Chronic Alcoholism and HHV-6 Infection Synergistically Promote Neuroinflammatory Microglial Phenotypes in the Substantia Nigra of the Adult Human Brain
Abstract
1. Introduction
2. Materials and Methods
2.1. Autopsy Brain Tissue Collection and Characteristics
2.2. Immunohistochemistry (IHC), Double Immunohistochemistry, and Immunofluorescence (IF)
2.3. DNA Extraction and HHV-6 Detection Using Nested Polymerase Chain Reaction (nPCR)
2.4. HHV-6A and HHV-6B Variant Detection Using HindIII Restriction Endonuclease
2.5. Viral Load Determination Using Real-Time PCR (RT-PCR)
2.6. Statistical Analysis
3. Results
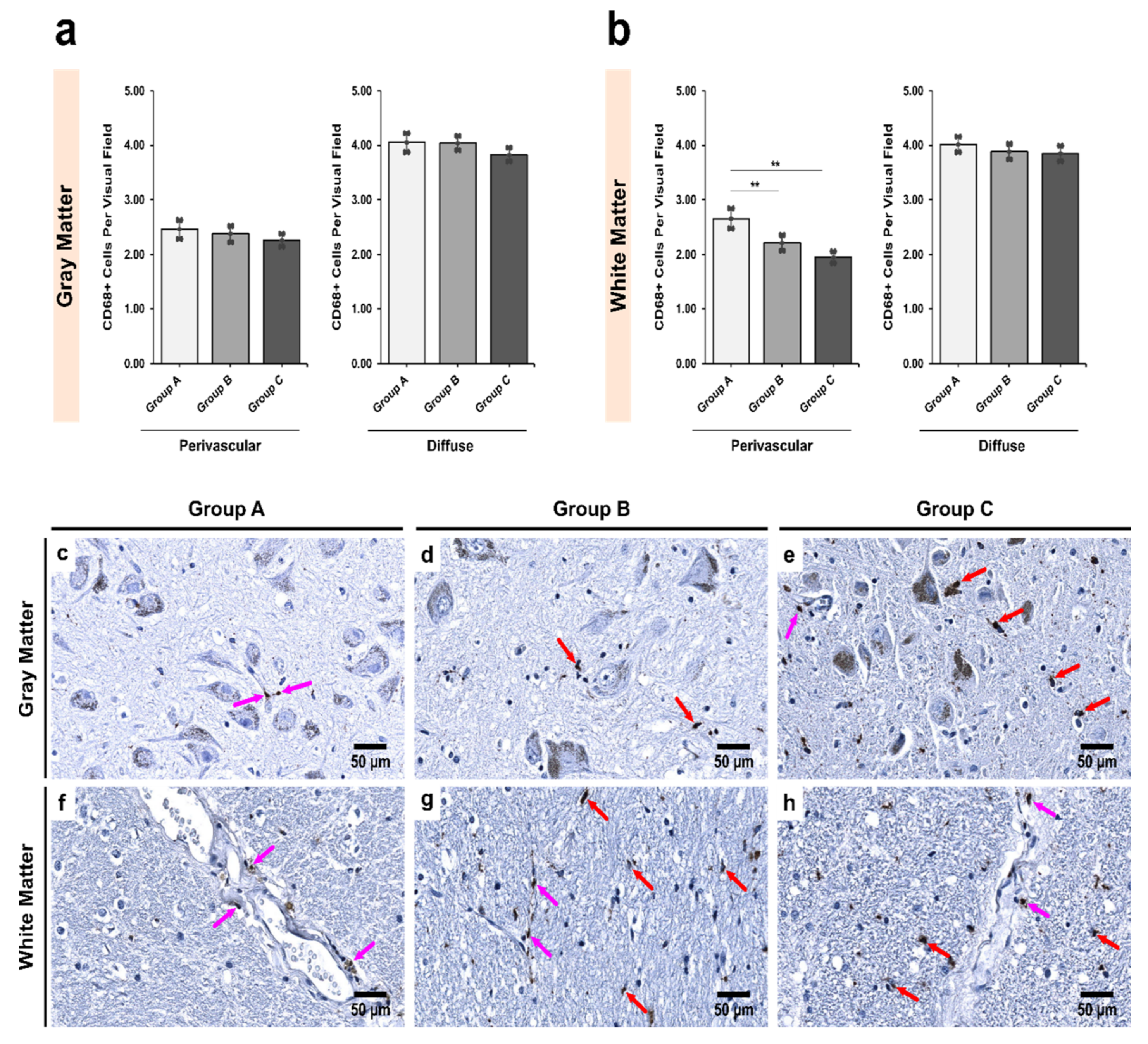
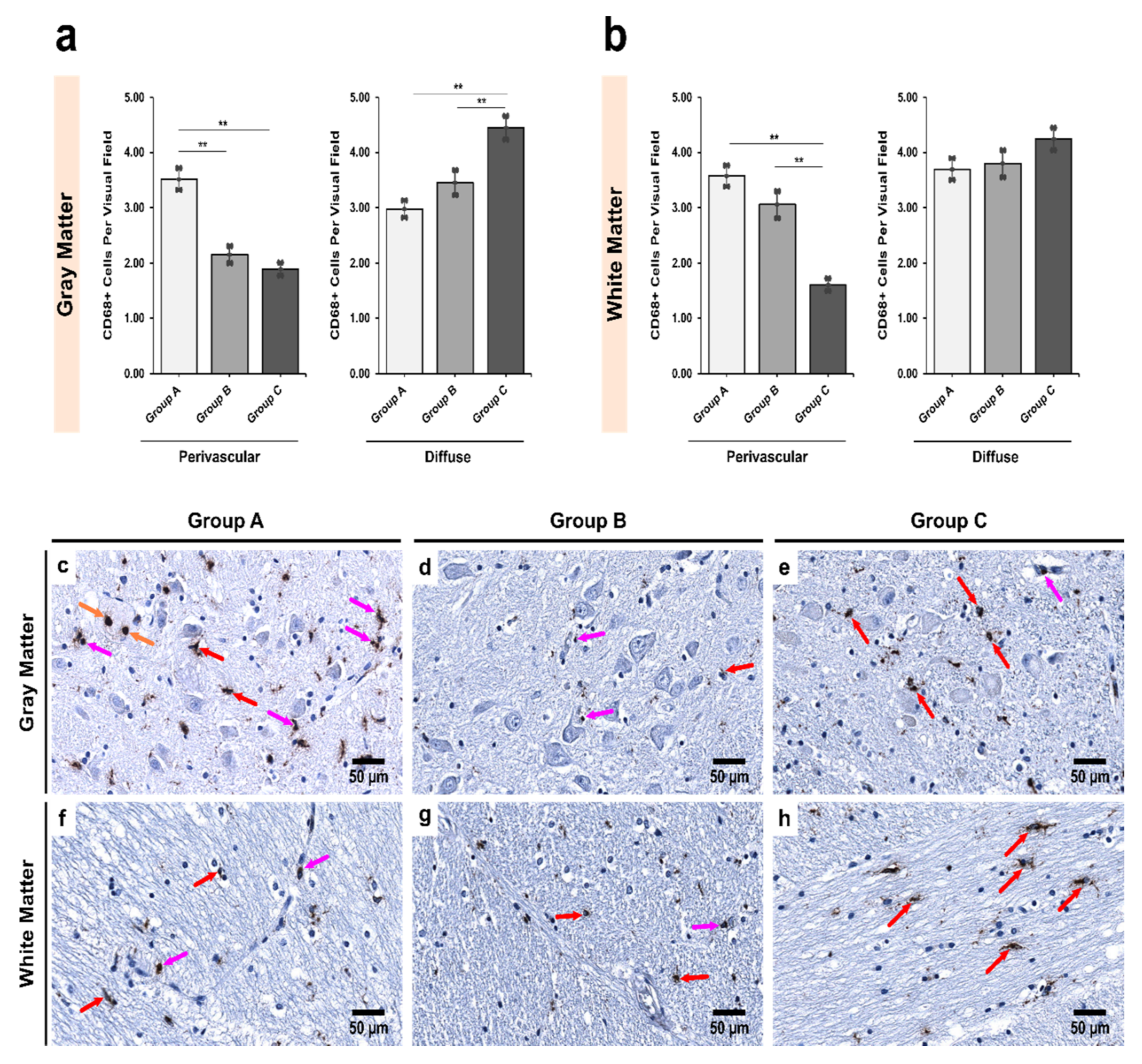
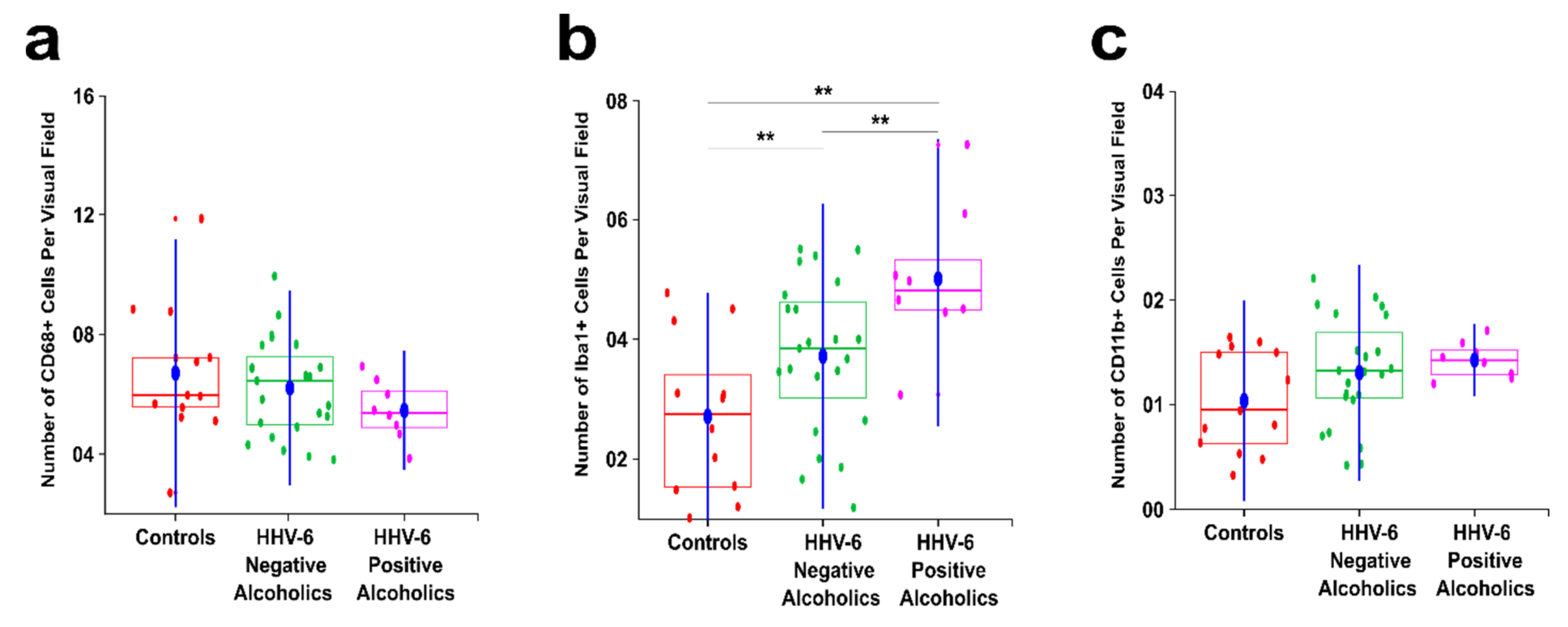
3.1. Alcoholics Showed a Significant Decrease in CD68+ Cells Than Controls in Both Gray and White Matter
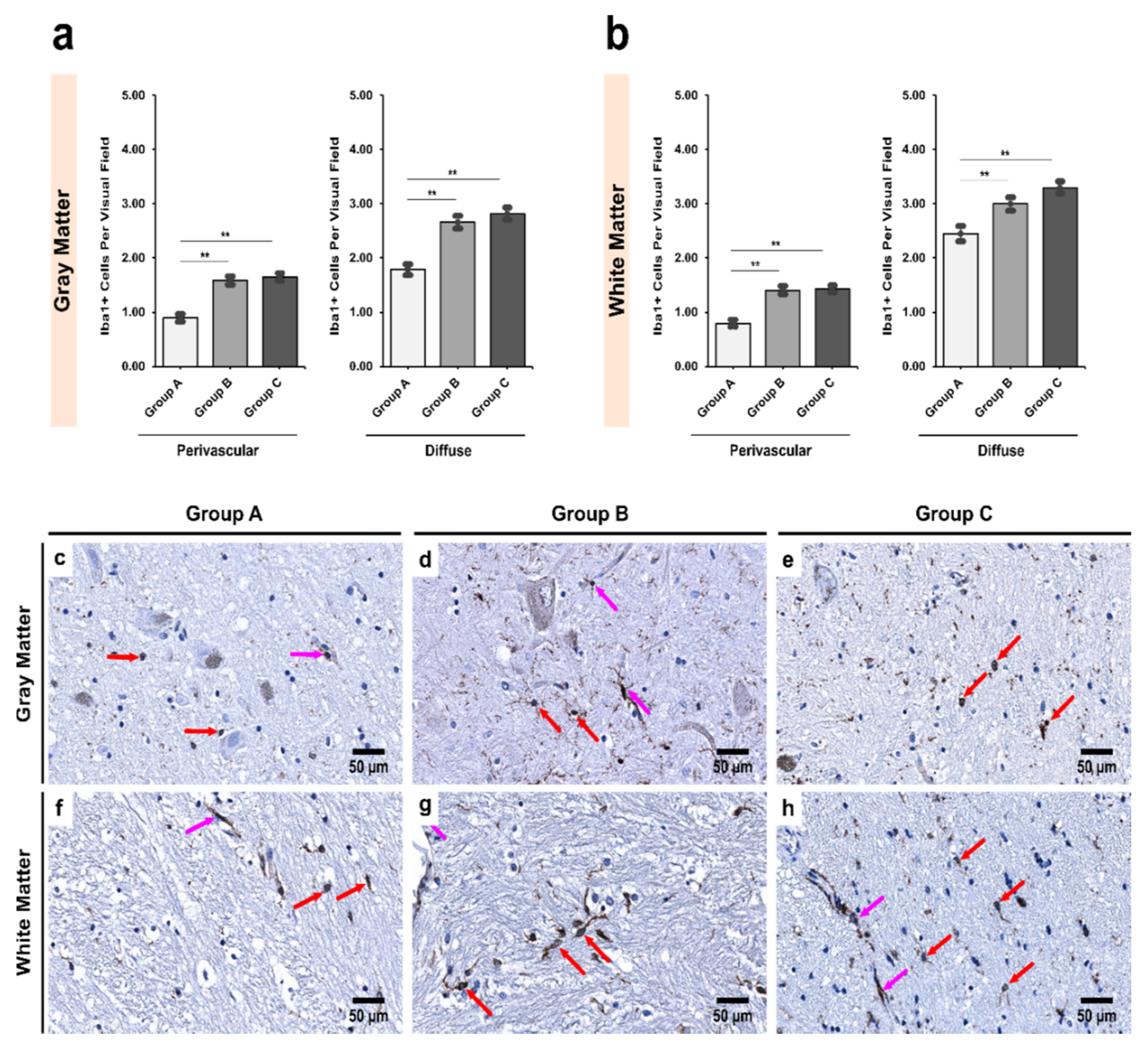
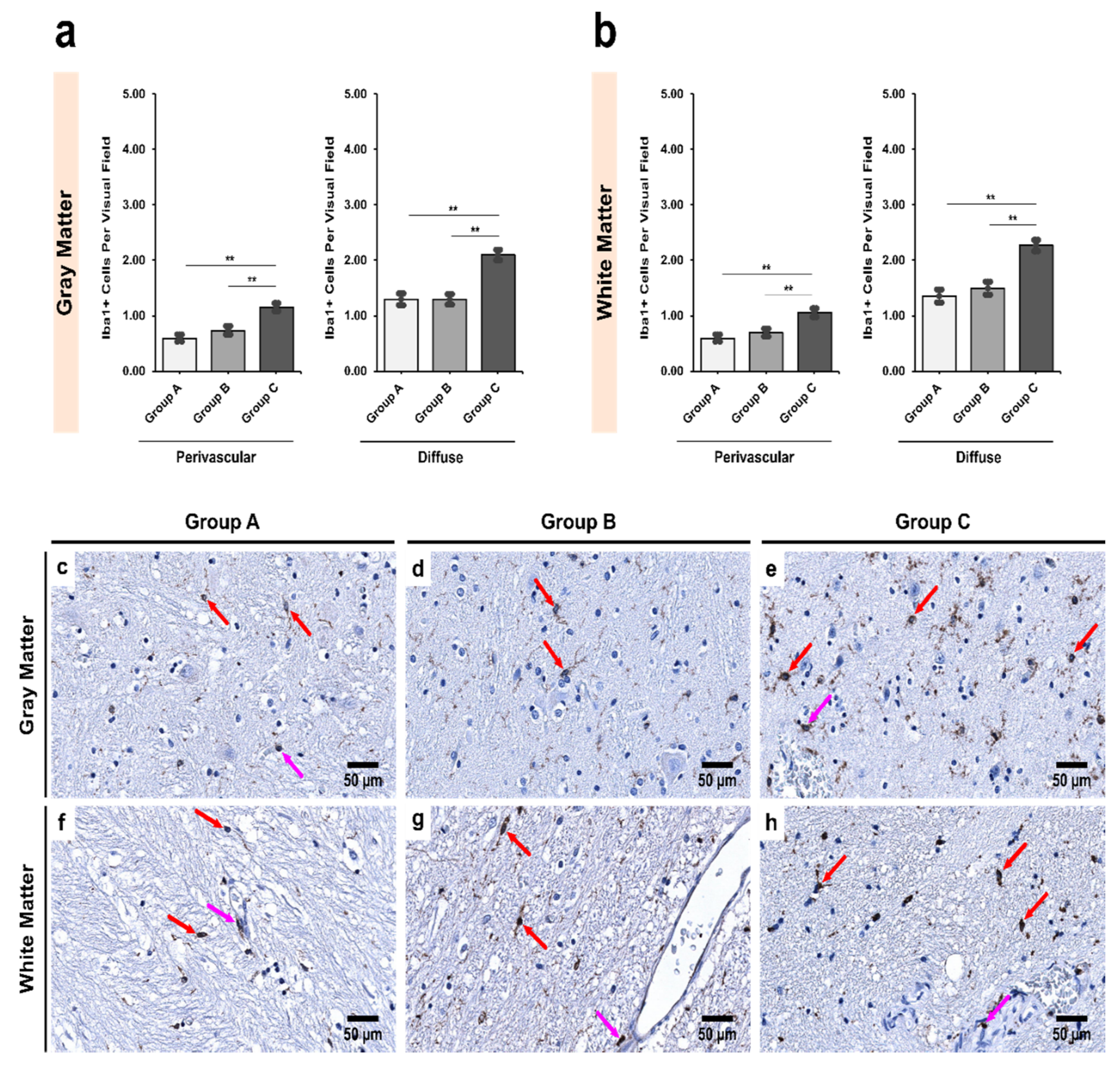
3.2. Alcoholics Showed a Significant Increase in Iba1+ Cells Compared to Controls in Both Gray and White Matter
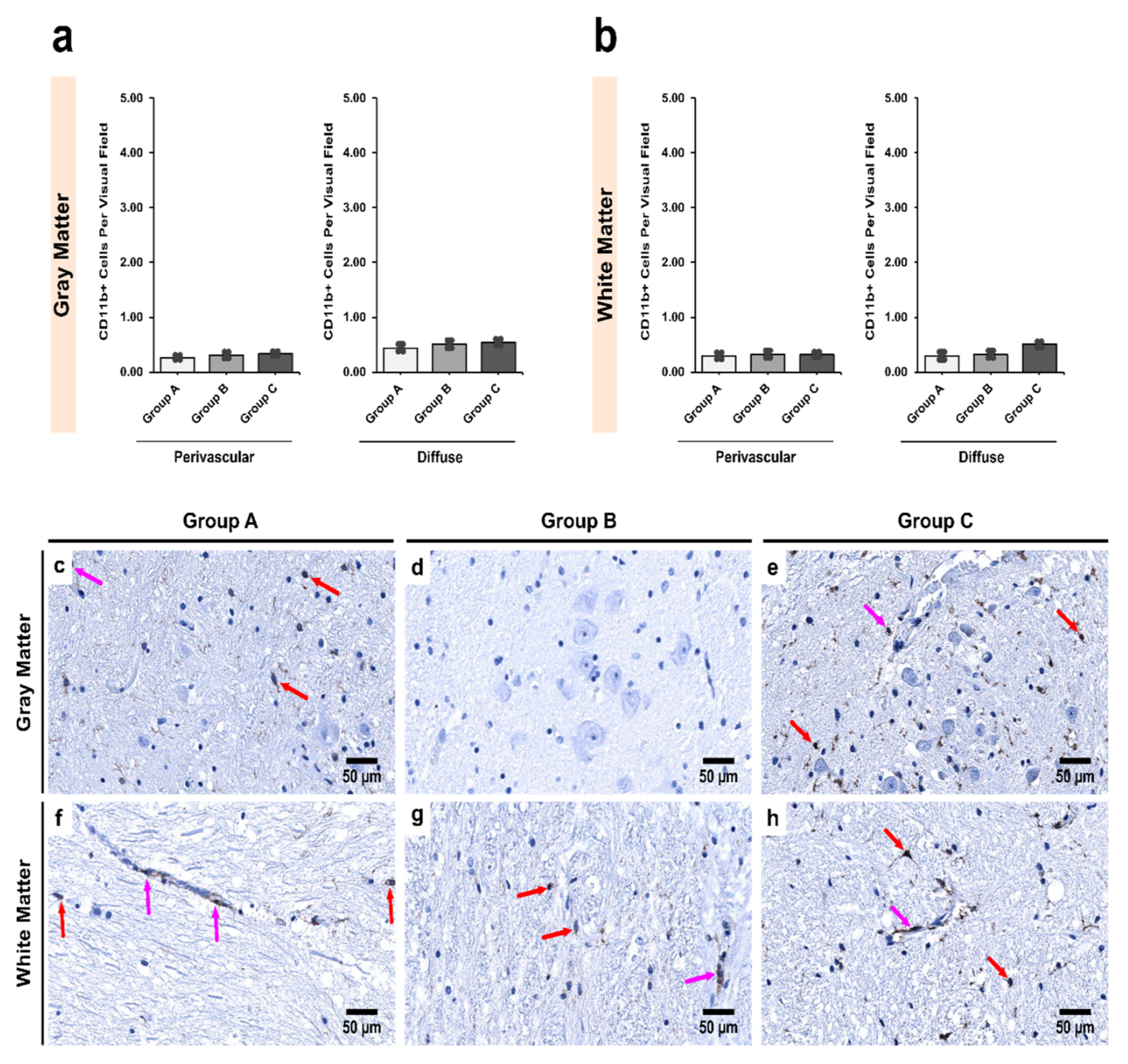
3.3. Alcoholics Showed Significantly More CD11b+ Cells Than Controls in Both Gray and White Matter
3.4. Exposure to Alcohol Induces Stronger Iba1 Expression Than CD11b in Microglial Cells
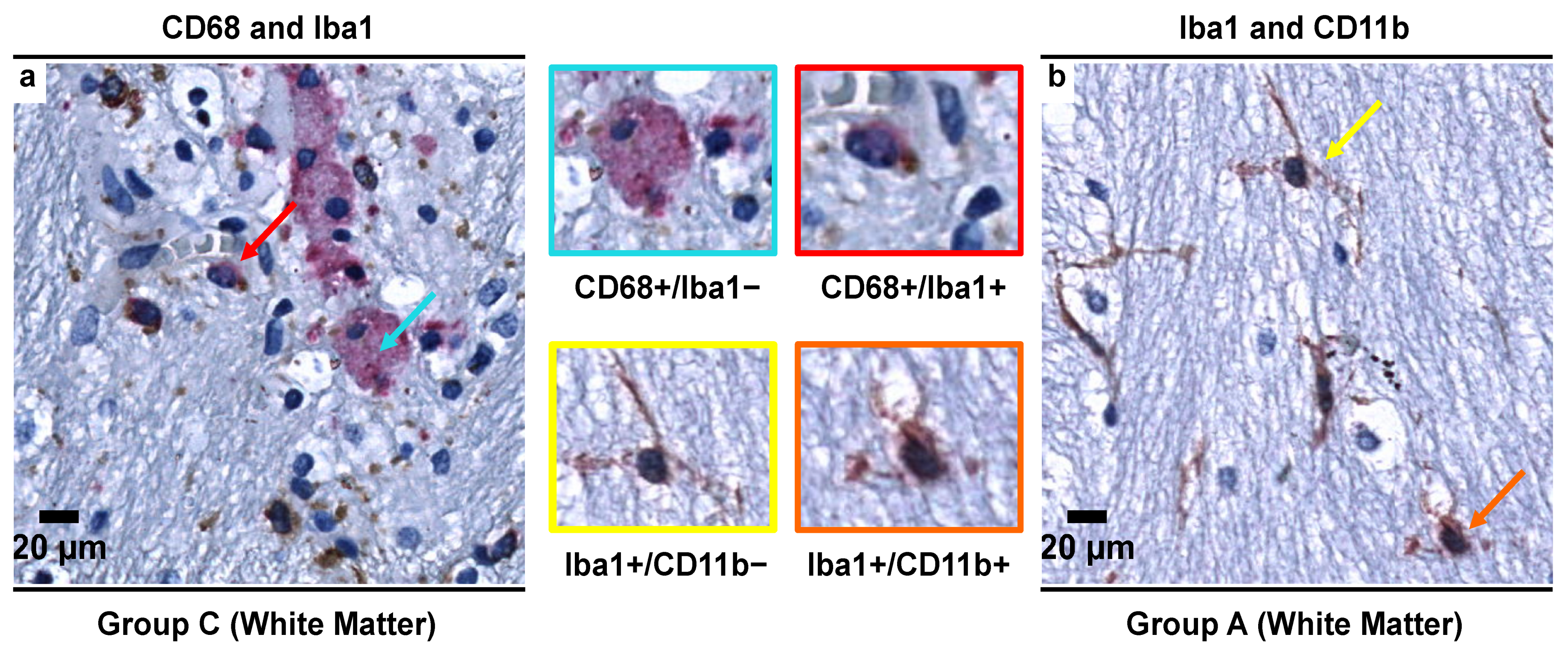
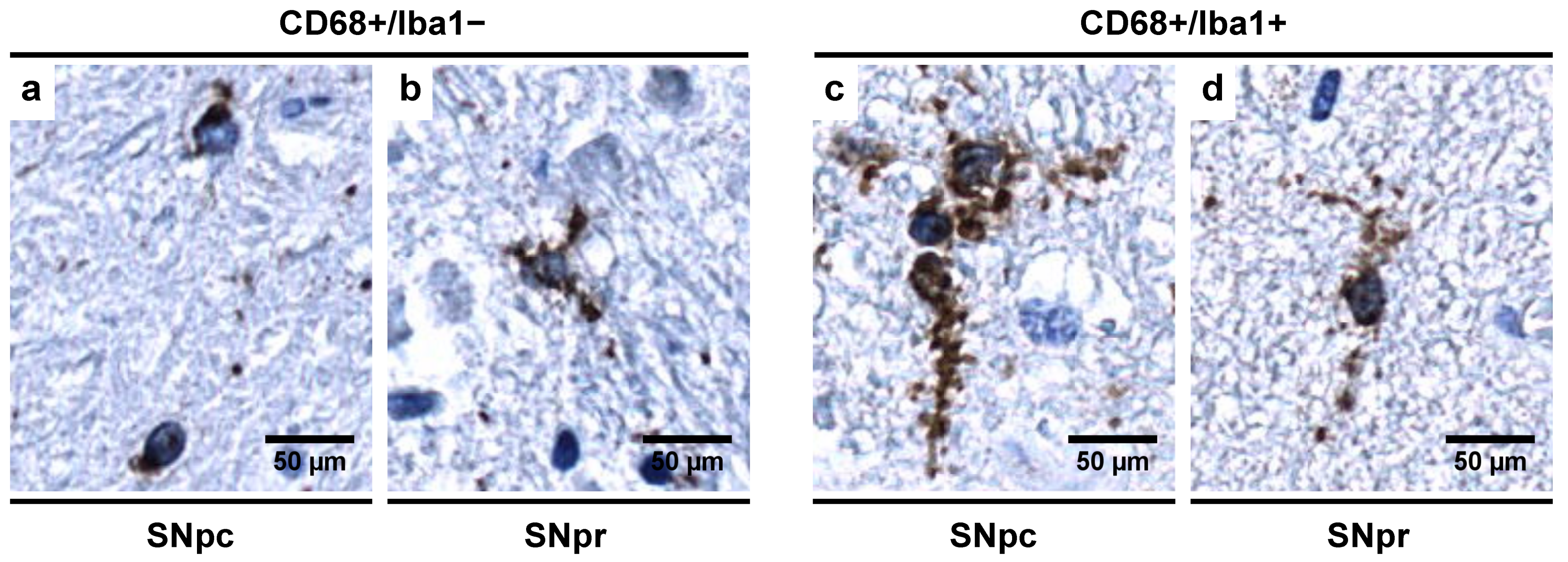
3.5. Morphological Characterization of Microglial Subpopulations
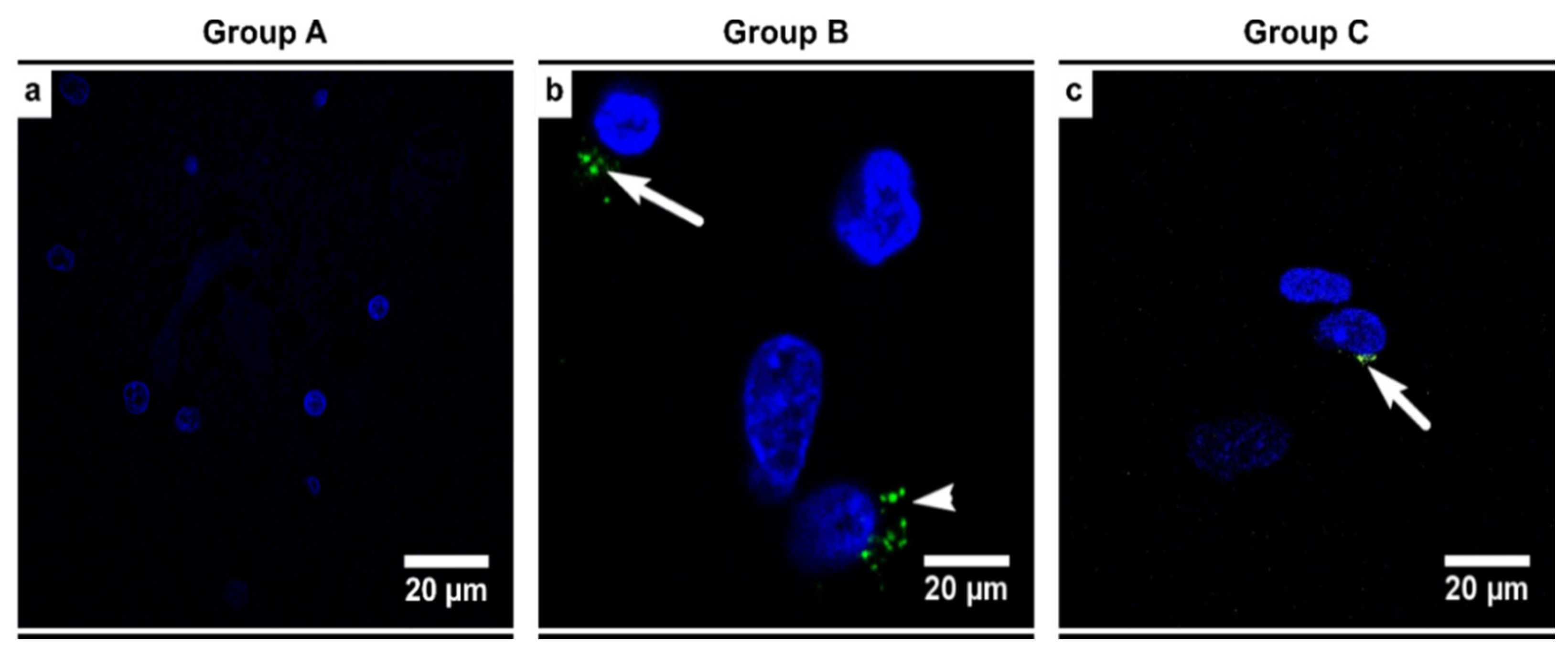
3.6. Alcoholics Showed Detectable HHV-6 Positivity and Viral Load
3.7. Alcohol and HHV-6 Infection in a Synergistic and Potentiating Relationship Cause Disruption of Homeostasis in the SN Region
4. Discussion
4.1. Abundance of CD68+/Iba1− Microglial Subpopulation in Controls Indicates a Special Physiological “Controlled Phagocytotic Activated State”
4.2. Decrease in the Number of CD68+ Microglia with Increase in Iba1+ Expression Shows Microglial Dystrophy Which Leads to Compensatory Mobility from Perivascular to Diffuse Locations
4.3. Alcohol and HHV-6 Infection Co-Induce and Accelerate Microglial Dystrophy
4.4. Dominance of Iba1+/CD11b− Microglial Subpopulation in Alcoholics Leads to Chronic Inflammation, Hyperalgesia, and Allodynia
4.5. Limitations of the Present Study
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Louten, J. Herpesviruses. In Essential Human Virology; Elsevier (Academic Press): Cambridge, MA, USA, 2016; pp. 235–256. ISBN 978-0-12-800947-5. [Google Scholar]
- Salahuddin, S.; Ablashi, D.; Markham, P.; Josephs, S.; Sturzenegger, S.; Kaplan, M.; Halligan, G.; Biberfeld, P.; Wong-Staal, F.; Kramarsky, B.; et al. Isolation of a New Virus, HBLV, in Patients with Lymphoproliferative Disorders. Science 1986, 234, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Collot, S.; Petit, B.; Bordessoule, D.; Alain, S.; Touati, M.; Denis, F.; Ranger-Rogez, S. Real-Time PCR for Quantification of Human Herpesvirus 6 DNA from Lymph Nodes and Saliva. J. Clin. Microbiol. 2002, 40, 2445–2451. [Google Scholar] [CrossRef]
- Di Luca, D.; Mirandola, P.; Ravaioli, T.; Frigatti, A.; Bovenzi, P.; Monini, P.; Cassai, E.; Sighinolfi, L.; Dolcetti, R. Human Herpesviruses 6 and 7 in Salivary Glands and Shedding in Saliva of Healthy and Human Immunodeficiency Virus Positive Individuals. J. Med. Virol. 1995, 45, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Yamamoto, T.; Kondo, T.; Kondo, K.; Okuno, T.; Kosuge, H.; Yamanishi, K. Molecular Epidemiological Studies of Human Herpesvirus 6 in Families. J. Med. Virol. 1994, 42, 224–227. [Google Scholar] [CrossRef]
