Quantitative Approaches to Study Retinal Neurogenesis
Abstract
1. Introduction
2. Methods
2.1. Animals
2.2. Sample Preparation
2.3. Immunostaining and Mounting
2.4. Image Acquisition
2.5. Object Density Analysis
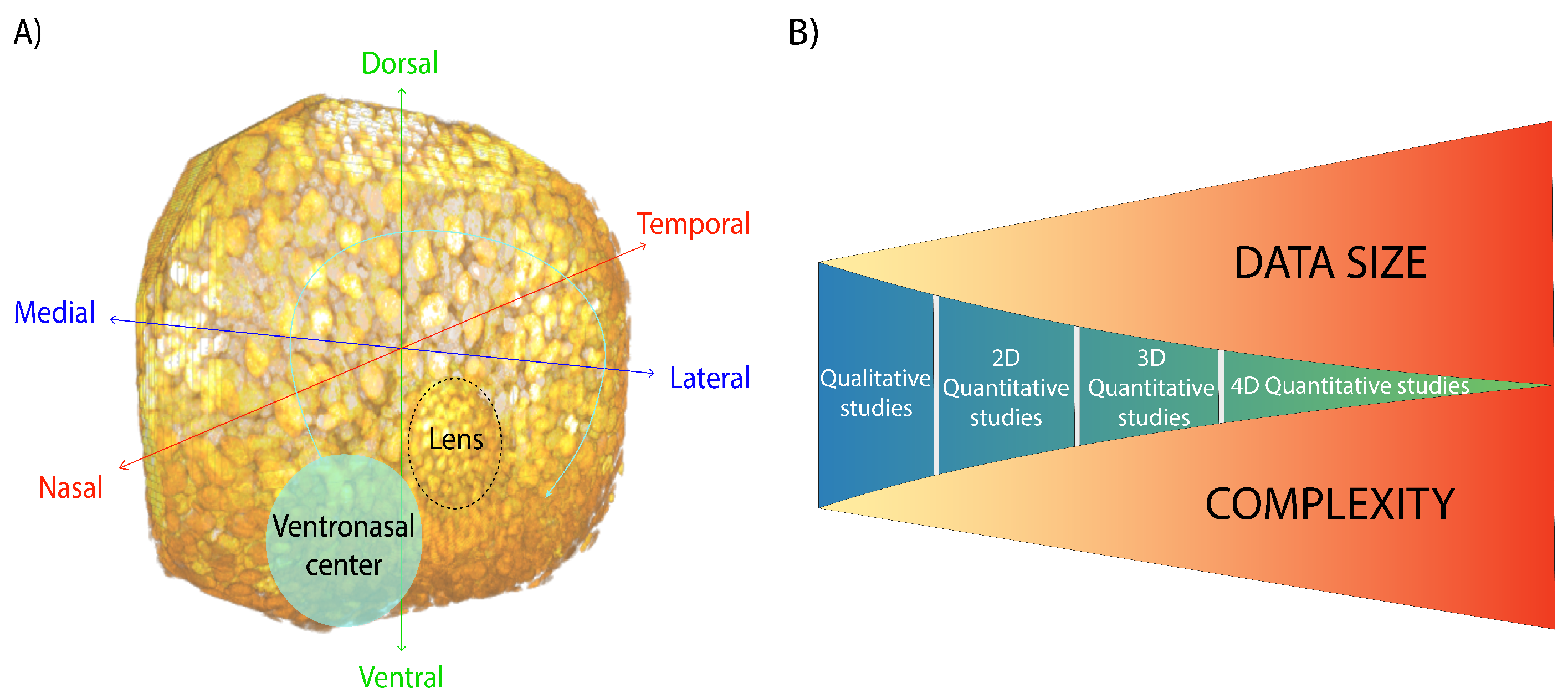
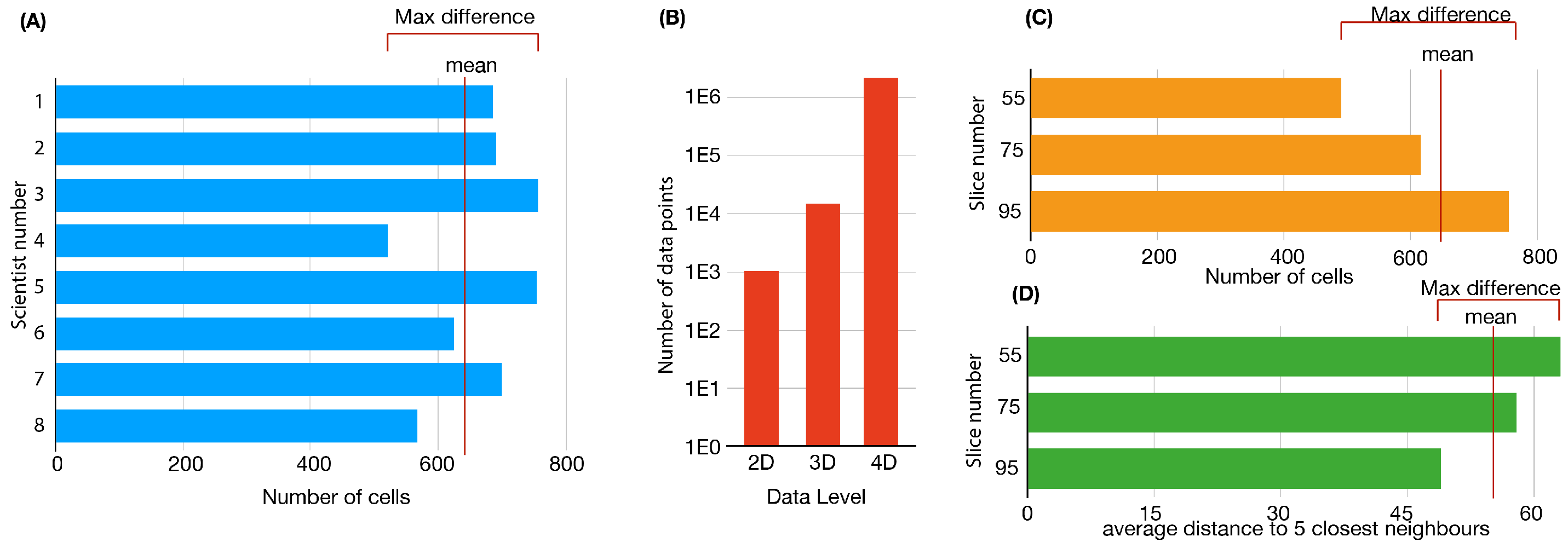
3. From Qualitative to Quantitative
4. 2D Quantitative Data
5. Automated Image Analysis
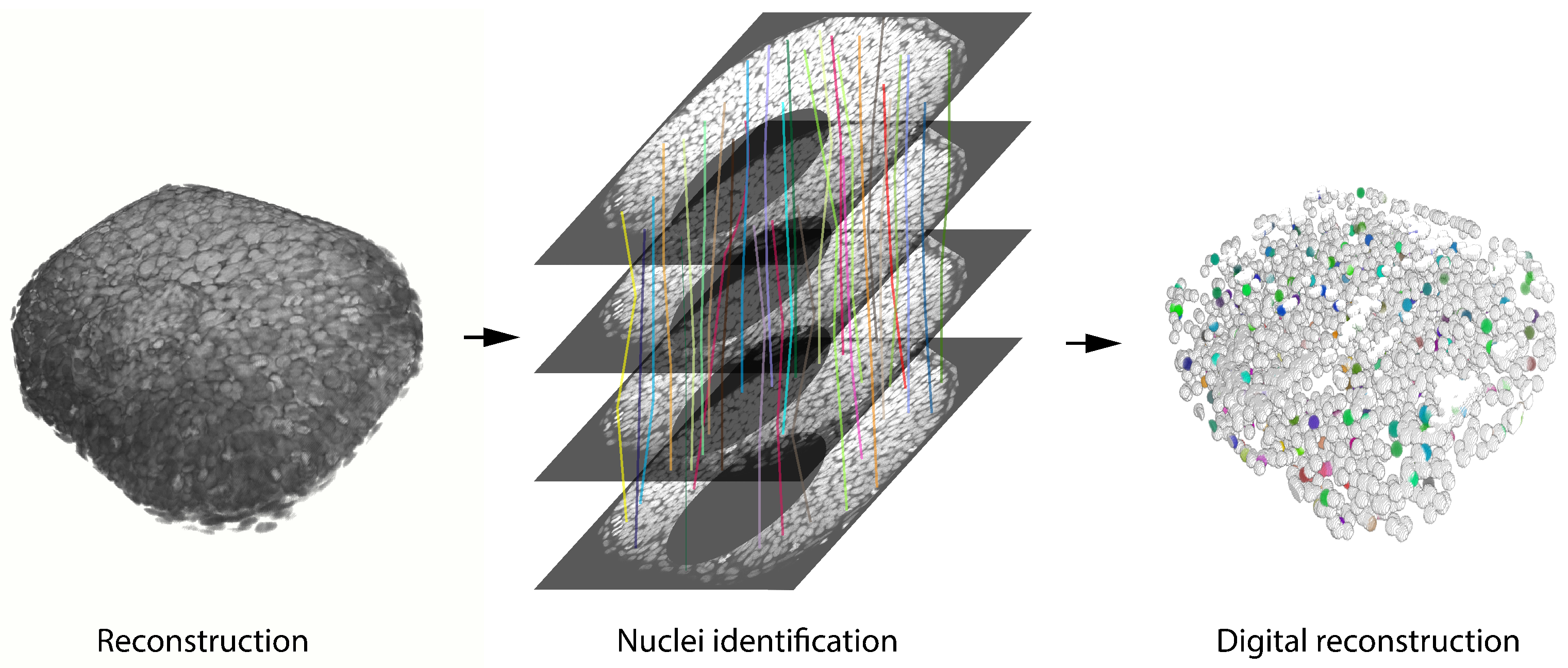
6. The Developing Vertebrate Retina in Three-Dimensions
7. The Developing Vertebrate Retina in Four-Dimensions
8. Discussion
9. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
Abbreviations
| RPC | Retinal progenitor cell |
| HPF | Hours post fertilization |
| mInsc | Mammalian homolog of Inscuteable |
| LSFM | Light sheet fluorescence microscopy |
| SPIM | Selective plane illumination microscopy |
| iPS | Induced pluripotent stem |
References
- Toma, K.; Wang, T.C.; Hanashima, C. Encoding and decoding time in neural development. Dev. Growth Differ. 2016, 58, 59–72. [Google Scholar] [CrossRef]
- Wallace, V.A. Concise review: Making a retina—From the building blocks to clinical applications. Stem Cells 2011, 29, 412–417. [Google Scholar] [CrossRef]
- Centanin, L.; Wittbrodt, J. Retinal neurogenesis. Development 2014, 141, 241–244. [Google Scholar] [CrossRef]
- Hamon, A.; Roger, J.E.; Yang, X.J.; Perron, M. Müller glial cell-dependent regeneration of the neural retina: An overview across vertebrate model systems. Dev. Dyn. 2016, 245, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.L.; Tang, S.B. Differentiation of retinal ganglion cells from induced pluripotent stem cells: A review. Int. J. Ophthalmol. 2019, 12, 152. [Google Scholar] [PubMed]
- Thoreson, W.B. The vertebrate retina. In Neuroimmune Pharmacology; Springer: New York, NY, USA, 2017; pp. 55–68. [Google Scholar]
- Baden, T.; Euler, T.; Berens, P. Understanding the retinal basis of vision across species. Nat. Rev. Neurosci. 2020, 21, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Valdivia, L.E.; Lamb, D.B.; Horner, W.; Wierzbicki, C.; Tafessu, A.; Williams, A.M.; Gestri, G.; Krasnow, A.M.; Vleeshouwer-Neumann, T.S.; Givens, M.; et al. Antagonism between Gdf6a and retinoic acid pathways controls timing of retinal neurogenesis and growth of the eye in zebrafish. Development 2016, 143, 1087–1098. [Google Scholar] [CrossRef]
- Wycliffe, R.; Plaisancie, J.; Leaman, S.; Santis, O.; Tucker, L.; Cavieres, D.; Fernandez, M.; Weiss-Garrido, C.; Sobarzo, C.; Gestri, G.; et al. Developmental delay during eye morphogenesis underlies optic cup and neurogenesis defects in mab21l2u517 zebrafish mutants. Int. J. Dev. Biol. 2020, 65, 289–299. [Google Scholar] [CrossRef]
