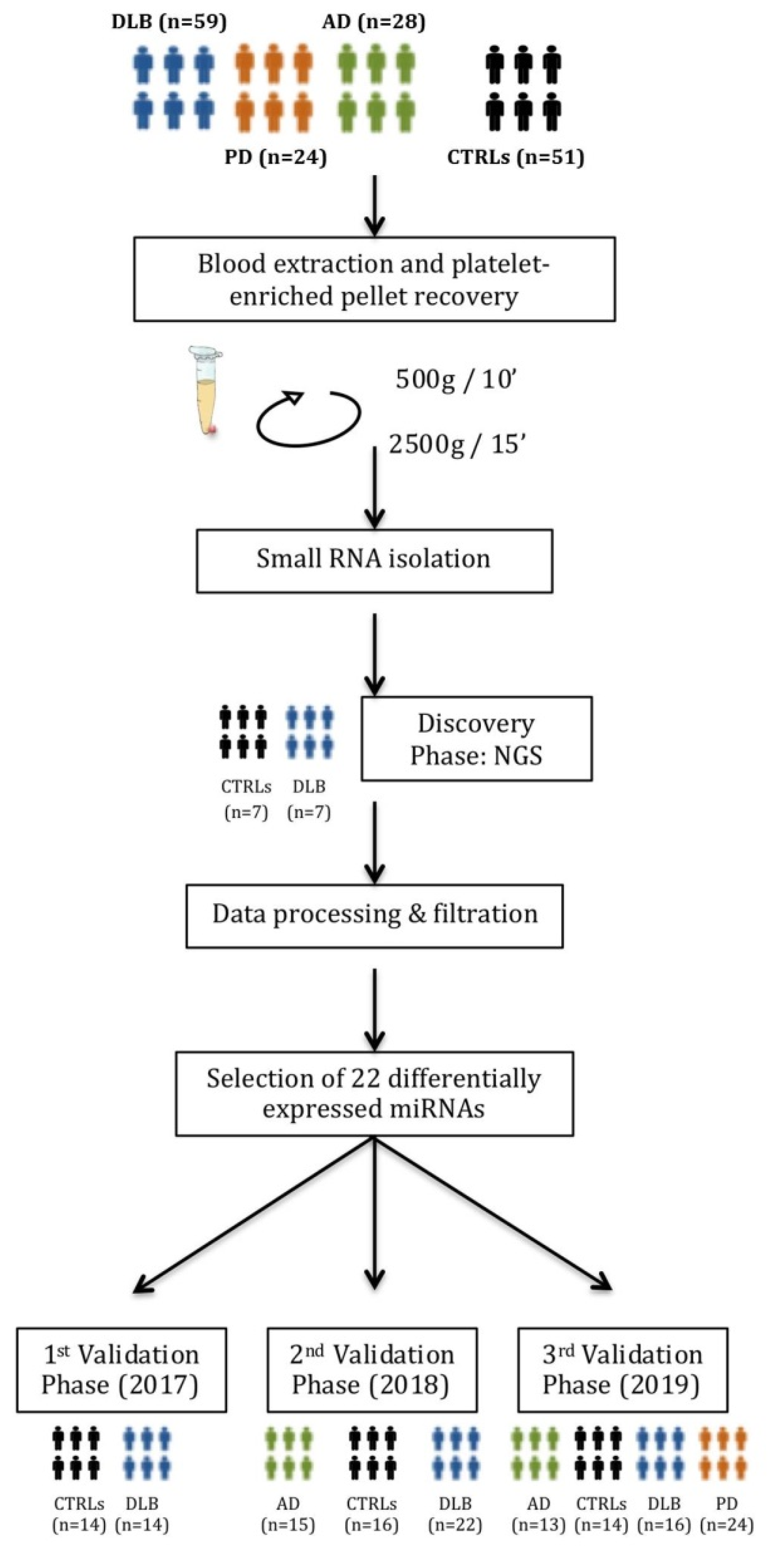
2.1. Participants
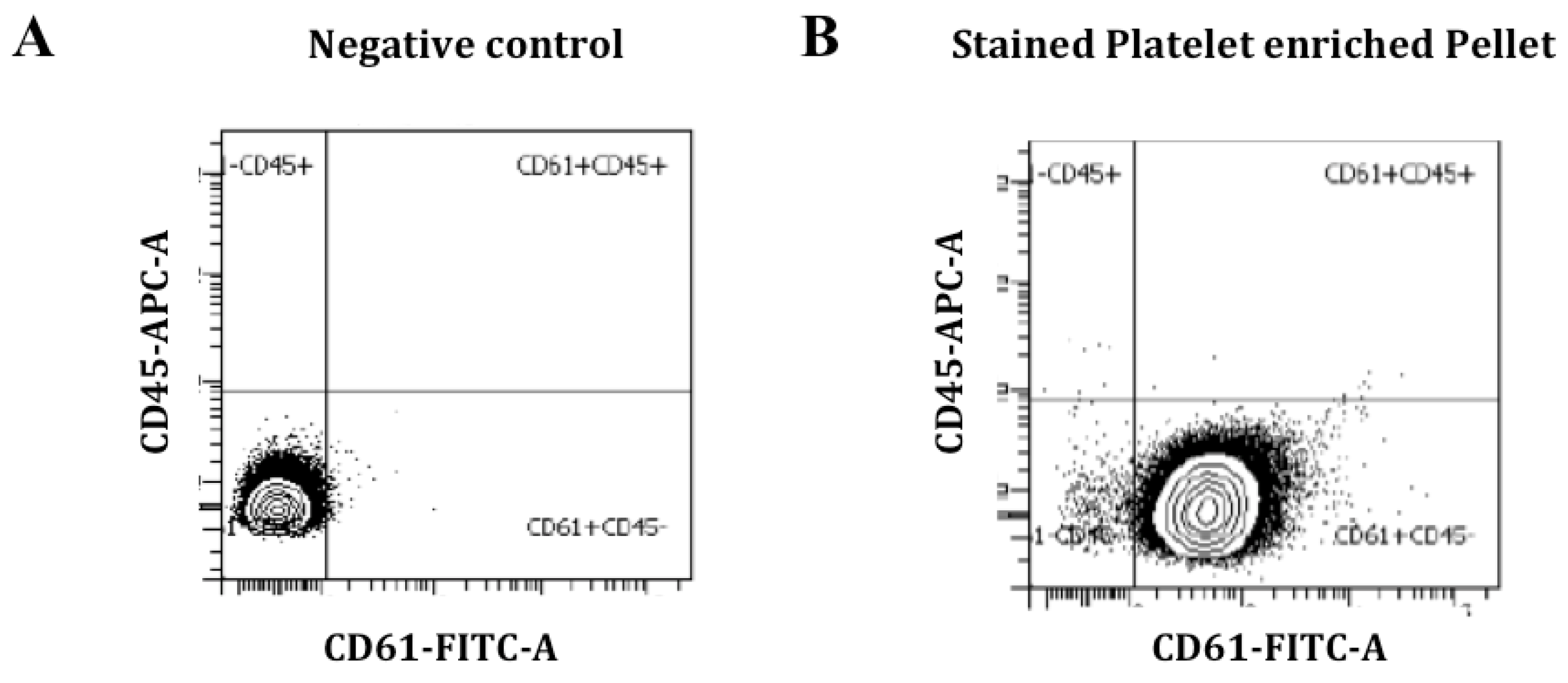
2.2. Blood Collection, Purification and Characterization of Platelets
2.3. Purification of Platelet-Derived Small RNA
2.4. MiRNA Isolation from Whole Blood
2.5. Discovery Phase: miRNA Sequencing and Sequencing Data Analysis
2.6. Validation Phase: Reverse Transcription and Quantitative Real-Time PCR
2.7. Statistical Analysis
2.8. miRNA Target Prediction and Analysis
3. Results
3.1. Demographic and Clinical Data
3.2. Platelet Characterization and miRNA Profile Discovery
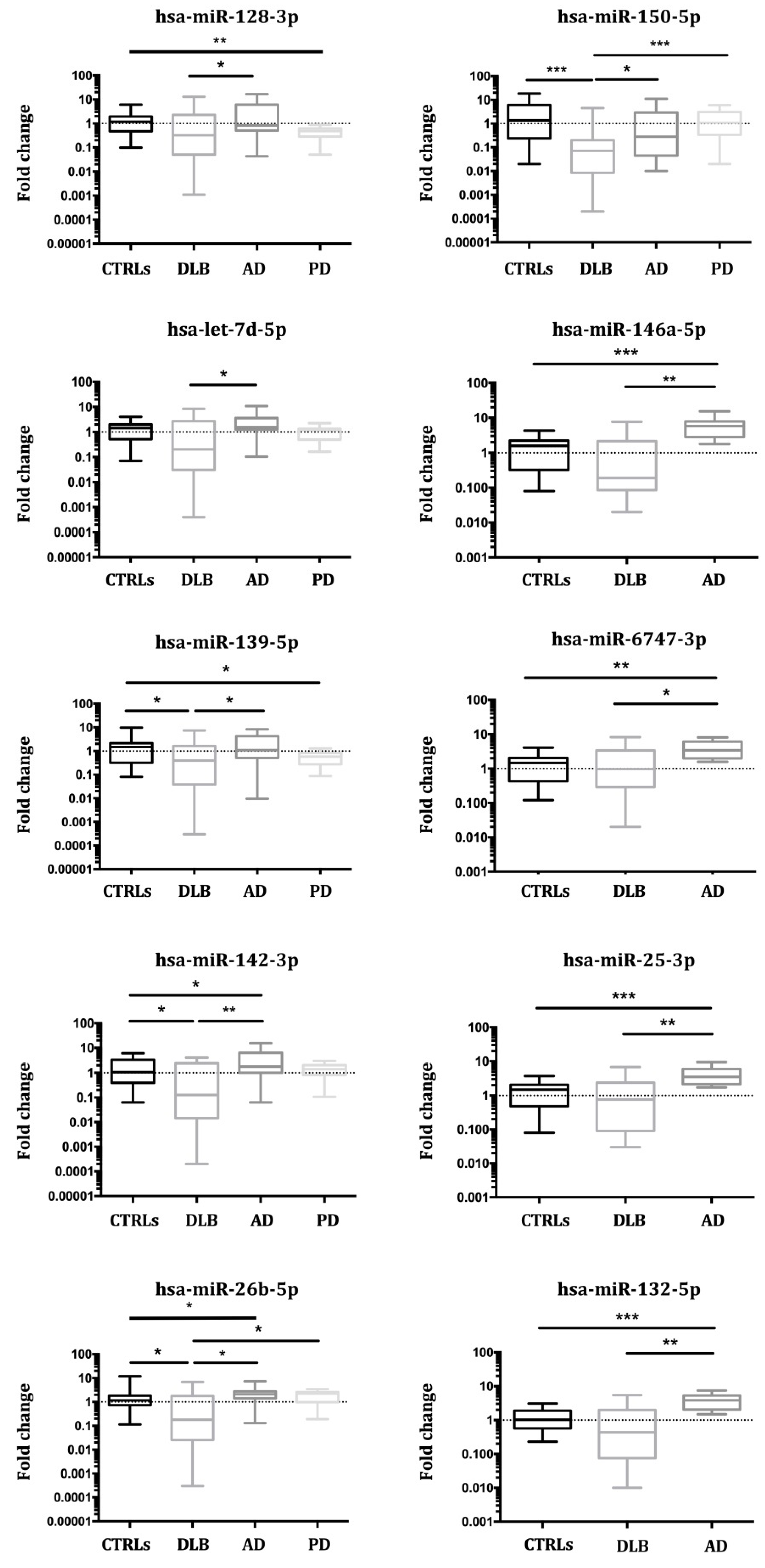
3.3. Validation of miRNA Expression
3.3.1. Study I (2017)
3.3.2. Study II (2018)
3.3.3. Study III (2019)
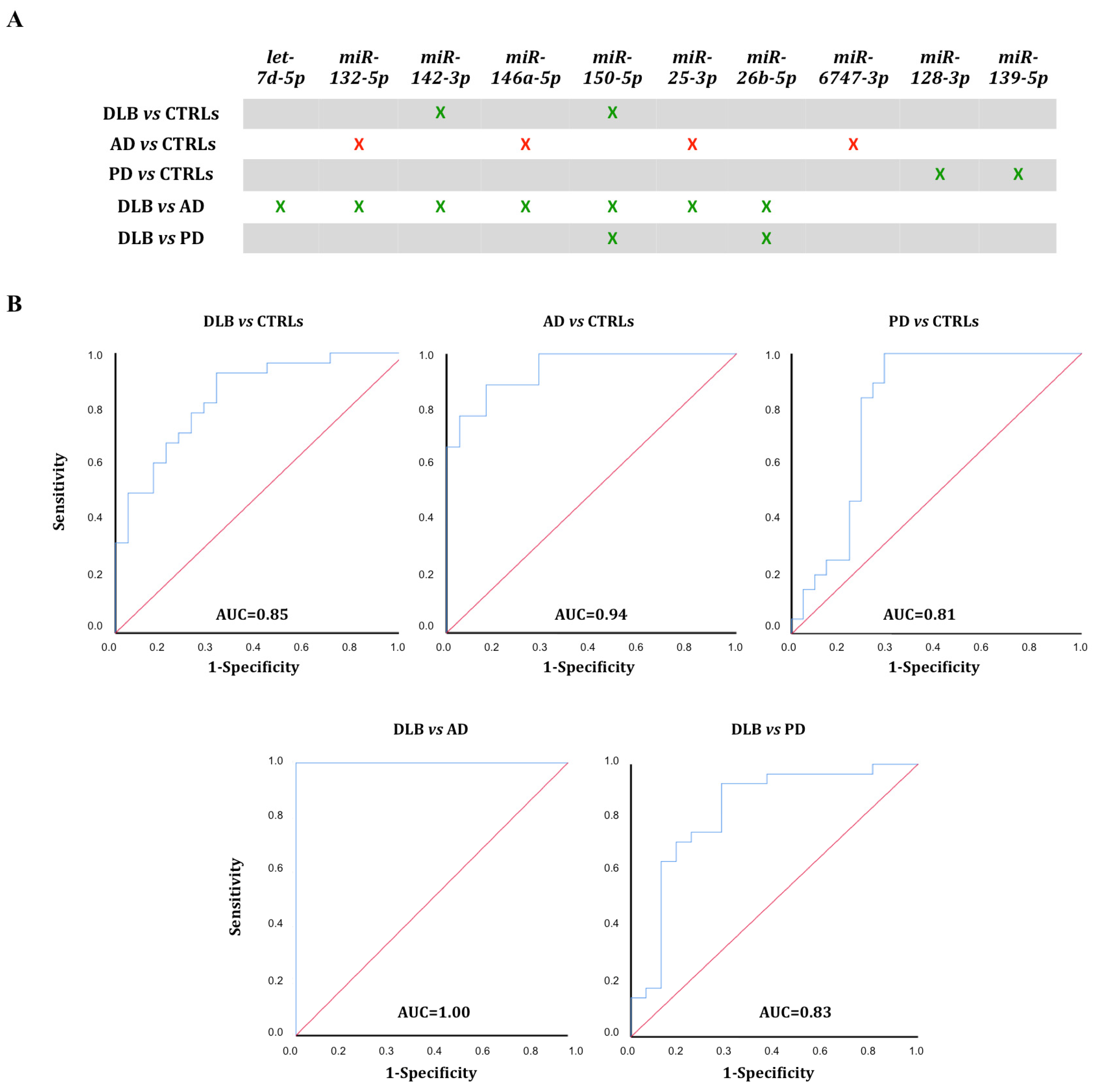
3.4. ROC Curve Analysis
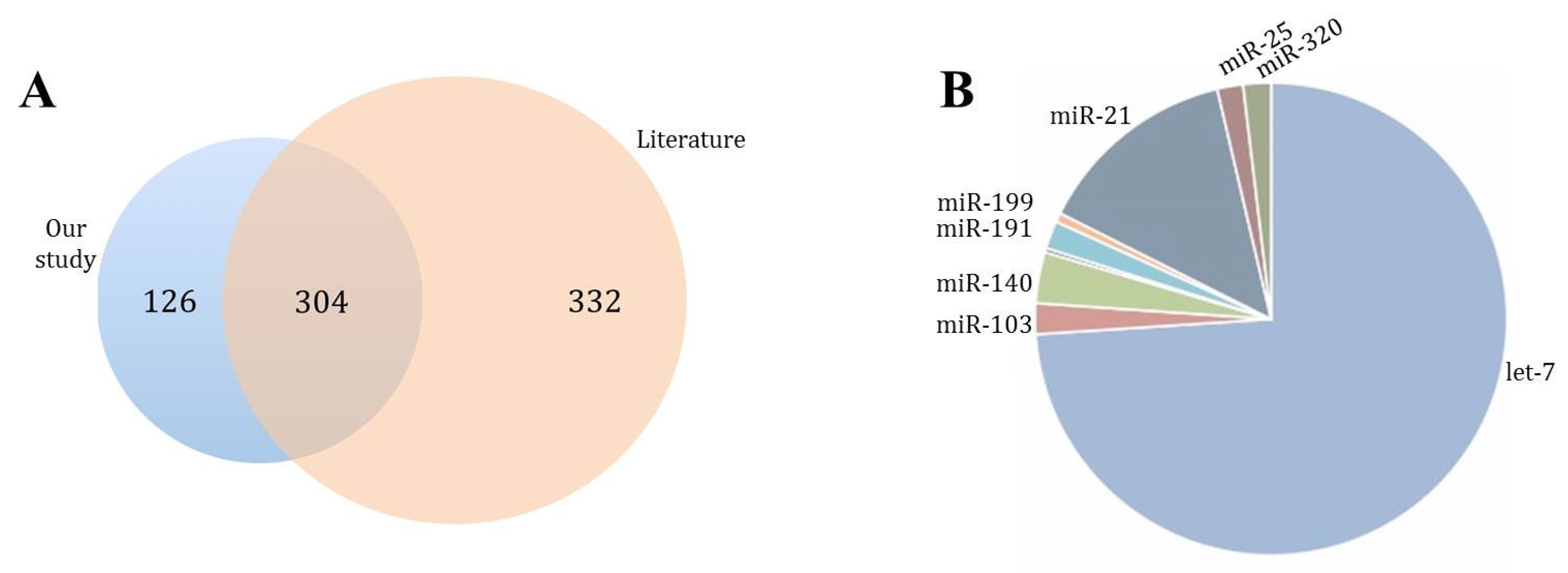
3.5. miRNA Expression in Whole Blood
3.6. miRNA Target Prediction
4. Discussion
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ingelsson, M. Alpha-synuclein Oligomers- Neurotoxic Molecules in Parkinson’s Disease and Other Lewy Body disorders. Front. Neurosci. 2016, 10, 408. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jellinger, K.A. Dementia with Lewy bodies and Parkinson’s disease-dementia: Current concepts and controversies. J. Neural. Transm. 2018, 125, 615–650. [Google Scholar] [CrossRef] [PubMed]
- Colom-Cadena, M.; Grau-Rivera, O.; Planellas, L.; Cerquera, C.; Morenas, E.; Helgueta, S.; Muñoz, L.; Kulisevsky, J.; Martí, M.J.; Tolosa, E.; et al. Regional overlap of pathologies in Lewy Body Disorders. J. Neuropathol. Exp. Neurol. 2017, 76, 216–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McKeith, I.G.; Boeve, B.F.; Dickson, D.W.; Halliday, G.; Taylor, J.P.; Weintraub, D.; Aarsland, D.; Galvin, J.; Attems, J.; Ballard, C.G.; et al. Diagnosis and management of dementia with Lewy bodies. Fourth consensus report of the DLB Consortium. Neurology 2017, 89, 88–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, R.M.; Paterson, R.W.; Warren, J.D.; Zetterberg, H.; O’Brien, J.T.; Fox, N.C.; Halliday, G.M.; Schott, J.M. Biomarkers in dementia, clinical utility and new directions. J. Neurol. Neurosurg. Psychiatry 2014, 85, 1426–1434. [Google Scholar] [CrossRef] [Green Version]
- Zetterberg, H.; Wilson, D.; Andreasson, U.; Minthon, L.; Blennow, K.; Randall, J.; Hansson, O. Plasma tau levels in Alzheimer’s disease. Alzheimers Res. Ther. 2013, 5, 9. [Google Scholar] [CrossRef]