- Tanaka-Taya, K.; Kondo, T.; Mukai, T.; Miyoshi, H.; Yamamoto, Y.; Okada, S.; Yamanishi, K. Seroepidemiological Study of Human Herpesvirus-6 and -7 in Children of Different Ages and Detection of These Two Viruses in Throat Swabs by Polymerase Chain Reaction. J. Med. Virol. 1996, 48, 88–94. [Google Scholar] [CrossRef]
- Boutolleau, D.; Cointe, D.; Dejean, A.G.; Mace, M.; Agut, H.; Keros, L.G.; Ingrand, D. No Evidence for a Major Risk of Roseolovirus Vertical Transmission during Pregnancy. Clin. Infect. Dis. 2003, 36, 1634–1635. [Google Scholar] [CrossRef] [PubMed]
- Dahl, H.; Fjaertoft, G.; Norsted, T.; Wang, F.; Mousavi-Jazi, M.; Linde, A. Reactivation of Human Herpesvirus 6 during Pregnancy. J. Infect. Dis. 1999, 180, 2035–2038. [Google Scholar] [CrossRef]
- De Bolle, L.; Naesens, L.; De Clercq, E. Update on Human Herpesvirus 6 Biology, Clinical Features, and Therapy. Clin. Microbiol. Rev. 2005, 18, 217–245. [Google Scholar] [CrossRef]
- Ablashi, D.; Agut, H.; Alvarez-Lafuente, R.; Clark, D.A.; Dewhurst, S.; DiLuca, D.; Flamand, L.; Frenkel, N.; Gallo, R.; Gompels, U.A.; et al. Classification of HHV-6A and HHV-6B as Distinct Viruses. Arch. Virol. 2014, 159, 863–870. [Google Scholar] [CrossRef]
- Caruso, A.; Rotola, A.; Comar, M.; Favilli, F.; Galvan, M.; Tosetti, M.; Campello, C.; Caselli, E.; Alessandri, G.; Grassi, M.; et al. HHV-6 Infects Human Aortic and Heart Microvascular Endothelial Cells, Increasing Their Ability to Secrete Proinflammatory Chemokines. J. Med. Virol. 2002, 67, 528–533. [Google Scholar] [CrossRef]
- Chan, P.K.S.; Ng, H.-K.; Hui, M.; Cheng, A.F. Prevalence and Distribution of Human Herpesvirus 6 Variants A and B in Adult Human Brain. J. Med. Virol. 2001, 64, 42–46. [Google Scholar] [CrossRef]
- Donati, D.; Akhyani, N.; Fogdell-Hahn, A.; Cermelli, C.; Cassiani-Ingoni, R.; Vortmeyer, A.; Heiss, J.D.; Cogen, P.; Gaillard, W.D.; Sato, S.; et al. Detection of Human Herpesvirus-6 in Mesial Temporal Lobe Epilepsy Surgical Brain Resections. Neurology 2003, 61, 1405–1411. [Google Scholar] [CrossRef]
- Ishiguro, N.; Yamada, S.; Takahashi, T.; Takahashi, Y.; Togashi, T.; Okuno, T.; Yamanishi, K. Meningo-Encephalitis Associated with HHV-6 Related Exanthem Subitum. Acta Paediatr. 1990, 79, 987–989. [Google Scholar] [CrossRef]
- Suga, S.; Yoshikawa, T.; Asano, Y.; Kozawa, T.; Nakashima, T.; Kobayashi, I.; Yazaki, T.; Yamamoto, H.; Kajita, Y.; Ozaki, T.; et al. Clinical and Virological Analyses of 21 Infants with Exanthem Subitum (Roseola Infantum) and Central Nervous System Complications. Ann. Neurol. 1993, 33, 597–603. [Google Scholar] [CrossRef]
- Eimer, W.A.; Vijaya Kumar, D.K.; Navalpur Shanmugam, N.K.; Rodriguez, A.S.; Mitchell, T.; Washicosky, K.J.; György, B.; Breakefield, X.O.; Tanzi, R.E.; Moir, R.D. Alzheimer’s Disease-Associated β-Amyloid Is Rapidly Seeded by Herpesviridae to Protect against Brain Infection. Neuron 2018, 99, 56–63.e3. [Google Scholar] [CrossRef] [PubMed]