- Viets, K.; Eldred, K.C.; Johnston, R.J., Jr. Mechanisms of photoreceptor patterning in vertebrates and invertebrates. Trends Genet. 2016, 32, 638–659. [Google Scholar] [CrossRef]
- He, J.; Zhang, G.; Almeida, A.D.; Cayouette, M.; Simons, B.D.; Harris, W.A. How variable clones build an invariant retina. Neuron 2012, 75, 786–798. [Google Scholar] [CrossRef]
- Hu, M.; Easter, S.S., Jr. Retinal neurogenesis: The formation of the initial central patch of postmitotic cells. Dev. Biol. 1999, 207, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Neumann, C.J.; Nuesslein-Volhard, C. Patterning of the zebrafish retina by a wave of sonic hedgehog activity. Science 2000, 289, 2137–2139. [Google Scholar] [CrossRef] [PubMed]
- Bone, C.R.; Starr, D.A. Nuclear migration events throughout development. J. Cell Sci. 2016, 129, 1951–1961. [Google Scholar] [CrossRef]
- Zhou, J.; Benito-Martin, A.; Mighty, J.; Chang, L.; Ghoroghi, S.; Wu, H.; Wong, M.; Guariglia, S.; Baranov, P.; Young, M.; et al. Retinal progenitor cells release extracellular vesicles containing developmental transcription factors, microRNA and membrane proteins. Sci. Rep. 2018, 8, 2823. [Google Scholar] [CrossRef] [PubMed]
- Damsgaard, C.; Lauridsen, H.; Funder, A.M.; Thomsen, J.S.; Desvignes, T.; Crossley, D.A., II; Møller, P.R.; Huong, D.T.; Phuong, N.T.; Detrich, H.W., III; et al. Retinal oxygen supply shaped the functional evolution of the vertebrate eye. Elife 2019, 8, e52153. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, S.K.; Ghosh, A. A novel retinal image segmentation using rSVM boosted convolutional neural network for exudates detection. Biomed. Signal Process. Control 2021, 68, 102785. [Google Scholar] [CrossRef]
- Seritrakul, P.; Gross, J.M. Genetic and epigenetic control of retinal development in zebrafish. Curr. Opin. Neurobiol. 2019, 59, 120–127. [Google Scholar] [CrossRef]
- Ledesma-Terrón, M.; Pérez-Dones, D.; Míguez, D.G. OSCAR: A framework to identify and quantify cells in densely packed three-dimensional biological samples. bioRxiv 2021. [Google Scholar] [CrossRef]
- Agathocleous, M.; Harris, W.A. From progenitors to differentiated cells in the vertebrate retina. Annu. Rev. Cell Dev. 2009, 25, 45–69. [Google Scholar] [CrossRef]
- Lamba, D.; Karl, M.; Reh, T. Neural regeneration and cell replacement: A view from the eye. Cell Stem Cell 2008, 2, 538–549. [Google Scholar] [CrossRef]
- Kay, J.N.; Link, B.A.; Baier, H. Staggered cell-intrinsic timing of ath5 expression underlies the wave of ganglion cell neurogenesis in the zebrafish retina. Development 2005, 132, 2573–2585. [Google Scholar] [CrossRef]