- Parnetti, L.; Paciotti, S.; Farotti, L.; Bellomo, G.; Sepe, F.N.; Eusebi, P. Parkinson’s and Lewy body dementia CSF biomarkers. Clin. Chim. Acta 2019, 495, 318–325. [Google Scholar] [CrossRef] [PubMed]
- Wojsiat, J.; Laskowska-Kaszub, K.; Mietelska-Porowska, A.; Wojda, U. Search for Alzheimer’s disease biomarkers in blood cells, hypotheses-driven approach. Biomark. Med. 2017, 11, 917–931. [Google Scholar] [CrossRef] [Green Version]
- Italiano, J.E., Jr.; Richardson, J.L.; Patel-Hett, S.; Battinelli, E.; Zaslavsky, A.; Short, S.; Ryeom, S.; Folkman, J.; Klement, G.L. Angiogenesis is regulated by a novel mechanism, pro- and antiangiogenic proteins are organized into separate platelet alpha granules and differentially released. Blood 2008, 111, 1227–1233. [Google Scholar] [CrossRef] [Green Version]
- Milioli, M.; Ibáñez-Vea, M.; Sidoli, S.; Palmisano, G.; Careri, M.; Larsen, M.R. Quantitative proteomics analysis of platelet-derived microparticles reveals distinct protein signatures when stimulated by different physiological agonists. J. Proteom. 2015, 121, 56–66. [Google Scholar] [CrossRef]
- Leiter, O.; Walker, T.L. Platelets, The missing link between the blood and brain? Prog. Neurobiol. 2019, 183, 101695. [Google Scholar] [CrossRef]
- Bruchova, H.; Merkerova, M.; Prchal, J.T. Aberrant expression of microRNA in polycythemia vera. Haematologica 2008, 93, 1009–1016. [Google Scholar] [CrossRef] [Green Version]
- Landry, P.; Plante, I.; Ouellet, D.L.; Perron, M.P.; Rousseau, G.; Provost, P. Existence of a microRNA pathway in anucleate platelets. Nat. Struct. Mol. Biol. 2009, 16, 961–966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edelstein, L.C.; Bray, P.F. MicroRNAs in platelet production and activation. Blood 2011, 117, 5289–5296. [Google Scholar] [CrossRef] [PubMed]
- Plé, H.; Landry, P.; Benham, A.; Coarfa, C.; Gunaratne, P.H.; Provost, P. The repertoire and features of human platelet microRNAs. PLoS ONE 2012, 7, e50746. [Google Scholar] [CrossRef] [PubMed]