- Readhead, B.; Haure-Mirande, J.-V.; Funk, C.C.; Richards, M.A.; Shannon, P.; Haroutunian, V.; Sano, M.; Liang, W.S.; Beckmann, N.D.; Price, N.D.; et al. Multiscale Analysis of Independent Alzheimer’s Cohorts Finds Disruption of Molecular, Genetic, and Clinical Networks by Human Herpesvirus. Neuron 2018, 99, 64–82.e7. [Google Scholar] [CrossRef] [PubMed]
- Dunn, N.; Kharlamova, N.; Fogdell-Hahn, A. The Role of Herpesvirus 6A and 6B in Multiple Sclerosis and Epilepsy. Scand. J. Immunol. 2020, 92. [Google Scholar] [CrossRef] [PubMed]
- Opsahl, M.L. Early and Late HHV-6 Gene Transcripts in Multiple Sclerosis Lesions and Normal Appearing White Matter. Brain 2005, 128, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Prusty, B.K.; Gulve, N.; Govind, S.; Krueger, G.R.F.; Feichtinger, J.; Larcombe, L.; Aspinall, R.; Ablashi, D.V.; Toro, C.T. Active HHV-6 Infection of Cerebellar Purkinje Cells in Mood Disorders. Front. Microbiol. 2018, 9, 1955. [Google Scholar] [CrossRef]
- Kobayashi, N.; Oka, N.; Takahashi, M.; Shimada, K.; Ishii, A.; Tatebayashi, Y.; Shigeta, M.; Yanagisawa, H.; Kondo, K. Human Herpesvirus 6B Greatly Increases Risk of Depression by Activating Hypothalamic-Pituitary -Adrenal Axis during Latent Phase of Infection. iScience 2020, 23, 101187. [Google Scholar] [CrossRef]
- Beović, B.; Pecaric-Meglic, N.; Marin, J.; Bedernjak, J.; Muzlovic, I.; Cizman, M. Fatal Human Herpesvirus 6-Associated Multifocal Meningoencephalitis in an Adult Female Patient. Scand. J. Infect. Dis. 2001, 33, 942–944. [Google Scholar] [CrossRef]
- Drobyski, W.R.; Knox, K.K.; Majewski, D.; Carrigan, D.R. Fatal Encephalitis Due to Variant B Human Herpesvirus-6 Infection in a Bone Marrow-Transplant Recipient. N. Engl. J. Med. 1994, 330, 1356–1360. [Google Scholar] [CrossRef] [PubMed]
- Novoa, L.J.; Nagra, R.M.; Nakawatase, T.; Edwards-Lee, T.; Tourtellotte, W.W.; Cornford, M.E. Fulminant Demyelinating Encephalomyelitis Associated with Productive HHV-6 Infection in an Immunocompetent Adult. J. Med. Virol. 1997, 52, 301–308. [Google Scholar] [CrossRef]
- Seeley, W.W.; Marty, F.M.; Holmes, T.M.; Upchurch, K.; Soiffer, R.J.; Antin, J.H.; Baden, L.R.; Bromfield, E.B. Post-Transplant Acute Limbic Encephalitis: Clinical Features and Relationship to HHV6. Neurology 2007, 69, 156–165. [Google Scholar] [CrossRef]
- Provenzale, J.M.; van Landingham, K.E.; Lewis, D.V.; Mukundan, S.; White, L.E. Extrahippocampal Involvement in Human Herpesvirus 6 Encephalitis Depicted at MR Imaging. Radiology 2008, 249, 955–963. [Google Scholar] [CrossRef] [PubMed]
- Webb, D.W.; Bjornson, B.H.; Sargent, M.A.; Hukin, J.; Thomas, E.E. Basal Ganglia Infarction Associated with HHV-6 Infection. Arch. Dis. Child. 1997, 76, 362–364. [Google Scholar] [CrossRef]
- Wainwright, M.S.; Martin, P.L.; Morse, R.P.; Lacaze, M.; Provenzale, J.M.; Coleman, R.E.; Morgan, M.A.; Hulette, C.; Kurtzberg, J.; Bushnell, C.; et al. Human Herpesvirus 6 Limbic Encephalitis after Stem Cell Transplantation. Ann. Neurol. 2001, 50, 612–619. [Google Scholar] [CrossRef]
- Singh, N.; Paterson, D.L. Encephalitis caused by human herpesvirus-6 in transplant recipients: Relevance of a Novel Neurotropic Virus. Transplantation 2000, 69, 2474–2479. [Google Scholar] [CrossRef]
- Pulickal, A.S.; Ramachandran, S.; Rizek, P.; Narula, P.; Schubert, R. Chorea and Developmental Regression Associated with Human Herpes Virus-6 Encephalitis. Pediatric Neurol. 2013, 48, 249–251. [Google Scholar] [CrossRef]