- Yang, Z.; Ding, K.; Pan, L.; Deng, M.; Gan, L. Math5 determines the competence state of retinal ganglion cell progenitors. Dev. Biol. 2003, 264, 240–254. [Google Scholar] [CrossRef]
- Locker, M.; Agathocleous, M.; Amato, M.A.; Parain, K.; Harris, W.A.; Perron, M. Hedgehog signaling and the retina: Insights into the mechanisms controlling the proliferative properties of neural precursors. Genes Dev. 2006, 20, 3036–3048. [Google Scholar] [CrossRef]
- Pandit, T.; Jidigam, V.K.; Patthey, C.; Gunhaga, L. Neural retina identity is specified by lens-derived BMP signals. Development 2015, 142, 1850–1859. [Google Scholar] [CrossRef] [PubMed]
- Steinfeld, J.; Steinfeld, I.; Bausch, A.; Coronato, N.; Hampel, M.L.; Depner, H.; Layer, P.G.; Vogel-Höpker, A. BMP-induced reprogramming of the neural retina into retinal pigment epithelium requires Wnt signalling. Biol. Open 2017, 6, 979–992. [Google Scholar] [CrossRef]
- Geng, Y.; Yu, Q.; Sicinska, E.; Das, M.; Bronson, R.T.; Sicinski, P. Deletion of the p27Kip1 gene restores normal development in cyclin D1-deficient mice. Proc. Natl. Acad. Sci. USA 2001, 98, 194–199. [Google Scholar] [CrossRef]
- Skapek, S.X.; Lin, S.C.J.; Jablonski, M.M.; McKeller, R.N.; Tan, M.; Hu, N.; Eva, Y.; Lee, H. Persistent expression of cyclin D1 disrupts normal photoreceptor differentiation and retina development. Oncogene 2001, 20, 6742–6751. [Google Scholar] [CrossRef] [PubMed]
- Bienvenu, F.; Jirawatnotai, S.; Elias, J.E.; Meyer, C.A.; Mizeracka, K.; Marson, A.; Frampton, G.M.; Cole, M.F.; Odom, D.T.; Odajima, J.; et al. Transcriptional role of cyclin D1 in development revealed by a genetic–proteomic screen. Nature 2010, 463, 374–378. [Google Scholar] [CrossRef]
- Mabaso, M.A.; Withey, D.J.; Twala, B. Spot detection methods in fluorescence microscopy imaging: A review. Image Anal. Stereol. 2018, 37, 173–190. [Google Scholar] [CrossRef]
- Meijering, E. Cell segmentation: 50 years down the road [life sciences]. IEEE Signal Process. Mag. 2012, 29, 140–145. [Google Scholar] [CrossRef]
- Chen, W.; Li, W.; Dong, X.; Pei, J. A review of biological image analysis. Curr. Bioinform. 2018, 13, 337–343. [Google Scholar] [CrossRef]
- Žigman, M.; Cayouette, M.; Charalambous, C.; Schleiffer, A.; Hoeller, O.; Dunican, D.; McCudden, C.R.; Firnberg, N.; Barres, B.A.; Siderovski, D.P.; et al. Mammalian inscuteable regulates spindle orientation and cell fate in the developing retina. Neuron 2005, 48, 539–545. [Google Scholar] [CrossRef]
- Shkumatava, A.; Neumann, C.J. Shh directs cell-cycle exit by activating p57Kip2 in the zebrafish retina. EMBO Rep. 2005, 6, 563–569. [Google Scholar] [CrossRef] [PubMed]