- Gowert, N.S.; Donner, L.; Chatterjee, M.; Eisele, Y.S.; Towhid, S.T.; Münzer, P.; Walker, B.; Ogorek, I.; Borst, O.; Grandoch, M.; et al. Blood platelets in the progression of Alzheimer’s disease. PLoS ONE 2014, 9, e90523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wojsiat, J.; Laskowska-Kaszub, K.; Alquézar, C.; Białopiotrowicz, E.; Esteras, N.; Zdioruk, M.; Martin-Requero, A.; Wojda, U. Familial Alzheimer’s disease lymphocytes respond differently than sporadic cells to oxidative stress, upregulated p53-p21 signaling linked with presenilin 1 mutants. Mol. Neurobiol. 2016, 54, 5683–5698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, S.; Zhao, J.; Zhang, T.; Guo, C. Increased apoptosis in the platelets of patients with Alzheimer’s disease and amnestic mild cognitive impairment. Clin. Neurol. Neurosurg. 2016, 143, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Michell, A.W.; Luheshi, L.M.; Barker, R.A. Skin and platelet alpha-synuclein as peripheral biomarkers of Parkinson’s disease. Neurosci. Lett. 2005, 381, 294–298. [Google Scholar] [CrossRef]
- Leiter, O.; Seidemann, S.; Overall, R.W.; Ramasz, B.; Rund, N.; Schallenberg, S.; Grinenko, T.; Wielockx, B.; Kempermann, G.; Walker, T.L. Exercise-Induced Activated Platelets Increase Adult Hippocampal Precursor Proliferation and Promote Neuronal Differentiation. Stem Cell Rep. 2019, 12, 667–679. [Google Scholar] [CrossRef] [Green Version]
- Behari, M.; Shrivastava, M. Role of platelets in neurodegenerative diseases, a universal pathophysiology. Int. J. Neurosci. 2013, 123, 287–299. [Google Scholar] [CrossRef] [PubMed]
- McKeith, I.G.; Dickson, D.W.; Lowe, J.; Emre, M.; O’Brien, J.T.; Feldman, H.; Cummings, J.; Duda, J.E.; Lippa, C.; Perry, E.K.; et al. Diagnosis and management of dementia with Lewy bodies, third report of the DLB Consortium. Neurology 2005, 65, 1863–1872. [Google Scholar] [CrossRef] [Green Version]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease, recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef] [Green Version]
- Hughes, A.J.; Daniel, S.E.; Kilford, L.; Lees, A.J. Accuracy of clinical diagnosis of idiopathic Parkinson’s disease, a clinic-pathological study of 100 cases. J. Neurol. Neurosurg. Psychiatry 1992, 55, 181–184. [Google Scholar] [CrossRef] [Green Version]
- Lynöe, N.; Sandlund, M.; Dahlqvist, G.; Jacobsson, L. Informed consent, study of quality of information given to participants in a clinical trial. BMJ 1991, 303, 610–613. [Google Scholar] [CrossRef] [Green Version]