- DeLong, M.; Wichmann, T. Changing Views of Basal Ganglia Circuits and Circuit Disorders. Clin. EEG Neurosci. 2010, 41, 61–67. [Google Scholar] [CrossRef]
- Sonne, J.; Reddy, V.; Beato, M.R. Neuroanatomy, Substantia Nigra; StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK536995/ (accessed on 25 June 2021).
- Fedorow, H.; Tribl, F.; Halliday, G.; Gerlach, M.; Riederer, P.; Double, K. Neuromelanin in Human Dopamine Neurons: Comparison with Peripheral Melanins and Relevance to Parkinson’s Disease. Prog. Neurobiol. 2005, 75, 109–124. [Google Scholar] [CrossRef]
- Javed, N.; Cascella, M. Neuroanatomy, Globus Pallidus; StatPearls Publishing: Treasure Island, FL, USA, 2021. Available online: https://www.ncbi.nlm.nih.gov/books/NBK557755/ (accessed on 25 June 2021).
- Phillips, T.J.; Shen, E.H.; McKinnon, C.S.; Burkhart-Kasch, S.; Lessov, C.N.; Palmer, A.A. Forward, Relaxed, and Reverse Selection for Reduced and Enhanced Sensitivity to Ethanol’s Locomotor Stimulant Effects in Mice. Alcohol. Clin. Exp. Res. 2002, 26, 593–602. [Google Scholar] [CrossRef] [PubMed]
- Dudek, B.C.; Phillips, T.J. Distinctions among Sedative, Disinhibitory, and Ataxic Properties of Ethanol in Inbred and Selectively Bred Mice. Psychopharmacology 1990, 101, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Arizzi-LaFrance, M.N.; Correa, M.; Aragon, C.M.G.; Salamone, J.D. Motor Stimulant Effects of Ethanol Injected into the Substantia Nigra Pars Reticulata: Importance of Catalase-Mediated Metabolism and the Role of Acetaldehyde. Neuropsychopharmacology 2006, 31, 997–1008. [Google Scholar] [CrossRef] [PubMed]
- Leclercq, S.; de Timary, P.; Delzenne, N.M.; Stärkel, P. The Link between Inflammation, Bugs, the Intestine and the Brain in Alcohol Dependence. Transl. Psychiatry 2017, 7, e1048. [Google Scholar] [CrossRef]
- Ostergren, A.; Fredriksson, A.; Brittebo, E.B. Norharman-induced motoric impairment in mice: Neurodegeneration and glial activation in Substantia Nigra. J. Neural. Transm. 2006, 113, 313–329. [Google Scholar] [CrossRef] [PubMed]
- Streit, W.J.; Mrak, R.E.; Griffin, W.S.T. Microglia and Neuroinflammation: A Pathological Perspective. J. Neuroinflamm. 2004, 1, 1–14. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bachiller, S.; Jiménez-Ferrer, I.; Paulus, A.; Yang, Y.; Swanberg, M.; Deierborg, T.; Boza-Serrano, A. Microglia in Neurological Diseases: A Road Map to Brain-Disease Dependent-Inflammatory Response. Front. Cell Neurosci. 2018, 12, 488. [Google Scholar] [CrossRef]
- Li, Q.; Barres, B.A. Microglia and Macrophages in Brain Homeostasis and Disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Heindl, S.; Gesierich, B.; Benakis, C.; Llovera, G.; Duering, M.; Liesz, A. Automated Morphological Analysis of Microglia after Stroke. Front. Cell. Neurosci. 2018, 12, 106. [Google Scholar] [CrossRef]
- Wolf, Y.; Yona, S.; Kim, K.-W.; Jung, S. Microglia, Seen from the CX3CR1 Angle. Front. Cell. Neurosci. 2013, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Bortolotti, D.; Gentili, V.; Rotola, A.; Caselli, E.; Rizzo, R. HHV-6A Infection Induces Amyloid-Beta Expression and Activation of Microglial Cells. Alzheimer’s Res. Ther. 2019, 11, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Le, W. Differential Roles of M1 and M2 Microglia in Neurodegenerative Diseases. Mol. Neurobiol. 2016, 53, 1181–1194. [Google Scholar] [CrossRef] [PubMed]