- Wall, D.S.; Mears, A.J.; McNeill, B.; Mazerolle, C.; Thurig, S.; Wang, Y.; Kageyama, R.; Wallace, V.A. Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell Biol. 2009, 184, 101–112. [Google Scholar] [CrossRef]
- Ma, L.; Cantrup, R.; Varrault, A.; Colak, D.; Klenin, N.; Götz, M.; McFarlane, S.; Journot, L.; Schuurmans, C. Zac1 functions through TGFβII to negatively regulate cell number in the developing retina. Neural Dev. 2007, 2, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Cayouette, M.; Raff, M. The orientation of cell division influences cell-fate choice in the developing mammalian retina. Development 2003, 130, 2329–2339. [Google Scholar] [CrossRef][Green Version]
- Poggi, L.; Vitorino, M.; Masai, I.; Harris, W.A. Influences on neural lineage and mode of division in the zebrafish retina in vivo. J. Cell Biol. 2005, 171, 991–999. [Google Scholar] [CrossRef]
- Riesenberg, A.N.; Liu, Z.; Kopan, R.; Brown, N.L. Rbpj cell autonomous regulation of retinal ganglion cell and cone photoreceptor fates in the mouse retina. J. Neurosci. 2009, 29, 12865–12877. [Google Scholar] [CrossRef]
- Schmidt, U.; Weigert, M.; Broaddus, C.; Myers, G. Cell detection with star-convex polygons. In International Conference on Medical Image Computing and Computer-Assisted Intervention; Springer: New York, NY, USA, 2018; pp. 265–273. [Google Scholar]
- Scherr, T.; Streule, K.; Bartschat, A.; Böhland, M.; Stegmaier, J.; Reischl, M.; Orian-Rousseau, V.; Mikut, R. BeadNet: Deep learning-based bead detection and counting in low-resolution microscopy images. Bioinformatics 2020, 36, 4668–4670. [Google Scholar] [CrossRef]
- Salinas-Navarro, M.; Jiménez-López, M.; Valiente-Soriano, F.; Alarcón-Martínez, L.; Avilés-Trigueros, M.; Mayor, S.; Holmes, T.; Lund, R.; Villegas-Pérez, M.; Vidal-Sanz, M. Retinal ganglion cell population in adult albino and pigmented mice: A computerized analysis of the entire population and its spatial distribution. Vis. Res. 2009, 49, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Navarro, M.; Mayor-Torroglosa, S.; Jimenez-Lopez, M.; Avilés-Trigueros, M.; Holmes, T.; Lund, R.; Villegas-Pérez, M.; Vidal-Sanz, M. A computerized analysis of the entire retinal ganglion cell population and its spatial distribution in adult rats. Vis. Res. 2009, 49, 115–126. [Google Scholar] [CrossRef] [PubMed]
- Matejčić, M.; Salbreux, G.; Norden, C. A non-cell-autonomous actin redistribution enables isotropic retinal growth. PLoS Biol. 2018, 16, e2006018. [Google Scholar] [CrossRef]
- Moreno-Mármol, T.; Ledesma-Terrón, M.; Tabanera, N.; Martin-Bermejo, M.J.; Cardozo, M.J.; Cavodeassi, F.; Bovolenta, P. Stretching of the retinal pigment epithelium contributes to zebrafish optic cup morphogenesis. bioRxiv 2020. [Google Scholar] [CrossRef]