- György, B.; Pálóczi, K.; Kovács, A.; Barabás, E.; Bekő, G.; Várnai, K.; Pállinger, É.; Szabó-Taylor, K.; Szabó, T.G.; Kiss, A.A.; et al. Improved circulating microparticle analysis in acid-citrate dextrose (ACD) anticoagulant tube. Thromb. Res. 2014, 133, 285–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sáenz-Cuesta, M.; Arbelaiz, A.; Oregi, A.; Irizar, H.; Osorio-Querejeta, I.; Muñoz-Culla, M.; Banales, J.M.; Falcón-Pérez, J.M.; Olascoaga, J.; Otaegui, D. Methods for extracellular vesicles isolation in a hospital setting. Front. Immunol. 2015, 6, 50. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bishop, C.M. Pattern Recognition and Machine Learning, 2nd ed.; Springer: New York, NY, USA, 2011. [Google Scholar]
- Madsen, H.; Thyregod, P. Introduction to General and Generalized Linear Models, 1st ed.; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Russell, S.; Norwig, P. Artificial Intelligence: A Modern Approach, 3rd ed.; Pearson Education: London, UK, 2010. [Google Scholar]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1118. [Google Scholar] [CrossRef]
- Chou, C.H.; Shrestha, S.; Yang, C.D.; Chang, N.W.; Lin, Y.L.; Liao, K.W.; Huang, W.C.; Sun, T.H.; Tu, S.J.; Lee, W.H.; et al. miRTarBase update 2018, a resource for experimentally validated microRNA-target interactions. Nucleic Acids Res. 2018, 46, D296–D302. [Google Scholar] [CrossRef]
- Andrés-León, E.; Gómez-López, G.; Pisano, D.G. Prediction of miRNA-mRNA Interactions Using miRGate. Methods Mol. Biol. 2017, 1580, 225–237. [Google Scholar]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017, quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Pundir, S.; Martin, M.J.; O’Donovan, C. UniProt Protein Knowledgebase. Methods Mol. Biol. 2017, 1558, 41–55. [Google Scholar]
- Osman, A.; Falker, K. Characterization of human platelet microRNA by quantitative PCR coupled with an annotation network for predicted target genes. Platelets 2011, 22, 433–441. [Google Scholar] [CrossRef]
- Nussbacher, J.K.; Tabet, R.; Yeo, G.W.; Lagier-Tourenne, C. Disruption of RNA Metabolism in Neurological Diseases and Emerging Therapeutic Interventions. Neuron 2019, 102, 294–320. [Google Scholar] [CrossRef] [PubMed]