- Jarrahi, A.; Braun, M.; Ahluwalia, M.; Gupta, R.V.; Wilson, M.; Munie, S.; Ahluwalia, P.; Vender, J.R.; Vale, F.L.; Dhandapani, K.M.; et al. Revisiting Traumatic Brain Injury: From Molecular Mechanisms to Therapeutic Interventions. Biomedicines 2020, 8, 389. [Google Scholar] [CrossRef] [PubMed]
- Dauber, W.; Feneis, H.; Feneis, H. Pocket Atlas of Human Anatomy: Founded by Heinz Feneis, 5th rev. ed.; Thieme Stuttgart: New York, NY, USA, 2007; ISBN 978-3-13-511205-3. [Google Scholar]
- Harper, C.; Dixon, G.; Sheedy, D.; Garrick, T. Neuropathological alterations in alcoholic brains. Studies arising from the New South Wales Tissue Resource Centre. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2003, 27, 951–961. [Google Scholar] [CrossRef]
- Skuja, S.; Groma, V.; Ravina, K.; Tarasovs, M.; Cauce, V.; Teteris, O. Protective reactivity and alteration of the brain tissue in alcoholics evidenced by SOD1, MMP9 immunohistochemistry, and electron microscopy. Ultrastruct. Pathol. 2013, 37, 346–355. [Google Scholar] [CrossRef]
- Skuja, S.; Svirskis, S.; Murovska, M. Human Herpesvirus-6 and -7 in the Brain Microenvironment of Persons with Neurological Pathology and Healthy People. Int. J. Mol. Sci. 2021, 22, 2364. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Killingsworth, M.C.; Myasoedova, V.A.; Orekhov, A.N.; Bobryshev, Y.V. CD68/Macrosialin: Not Just a Histochemical Marker. Lab. Investig. 2017, 97, 4–13. [Google Scholar] [CrossRef]
- Holness, C.; Simmons, D. Molecular Cloning of CD68, a Human Macrophage Marker Related to Lysosomal Glycoproteins. Blood 1993, 81, 1607–1613. [Google Scholar] [CrossRef]
- Gottfried, E.; Kunz-Schughart, L.A.; Weber, A.; Rehli, M.; Peuker, A.; Müller, A.; Kastenberger, M.; Brockhoff, G.; Andreesen, R.; Kreutz, M. Expression of CD68 in Non-Myeloid Cell Types. Scand. J. Immunol. 2008, 67, 453–463. [Google Scholar] [CrossRef]
- Hopperton, K.E.; Mohammad, D.; Trépanier, M.O.; Giuliano, V.; Bazinet, R.P. Markers of Microglia in Post-Mortem Brain Samples from Patients with Alzheimer’s Disease: A Systematic Review. Mol. Psychiatry 2018, 23, 177–198. [Google Scholar] [CrossRef]
- Walker, D.G.; Lue, L.-F. Immune Phenotypes of Microglia in Human Neurodegenerative Disease: Challenges to Detecting Microglial Polarization in Human Brains. Alzheimer Res. Ther. 2015, 7, 56. [Google Scholar] [CrossRef]
- Perego, C.; Fumagalli, S.; De Simoni, M.-G. Temporal Pattern of Expression and Colocalization of Microglia/Macrophage Phenotype Markers Following Brain Ischemic Injury in Mice. J. Neuroinflamm. 2011, 8, 174. [Google Scholar] [CrossRef]
- Fischer, H.-G.; Reichmann, G. Brain Dendritic Cells and Macrophages/Microglia in Central Nervous System Inflammation. J. Immunol. 2001, 166, 2717–2726. [Google Scholar] [CrossRef]
- Bachstetter, A.D.; Van Eldik, L.J.; Schmitt, F.A.; Neltner, J.H.; Ighodaro, E.T.; Webster, S.J.; Patel, E.; Abner, E.L.; Kryscio, R.J.; Nelson, P.T. Disease-Related Microglia Heterogeneity in the Hippocampus of Alzheimer’s Disease, Dementia with Lewy Bodies, and Hippocampal Sclerosis of Aging. Acta Neuropathol. Commun. 2015, 3, 32. [Google Scholar] [CrossRef]