- Carreño, H.; Santos-Ledo, A.; Velasco, A.; Lara, J.M.; Aijón, J.; Arévalo, R. Effects of retinoic acid exposure during zebrafish retinogenesis. Neurotoxicol. Teratol. 2013, 40, 35–45. [Google Scholar] [CrossRef]
- Kashyap, B.; Frederickson, L.C.; Stenkamp, D.L. Mechanisms for persistent microphthalmia following ethanol exposure during retinal neurogenesis in zebrafish embryos. Vis. Neurosci. 2007, 24, 409. [Google Scholar] [CrossRef] [PubMed]
- Santos-Ledo, A.; Arenzana, F.; Porteros, A.; Lara, J.; Velasco, A.; Aijón, J.; Arévalo, R. Cytoarchitectonic and neurochemical differentiation of the visual system in ethanol-induced cyclopic zebrafish larvae. Neurotoxicol. Teratol. 2011, 33, 686–697. [Google Scholar] [CrossRef]
- Abdel-Khalek, S.; Ishak, A.B.; Omer, O.A.; Obada, A.S. A two-dimensional image segmentation method based on genetic algorithm and entropy. Optik 2017, 131, 414–422. [Google Scholar] [CrossRef]
- Chen, J.; Zheng, H.; Lin, X.; Wu, Y.; Su, M. A novel image segmentation method based on fast density clustering algorithm. Eng. Appl. Artif. Intell. 2018, 73, 92–110. [Google Scholar] [CrossRef]
- Shan, P. Image segmentation method based on K-mean algorithm. EURASIP J. Image Video Process. 2018, 2018, 1–9. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, X.; Zhu, Y.; Su, X. NSST and vector-valued C–V model based image segmentation algorithm. IET Image Process. 2020, 14, 1614–1620. [Google Scholar] [CrossRef]
- Choy, S.K.; Ng, T.C.; Yu, C. Unsupervised fuzzy model-based image segmentation. Signal Process. 2020, 171, 107483. [Google Scholar] [CrossRef]
- Zhang, S. Beyond the Petri dish. Nat. Biotechnol. 2004, 22, 151–152. [Google Scholar] [CrossRef]
- Vigouroux, R.J.; Belle, M.; Chédotal, A. Neuroscience in the third dimension: Shedding new light on the brain with tissue clearing. Mol. Brain 2017, 10, 33. [Google Scholar] [CrossRef]
- Susaki, E.A.; Ueda, H.R. Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: Toward organism-level systems biology in mammals. Cell Chem. Biol. 2016, 23, 137–157. [Google Scholar] [CrossRef]
- Yeh, A.T.; Hirshburg, J. Molecular interactions of exogenous chemical agents with collagen—Implications for tissue optical clearing. J. Biomed. Opt. 2006, 11, 014003. [Google Scholar] [CrossRef]
- Yu, T.; Wen, X.; Luo, Q.; Zhu, D.; Tuchin, V.V. Quantitative analysis of dehydration in porcine skin for assessing mechanism of optical clearing. J. Biomed. Opt. 2011, 16, 095002. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ptak, D.; Zhang, L.; Walls, E.K.; Zhong, W.; Leung, Y.F. Phenylthiourea specifically reduces zebrafish eye size. PLoS ONE 2012, 7, e40132. [Google Scholar] [CrossRef]