- Pantano, L.; Friedländer, M.R.; Escaramís, G.; Lizano, E.; Pallarès-Albanell, J.; Ferrer, I.; Estivill, X.; Martí, E. Specific small-RNA signatures in the amygdala at premotor and motor stages of Parkinson’s disease revealed by deep sequencing analysis. Bioinformatics 2016, 32, 673–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Apicco, D.J.; Ash, P.E.A.; Maziuk, B.; LeBlang, C.; Medalla, M.; Al Abdullatif, A.; Ferragud, A.; Botelho, E.; Balance, H.I.; Dhawan, U.; et al. Reducing the RNA binding protein TIA1 protects against tau-mediated neurodegeneration in vivo. Nat. Neurosci. 2018, 21, 72–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lachke, S.A.; Alkuraya, F.S.; Kneeland, S.C.; Ohn, T.; Aboukhalil, A.; Howell, G.R.; Saadi, I.; Cavallesco, R.; Yue, Y.; Tsaim, A.C. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science 2011, 331, 1571–1576. [Google Scholar] [CrossRef] [Green Version]
- Wolozin, B.; Ivanov, P. Stress granules and neurodegeneration. Nat. Rev. Neurosci. 2019, 20, 649–666. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Santiago, R.; Martín-Flores, N.; Antonelli, F.; Cerquera, C.; Moreno, V.; Bandres-Ciga, S.; Manduchi, E.; Tolosa, E.; Singleton, A.B.; Moore, J.H.; et al. SNCA and mTOR Pathway Single Nucleotide Polymorphisms Interact to Modulate the Age at Onset of Parkinson’s Disease. Mov. Disord. 2019, 34, 1333–1344. [Google Scholar] [CrossRef] [PubMed]
- Martín-Flores, N.; Fernández-Santiago, R.; Antonelli, F.; Cerquera, C.; Moreno, V.; Martí, M.J.; Ezquerra, M.; Malagelada, C. MTOR Pathway-Based Discovery of Genetic Susceptibility to L-DOPA-Induced Dyskinesia in Parkinson’s Disease Patients. Mol. Neurobiol. 2019, 56, 2092–2100. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Yang, C.; Iyaswamy, A.; Krishnamoorthi, S.; Sreenivasmurthy, S.G.; Liu, J.; Wang, Z.; Tong, B.C.; Song, J.; Lu, J.; et al. Balancing mTOR Signaling and Autophagy in the Treatment of Parkinson’s Disease. Int. J. Mol. Sci. 2019, 20, 728. [Google Scholar] [CrossRef] [Green Version]
- Bram, J.M.F.; Talib, L.L.; Joaquim, H.P.G.; Sarno, T.A.; Gattaz, W.F.; Forlenza, O.V. Protein levels of ADAM10, BACE1, and PSEN1 in platelets and leukocytes of Alzheimer’s disease patients. Eur. Arch. Psychiatry Clin. Neurosci. 2019, 269, 963–972. [Google Scholar] [CrossRef] [PubMed]
- Koçer, A.; Yaman, A.; Niftaliyev, E.; Dürüyen, H.; Eryılmaz, M.; Koçer, E. Assessment of platelet indices in patients with neurodegenerative diseases, mean platelet volume was increased in patients with Parkinson’s disease. Curr. Gerontol. Geriatr. Res. 2013, 2013, 986254. [Google Scholar] [CrossRef]