- Streit, W.J.; Braak, H.; Xue, Q.-S.; Bechmann, I. Dystrophic (Senescent) Rather than Activated Microglial Cells Are Associated with Tau Pathology and Likely Precede Neurodegeneration in Alzheimer’s Disease. Acta Neuropathol. 2009, 118, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Santpere, G.; Telford, M.; Andrés-Benito, P.; Navarro, A.; Ferrer, I. The Presence of Human Herpesvirus 6 in the Brain in Health and Disease. Biomolecules 2020, 10, 1520. [Google Scholar] [CrossRef]
- Siddon, A.; Lozovatsky, L.; Mohamed, A.; Hudnall, S.D. Human Herpesvirus 6 Positive Reed-Sternberg Cells in Nodular Sclerosis Hodgkin Lymphoma. Br. J. Haematol. 2012, 158, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Vandamme, A.-M.; Fransen, K.; Debaisieux, L.; Marissens, D.; Sprecher, S.; Vaira, D.; Vandenbroucke, A.T.; Verhofstede, C.; Van Dooren, S.; Goubau, P.; et al. Standardisation of Primers and an Algorithm for HIV-1 Diagnostic PCR Evaluated in Patients Harbouring Strains of Diverse Geographical Origin. J. Virol. Methods 1995, 51, 305–316. [Google Scholar] [CrossRef]
- Sultanova, A.; Cistjakovs, M.; Gravelsina, S.; Chapenko, S.; Roga, S.; Cunskis, E.; Nora-Krukle, Z.; Groma, V.; Ventina, I.; Murovska, M. Association of Active Human Herpesvirus-6 (HHV-6) Infection with Autoimmune Thyroid Gland Diseases. Clin. Microbiol. Infect. 2017, 23, e1–e5. [Google Scholar] [CrossRef] [PubMed]
- Lyall, E.G.H.; Cubie, H.A. Human Herpesvirus-6 DNA in the Saliva of Paediatric Oncology Patients and Controls. J. Med. Virol. 1995, 47, 317–322. [Google Scholar] [CrossRef]
- Carlson, S.L.; Parrish, M.E.; Springer, J.E.; Doty, K.; Dossett, L. Acute Inflammatory Response in Spinal Cord Following Impact Injury. Exp. Neurol. 1998, 151, 77–88. [Google Scholar] [CrossRef] [PubMed]
- Zrzavy, T.; Machado-Santos, J.; Christine, S.; Baumgartner, C.; Weiner, H.L.; Butovsky, O.; Lassmann, H. Dominant Role of Microglial and Macrophage Innate Immune Responses in Human Ischemic Infarcts: Inflammation in Ischemic Lesions. Brain Pathol. 2018, 28, 791–805. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Hamanaka, G.; Lo, E.H.; Arai, K. Heterogeneity of Microglia and Their Differential Roles in White Matter Pathology. CNS Neurosci. Ther. 2019, 25, 1290–1298. [Google Scholar] [CrossRef] [PubMed]
- Ohsawa, K.; Imai, Y.; Kanazawa, H.; Sasaki, Y.; Kohsaka, S. Involvement of Iba1 in Membrane Ruffling and Phagocytosis of Macrophages/Microglia. J. Cell Sci. 2000, 113, 3073–3084. [Google Scholar] [CrossRef] [PubMed]
- Ito, D.; Imai, Y.; Ohsawa, K.; Nakajima, K.; Fukuuchi, Y.; Kohsaka, S. Microglia-Specific Localisation of a Novel Calcium Binding Protein, Iba1. Mol. Brain Res. 1998, 57, 1–9. [Google Scholar] [CrossRef]
- Waller, R.; Baxter, L.; Fillingham, D.J.; Coelho, S.; Pozo, J.M.; Mozumder, M.; Frangi, A.F.; Ince, P.G.; Simpson, J.E.; Highley, J.R. Iba1−/CD68+ Microglia are a Prominent Feature of Age-Associated Deep Subcortical White Matter Lesions. PLoS ONE 2019, 14, e0210888. [Google Scholar] [CrossRef] [PubMed]
- Gheorghe, R.-O.; Deftu, A.; Filippi, A.; Grosu, A.; Bica-Popi, M.; Chiritoiu, M.; Chiritoiu, G.; Munteanu, C.; Silvestro, L.; Ristoiu, V. Silencing the Cytoskeleton Protein Iba1 (Ionized Calcium Binding Adapter Protein 1) Interferes with BV2 Microglia Functioning. Cell. Mol. Neurobiol. 2020, 40, 1011–1027. [Google Scholar] [CrossRef] [PubMed]