- Keller, P.J.; Ahrens, M.B. Visualizing whole-brain activity and development at the single-cell level using light-sheet microscopy. Neuron 2015, 85, 462–483. [Google Scholar] [CrossRef] [PubMed]
- Lazzari, G.; Vinciguerra, D.; Balasso, A.; Nicolas, V.; Goudin, N.; Garfa-Traore, M.; Fehér, A.; Dinnyés, A.; Nicolas, J.; Couvreur, P.; et al. Light sheet fluorescence microscopy versus confocal microscopy: In quest of a suitable tool to assess drug and nanomedicine penetration into multicellular tumor spheroids. Eur. J. Pharm. Biopharm. 2019, 142, 195–203. [Google Scholar] [CrossRef]
- Keller, P.J.; Schmidt, A.D.; Wittbrodt, J.; Stelzer, E.H. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 2008, 322, 1065–1069. [Google Scholar] [CrossRef] [PubMed]
- Schmid, B.; Shah, G.; Scherf, N.; Weber, M.; Thierbach, K.; Campos, C.P.; Roeder, I.; Aanstad, P.; Huisken, J. High-speed panoramic light-sheet microscopy reveals global endodermal cell dynamics. Nat. Commun. 2013, 4, 2207. [Google Scholar] [CrossRef]
- Meijering, E. A bird’s-eye view of deep learning in bioimage analysis. Comput. Struct. Biotechnol. J. 2020, 18, 2312. [Google Scholar] [CrossRef]
- Xie, Y.; Xing, F.; Shi, X.; Kong, X.; Su, H.; Yang, L. Efficient and robust cell detection: A structured regression approach. Med. Image Anal. 2018, 44, 245–254. [Google Scholar] [CrossRef]
- Yuan, P.; Rezvan, A.; Li, X.; Varadarajan, N.; Van Nguyen, H. Phasetime: Deep learning approach to detect nuclei in time lapse phase images. J. Clin. Med. 2019, 8, 1159. [Google Scholar] [CrossRef] [PubMed]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 39, 2481–2495. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Anal. Mach. Intell. 2017, 40, 834–848. [Google Scholar] [CrossRef]
- Arts, M.; Smal, I.; Paul, M.W.; Wyman, C.; Meijering, E. Particle mobility analysis using deep learning and the moment scaling spectrum. Sci. Rep. 2019, 9, 17160. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Su, X.; Zhao, L.; Zhang, J. Deep Reinforcement Learning for Data Association in Cell Tracking. Front. Bioeng. Biotechnol. 2020, 8, 298. [Google Scholar] [CrossRef]
- Inés, A.; Domínguez, C.; Heras, J.; Mata, E.; Pascual, V. DeepClas4Bio: Connecting bioimaging tools with deep learning frameworks for image classification. Comput. Biol. Med. 2019, 108, 49–56. [Google Scholar] [CrossRef]
- Phan, H.T.H.; Kumar, A.; Feng, D.; Fulham, M.; Kim, J. Unsupervised two-path neural network for cell event detection and classification using spatiotemporal patterns. IEEE Trans. Med. Imaging 2018, 38, 1477–1487. [Google Scholar] [CrossRef]