- Kucheryavykh, L.Y.; Dávila-Rodríguez, J.; Rivera-Aponte, D.E.; Zueva, L.V.; Washington, A.V.; Sanabria, P.; Inyushin, M.Y. Platelets are responsible for the accumulation of β-amyloid in blood clots inside and around blood vessels in mouse brain after thrombosis. Brain Res. Bull. 2017, 128, 98–105. [Google Scholar] [CrossRef] [PubMed]

| DLB (n = 59) | AD (n = 28) | PD (n = 24) | CTRLs (n = 51) | p 1 | |
|---|---|---|---|---|---|
| Mean age, y 2 (age range, y) | 75.1 (57–89) | 73.9 (65–82) | 66.9 (42–87) | 72.0 (61–85) | 0.0067 |
| Gender (male/female ratio) | 1.5:1 | 1:1.5 | 1.5:1 | 1:1.5 | 0.493 |
| Disease duration 3, y (range, y) | 4.2 (1.2–10.1) | 3.4 (0.8–5.6) | 5.2 (1.8–7.6) | - | 0.068 |
| MMSE 4, mean (range) | 15.1 (3–28) | 20.3 (6–28) | - | - | 0.189 |
| UPDRS-III 5, mean (range) | - | - | 20.9 (5–39) | - | - |
| GDS 6, mean (range) | - | 4.1 (3–6) | - | - | - |
| Parkinsonism, n (%) | 47 (79.7%) | - | - | - | - |
| Positive DAT imaging, n (%) | 55 (93.2%) | - | - | - | - |
| miRNA | Expr Change 1 | Dev Range 2 | p-Value 3 |
|---|---|---|---|
| 1343-3p | 0.31 | 0.11–0.88 | 0.24 |
| 191-3p | 0.34 | 0.13–0.89 | 0.25 |
| 6747-3p | 0.33 | 0.14–0.74 | 0.033 |
| 504-5p | 0.6 | 0.55–0.66 | 0.25 |
| 6741-3p | 0.32 | 0.16–0.66 | 0.17 |
| 128-3p | 0.17 | 0.03–0.71 | 0.039 |
| 1468-5p | 0.37 | 0.16–0.89 | 0.28 |
| 139-5p | 0.26 | 0.09–0.69 | 0.029 |
| 7d-5p | 0.16 | 0.05–0.55 | 0.015 |
| 7d-3p | 0.34 | 0.12–0.95 | 0.21 |
| 142-3p | 0.11 | 0.03–0.47 | 0.015 |
| 132-5p | 0.21 | 0.06–0.70 | 0.018 |
| 150-5p | 0.03 | 0.02–0.04 | <0.0001 |
| 23a-5p | 0.58 | 0.41–0.81 | 0.31 |
| 26b-5p | 0.16 | 0.04–0.62 | 0.017 |
| 1301-3p | 0.28 | 0.06–1.28 | 0.24 |
| 625-3p | 0.32 | 0.07–0.41 | 0.14 |
| 146a-5p | 0.16 | 0.05–0.53 | 0.017 |
| 25-3p | 0.20 | 0.06–0.66 | 0.034 |
| 877-3p | 0.34 | 0.14–0.84 | 0.22 |
| 1908-5p | 0.30 | 0.09–094 | 0.21 |
| 744-5p | 0.21 | 0.08–0.59 | 0.19 |
| miRNA | DLB vs. CTRLs | AD vs. CTRLs | DLB vs. AD |
|---|---|---|---|
| n | 22 vs. 16 | 15 vs. 16 | 22 vs. 15 |
| let-7d-5p | |||
| expr change 1 | 0.21 | 1.40 | 0.14 |
| dev range 2 | 0.024–1.73 | 1.12–1.75 | 0.02–0.98 |
| p-value 3 | 0.20 | 0.19 | 0.042 |
| miR-128-3p | |||
| expr change | 0.16 | 1.25 | 0.13 |
| dev range | 0.013–2.07 | 1.07–1.47 | 0.012–1.41 |