- De Biase, L.M.; Schuebel, K.E.; Fusfeld, Z.H.; Jair, K.; Hawes, I.A.; Cimbro, R.; Zhang, H.-Y.; Liu, Q.-R.; Shen, H.; Xi, Z.-X.; et al. Local Cues Establish and Maintain Region-Specific Phenotypes of Basal Ganglia Microglia. Neuron 2017, 95, 341–356.e6. [Google Scholar] [CrossRef]
- Lawson, L.J.; Perry, V.H.; Dri, P.; Gordon, S. Heterogeneity in the Distribution and Morphology of Microglia in the Normal Adult Mouse Brain. Neuroscience 1990, 39, 151–170. [Google Scholar] [CrossRef]
- Ayata, P.; Badimon, A.; Strasburger, H.J.; Duff, M.K.; Montgomery, S.E.; Loh, Y.-H.E.; Ebert, A.; Pimenova, A.A.; Ramirez, B.R.; Chan, A.T.; et al. Epigenetic Regulation of Brain Region-Specific Microglia Clearance Activity. Nat. Neurosci. 2018, 21, 1049–1060. [Google Scholar] [CrossRef] [PubMed]
- Sha’fie, M.S.A.; Rathakrishnan, S.; Hazanol, I.N.; Dali, M.H.I.; Khayat, M.E.; Ahmad, S.; Hussin, Y.; Alitheen, N.B.; Jiang, L.-H.; Syed Mortadza, S.A. Ethanol Induces Microglial Cell Death via the NOX/ROS/PARP/TRPM2 Signalling Pathway. Antioxidants 2020, 9, 1253. [Google Scholar] [CrossRef]
- Marshall, S.A.; McClain, J.A.; Wooden, J.I.; Nixon, K. Microglia Dystrophy Following Binge-Like Alcohol Exposure in Adolescent and Adult Male Rats. Front. Neuroanat. 2020, 14. [Google Scholar] [CrossRef] [PubMed]
- Marshall, S.; Geil, C.; Nixon, K. Prior Binge Ethanol Exposure Potentiates the Microglial Response in a Model of Alcohol-Induced Neurodegeneration. Brain Sci. 2016, 6, 16. [Google Scholar] [CrossRef]
- Peng, H.; Geil Nickell, C.R.; Chen, K.Y.; McClain, J.A.; Nixon, K. Increased Expression of M1 and M2 Phenotypic Markers in Isolated Microglia after Four-Day Binge Alcohol Exposure in Male Rats. Alcohol 2017, 62, 29–40. [Google Scholar] [CrossRef]
- Streit, W.J.; Xue, Q.-S.; Tischer, J.; Bechmann, I. Microglial Pathology. Acta neuropathol. Commun. 2014, 2, 142. [Google Scholar] [CrossRef]
- Baron, R.; Babcock, A.A.; Nemirovsky, A.; Finsen, B.; Monsonego, A. Accelerated Microglial Pathology Is Associated with A β Plaques in Mouse Models of A Lzheimer’s Disease. Aging Cell 2014, 13, 584–595. [Google Scholar] [CrossRef]
- Johnson, E.A.; Dao, T.L.; Guignet, M.A.; Geddes, C.E.; Koemeter-Cox, A.I.; Kan, R.K. Increased Expression of the Chemokines CXCL1 and MIP-1α by Resident Brain Cells Precedes Neutrophil Infiltration in the Brain Following Prolonged Soman-Induced Status Epilepticus in Rats. J. Neuroinflamm. 2011, 8, 41. [Google Scholar] [CrossRef]
- Tremblay, M.-E.; Stevens, B.; Sierra, A.; Wake, H.; Bessis, A.; Nimmerjahn, A. The Role of Microglia in the Healthy Brain. J. Neurosci. 2011, 31, 16064–16069. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Crews, F.T. Increased MCP-1 and Microglia in Various Regions of the Human Alcoholic Brain. Exp. Neurol. 2008, 210, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Patt, S.; Gertz, H.J.; Gerhard, L.; Cervós-Navarro, J. Pathological Changes in Dendrites of Substantia Nigra Neurons in Parkinson’s Disease: A Golgi Study. Histol. Histopathol. 1991, 6, 373–380. [Google Scholar] [PubMed]
- Albright, A.V.; Lavi, E.; Black, J.B.; Goldberg, S.; O’Connor, M.J.; Gonzalez-Scarano, F. The Effect of Human Herpesvirus-6 (HHV-6) on Cultured Human Neural Cells: Oligodendrocytes and Microglia. J. Neurovirol. 1998, 4, 486–494. [Google Scholar] [CrossRef] [PubMed]