- Oktay, A.B.; Gurses, A. Automatic detection, localization and segmentation of nano-particles with deep learning in microscopy images. Micron 2019, 120, 113–119. [Google Scholar] [CrossRef]
- Wang, E.K.; Zhang, X.; Pan, L.; Cheng, C.; Dimitrakopoulou-Strauss, A.; Li, Y.; Zhe, N. Multi-path dilated residual network for nuclei segmentation and detection. Cells 2019, 8, 499. [Google Scholar] [CrossRef]
- Falk, T.; Mai, D.; Bensch, R.; Çiçek, Ö.; Abdulkadir, A.; Marrakchi, Y.; Böhm, A.; Deubner, J.; Jäckel, Z.; Seiwald, K.; et al. U-Net: Deep learning for cell counting, detection, and morphometry. Nat. Methods 2019, 16, 67–70. [Google Scholar] [CrossRef]
- Lugagne, J.B.; Lin, H.; Dunlop, M.J. DeLTA: Automated cell segmentation, tracking, and lineage reconstruction using deep learning. PLoS Comput. Biol. 2020, 16, e1007673. [Google Scholar] [CrossRef]
- Götz, M.; Huttner, W.B. The cell biology of neurogenesis. Nat. Rev. Mol. Cell Biol. 2005, 6, 777–788. [Google Scholar] [CrossRef]
- Simó, R.; Villarroel, M.; Corraliza, L.; Hernández, C.; Garcia-Ramírez, M. The retinal pigment epithelium: Something more than a constituent of the blood-retinal barrier—Implications for the pathogenesis of diabetic retinopathy. J. Biomed. Biotechnol. 2010, 2010, 190724. [Google Scholar] [CrossRef]
- Malicki, J.; Pooranachandran, N.; Nikolaev, A.; Fang, X.; Avanesov, A. Analysis of the retina in the zebrafish model. Methods Cell Biol. 2016, 134, 257–334. [Google Scholar]
- Clark, B.S.; Stein-O’Brien, G.L.; Shiau, F.; Cannon, G.H.; Davis-Marcisak, E.; Sherman, T.; Santiago, C.P.; Hoang, T.V.; Rajaii, F.; James-Esposito, R.E.; et al. Single-cell RNA-seq analysis of retinal development identifies NFI factors as regulating mitotic exit and late-born cell specification. Neuron 2019, 102, 1111–1126. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Shiau, F.; Yi, W.; Lu, S.; Wu, Q.; Pearson, J.D.; Kallman, A.; Zhong, S.; Hoang, T.; Zuo, Z.; et al. Single-cell analysis of human retina identifies evolutionarily conserved and species-specific mechanisms controlling development. Dev. Cell 2020, 53, 473–491. [Google Scholar] [CrossRef] [PubMed]
- Oliveira-Valença, V.M.; Bosco, A.; Vetter, M.L.; Silveira, M.S. On the generation and regeneration of retinal ganglion cells. Front. Cell Dev. Biol. 2020, 8, 970. [Google Scholar] [CrossRef] [PubMed]
- Azizi, A.; Herrmann, A.; Wan, Y.; Buse, S.J.; Keller, P.J.; Goldstein, R.E.; Harris, W.A. Nuclear crowding and nonlinear diffusion during interkinetic nuclear migration in the zebrafish retina. Elife 2020, 9, e58635. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo-Valdés, M.; Sanchez-Ortiz, G.I.; Elkington, A.G.; Mohiaddin, R.H.; Rueckert, D. Segmentation of 4D cardiac MR images using a probabilistic atlas and the EM algorithm. Med. Image Anal. 2004, 8, 255–265. [Google Scholar] [CrossRef]