| p-value | 0.043 | 0.19 | 0.009 |
| miR-132-5p | |||
| expr change | 0.23 | 1.50 | 0.15 |
| dev range | 0.041–1.21 | 1.49–1.51 | 0.028–0.80 |
| p-value | 0.14 | 0.072 | 0.011 |
| miR-139-5p | |||
| expr change | 0.18 | 1.17 | 0.15 |
| dev range | 0.019–1.68 | 0.88–1.56 | 0.022–1.08 |
| p-value | 0.03 | 0.68 | 0.014 |
| miR-142-3p | |||
| expr change | 0.16 | 1.73 | 0.09 |
| dev range | 0.02–1.26 | 1.48–2.02 | 0.009–0.85 |
| p-value | 0.05 | 0.07 | 0.0026 |
| miR-146a-5p | |||
| expr change | 0.18 | 1.51 | 0.12 |
| dev range | 0.017–1.87 | 1.37–1.66 | 0.012–1.12 |
| p-value | 0.069 | 0.11 | 0.0089 |
| miR-150-5p | |||
| expr change | 0.011 | 0.34 | 0.031 |
| dev range | 0.003–0.034 | 0.29–0.40 | 0.008–0.118 |
| p-value | <0.0001 | 0.029 | <0.0001 |
| miR-25-3p | |||
| expr change | 0.20 | 1.15 | 0.17 |
| dev range | 0.026–1.52 | 0.94–1.41 | 0.02–1.08 |
| p-value | 0.03 | 0.85 | 0.025 |
| miR-26b-5p | |||
| expr change | 0.19 | 1.59 | 0.12 |
| dev range | 0.02–1.83 | 1.49–1.70 | 0.013–1.07 |
| p-value | 0.092 | 0.075 | 0.0071 |
| miR-6747-3p | |||
| expr change | 0.62 | 1.39 | 0.44 |
| dev range | 0.24–1.55 | 1.23–1.56 | 0.20–0.99 |
| p-value | 0.92 | 0.25 | 0.25 |
| miRNA | DLB vs. CTRLs | AD vs. CTRLs | PD vs. CTRLs | DLB vs. AD | DLB vs. PD |
|---|---|---|---|---|---|
| n | 59 vs. 51 | 28 vs. 51 | 24 vs. 51 | 59 vs. 28 | 59 vs. 24 |
| let-7d-5p | |||||
| expr change 1 | 0.25 | 1.81 | 0.98 | 0.14 | 0.28 |
| dev range 2 | 0.05–1.16 | 1.74–1.84 | 0.61–1.29 | 0.03–0.62 | 0.04–1.91 |
| p-value 3 | 0.08 | 0.102 | 0.17 | 0.006 | 0.15 |
| miR-128-3p | |||||
| expr change | 0.26 | 1.38 | 0.38 | 0.19 | 0.87 |
| dev range | 0.05–1.45 | 0.79–2.4 | 0.3–0.49 | 0.06–0.61 | 0.69–1.13 |
| p-value | 0.106 | 0.57 | 0.0007 | 0.05 | 0.89 |
| miR-132-5p | |||||
| expr change | 0.43 | 3.50 | 0.58 | 0.12 | 0.89 |
| dev range | 0.15–0.81 | 2.92–4.20 | 0.27–0.97 | 0.04–0.42 | 0.76–1.01 |
| p-value | 0.201 | <0.0001 | 0.18 | 0.001 | 0.81 |
| miR-139-5p | |||||
| expr change | 0.20 | 1.22 | 0.48 | 0.16 | 0.41 |
| dev range | 0.04–0.91 | 1.14–1.3 | 0.30–0.78 | 0.04–0.70 | 0.06–3.04 |
| p-value | 0.05 | 0.96 | 0.008 | 0.05 | 0.68 |
| miR-142-3p | |||||
| expr change | 0.14 | 2.09 | 1.10 | 0.07 | 0.12 |
| dev range | 0.02–0.78 | 1.99–2.21 | 0.80–1.52 | 0.01–0.35 | 0.02–0.97 |
| p-value | 0.007 | 0.05 | 0.65 | 0.0005 | 0.05 |
| miR-146a-5p | |||||