- Megason, S.G. In toto imaging of embryogenesis with confocal time-lapse microscopy. In Zebrafish; Springer: New York, NY, USA, 2009; pp. 317–332. [Google Scholar]
- Bensch, R.; Ronneberger, O.; Greese, B.; Fleck, C.; Wester, K.; Hulskamp, M.; Burkhardt, H. Image analysis of arabidopsis trichome patterning in 4D confocal datasets. In Proceedings of the 2009 IEEE International Symposium on Biomedical Imaging: From Nano to Macro, Boston, MA, USA, 28 June–1 July 2009; pp. 742–745. [Google Scholar]
- Kelley, L.C.; Wang, Z.; Hagedorn, E.J.; Wang, L.; Shen, W.; Lei, S.; Johnson, S.A.; Sherwood, D.R. Live-cell confocal microscopy and quantitative 4D image analysis of anchor-cell invasion through the basement membrane in Caenorhabditis elegans. Nat. Protoc. 2017, 12, 2081–2096. [Google Scholar] [CrossRef] [PubMed]
- Sakaue-Sawano, A.; Kurokawa, H.; Morimura, T.; Hanyu, A.; Hama, H.; Osawa, H.; Kashiwagi, S.; Fukami, K.; Miyata, T.; Miyoshi, H.; et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell 2008, 132, 487–498. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, M.; Sakaue-Sawano, A.; Iimura, T.; Fukami, K.; Kitaguchi, T.; Kawakami, K.; Okamoto, H.; Higashijima, S.I.; Miyawaki, A. Illuminating cell-cycle progression in the developing zebrafish embryo. Proc. Natl. Acad. Sci. USA 2009, 106, 20812–20817. [Google Scholar] [CrossRef] [PubMed]
- Saade, M.; Gutiérrez-Vallejo, I.; Le Dréau, G.; Rabadán, M.A.; Miguez, D.G.; Buceta, J.; Martí, E. Sonic hedgehog signaling switches the mode of division in the developing nervous system. Cell Rep. 2013, 4, 492–503. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pérez-Dones, D.; Ledesma-Terrón, M.; Míguez, D.G. Quantitative Approaches to Study Retinal Neurogenesis. Biomedicines 2021, 9, 1222. https://doi.org/10.3390/biomedicines9091222
Pérez-Dones D, Ledesma-Terrón M, Míguez DG. Quantitative Approaches to Study Retinal Neurogenesis. Biomedicines. 2021; 9(9):1222. https://doi.org/10.3390/biomedicines9091222
Chicago/Turabian StylePérez-Dones, Diego, Mario Ledesma-Terrón, and David G. Míguez. 2021. "Quantitative Approaches to Study Retinal Neurogenesis" Biomedicines 9, no. 9: 1222. https://doi.org/10.3390/biomedicines9091222
APA StylePérez-Dones, D., Ledesma-Terrón, M., & Míguez, D. G. (2021). Quantitative Approaches to Study Retinal Neurogenesis. Biomedicines, 9(9), 1222. https://doi.org/10.3390/biomedicines9091222