| expr change | 0.34 | 5.00 | 0.67 | 0.07 | 0.72 |
| dev range | 0.18–0.66 | 3.03–8.27 | 0.51–0.84 | 0.02–0.22 | 0.46–1.02 |
| p-value | 0.05 | 0.00013 | 0.41 | 0.0004 | 0.45 |
| miR-150-5p | |||||
| expr change | 0.04 | 0.36 | 0.87 | 0.10 | 0.04 |
| dev range | 0.02–0.07 | 0.28–0.46 | 0.49–1.53 | 0.07–0.17 | 0.01–0.14 |
| p-value | <0.0001 | 0.1 | 0.69 | 0.01 | <0.0001 |
| miR-25-3p | |||||
| expr change | 0.47 | 3.76 | 0.71 | 0.13 | 0.83 |
| dev range | 0.22–1.00 | 2.38–5.93 | 0.49–1.1 | 0.04–0.42 | 0.67–1.09 |
| p-value | 0.23 | 0.0003 | 0.76 | 0.002 | 0.49 |
| miR-26b-5p | |||||
| expr change | 0.18 | 1.65 | 1.54 | 0.09 | 0.11 |
| dev range | 0.03–1.07 | 1.12–1.80 | 1.24–1.92 | 0.01–0.68 | 0.02–0.86 |
| p-value | 0.05 | 0.05 | 0.05 | 0.001 | 0.008 |
| miR-6747-3p | |||||
| expr change | 0.76 | 3.50 | 1.23 | 0.22 | 0.56 |
| dev range | 0.35–1.63 | 2.40–5.10 | 1.02–1.46 | 0.07–0.68 | 0.52–0.64 |
| p-value | 0.71 | 0.0006 | 0.29 | 0.05 | 0.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gámez-Valero, A.; Campdelacreu, J.; Vilas, D.; Ispierto, L.; Gascón-Bayarri, J.; Reñé, R.; Álvarez, R.; Armengol, M.P.; Borràs, F.E.; Beyer, K. Platelet miRNA Biosignature Discriminates between Dementia with Lewy Bodies and Alzheimer’s Disease. Biomedicines 2021, 9, 1272. https://doi.org/10.3390/biomedicines9091272
Gámez-Valero A, Campdelacreu J, Vilas D, Ispierto L, Gascón-Bayarri J, Reñé R, Álvarez R, Armengol MP, Borràs FE, Beyer K. Platelet miRNA Biosignature Discriminates between Dementia with Lewy Bodies and Alzheimer’s Disease. Biomedicines. 2021; 9(9):1272. https://doi.org/10.3390/biomedicines9091272
Chicago/Turabian StyleGámez-Valero, Ana, Jaume Campdelacreu, Dolores Vilas, Lourdes Ispierto, Jordi Gascón-Bayarri, Ramón Reñé, Ramiro Álvarez, Maria P. Armengol, Francesc E. Borràs, and Katrin Beyer. 2021. "Platelet miRNA Biosignature Discriminates between Dementia with Lewy Bodies and Alzheimer’s Disease" Biomedicines 9, no. 9: 1272. https://doi.org/10.3390/biomedicines9091272
APA StyleGámez-Valero, A., Campdelacreu, J., Vilas, D., Ispierto, L., Gascón-Bayarri, J., Reñé, R., Álvarez, R., Armengol, M. P., Borràs, F. E., & Beyer, K. (2021). Platelet miRNA Biosignature Discriminates between Dementia with Lewy Bodies and Alzheimer’s Disease. Biomedicines, 9(9), 1272. https://doi.org/10.3390/biomedicines9091272