- Leibovitch, E.C.; Caruso, B.; Ha, S.K.; Schindler, M.K.; Lee, N.J.; Luciano, N.J.; Billioux, B.J.; Guy, J.R.; Yen, C.; Sati, P.; et al. Herpesvirus Trigger Accelerates Neuroinflammation in a Nonhuman Primate Model of Multiple Sclerosis. Proc. Natl. Acad. Sci. USA 2018, 115, 11292–11297. [Google Scholar] [CrossRef] [PubMed]
- von Herrath, M.G.; Fujinami, R.S.; Whitton, J.L. Microorganisms and Autoimmunity: Making the Barren Field Fertile? Nat. Rev. Microbiol. 2003, 1, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Xu, W.; Wang, Y.; Jiang, X.; Li, Y.; Yang, Y.; Yuan, H. CD11b−Activated Src Signal Attenuates Neuroinflammatory Pain by Orchestrating Inflammatory and Anti-Inflammatory Cytokines in Microglia. Mol. Pain 2018, 14, 174480691880815. [Google Scholar] [CrossRef]
- Han, C.; Jin, J.; Xu, S.; Liu, H.; Li, N.; Cao, X. Integrin CD11b Negatively Regulates TLR-Triggered Inflammatory Responses by Activating Syk and Promoting Degradation of MyD88 and TRIF via Cbl-B. Nat. Immunol. 2010, 11, 734–742. [Google Scholar] [CrossRef]
- Coizet, V.; Dommett, E.J.; Klop, E.M.; Redgrave, P.; Overton, P.G. The Parabrachial Nucleus Is a Critical Link in the Transmission of Short Latency Nociceptive Information to Midbrain Dopaminergic Neurons. Neuroscience 2010, 168, 263–272. [Google Scholar] [CrossRef]
- Alexander, G.E.; DeLong, M.R.; Strick, P.L. Parallel Organization of Functionally Segregated Circuits Linking Basal Ganglia and Cortex. Annu. Rev. Neurosci. 1986, 9, 357–381. [Google Scholar] [CrossRef]
- Gurney, K.; Prescott, T.J.; Redgrave, P. A Computational Model of Action Selection in the Basal Ganglia. I. A New Functional Anatomy. Biol. Cybern. 2001, 84, 401–410. [Google Scholar] [CrossRef]
- Lardeux, S.; Baunez, C. Alcohol Preference Influences the Subthalamic Nucleus Control on Motivation for Alcohol in Rats. Neuropsychopharmacology 2008, 33, 634–642. [Google Scholar] [CrossRef][Green Version]
- Pelloux, Y.; Baunez, C. Targeting the Subthalamic Nucleus in a Preclinical Model of Alcohol Use Disorder. Psychopharmacology 2017, 234, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- Pautrat, A.; Rolland, M.; Barthelemy, M.; Baunez, C.; Sinniger, V.; Piallat, B.; Savasta, M.; Overton, P.G.; David, O.; Coizet, V. Revealing a Novel Nociceptive Network That Links the Subthalamic Nucleus to Pain Processing. eLife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- de Tommaso, M.; Arendt-Nielsen, L.; Defrin, R.; Kunz, M.; Pickering, G.; Valeriani, M. Pain in Neurodegenerative Disease: Current Knowledge and Future Perspectives. Behav. Neurol. 2016, 2016, 7576292. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Angst, M.S.; Clark, J.D. Opioid-Induced Hyperalgesia. Anesthesiology 2006, 104, 570–587. [Google Scholar] [CrossRef]
- Avegno, E.M.; Lobell, T.D.; Itoga, C.A.; Baynes, B.B.; Whitaker, A.M.; Weera, M.M.; Edwards, S.; Middleton, J.W.; Gilpin, N.W. Central Amygdala Circuits Mediate Hyperalgesia in Alcohol-Dependent Rats. J. Neurosci. 2018, 38, 7761–7773. [Google Scholar] [CrossRef]


| Group A (Control Group) | Group B (Age-Matched Alcoholics) | Group C (Non-Age-Matched Alcoholics) | ||||||
|---|---|---|---|---|---|---|---|---|
| Individual Code | Age (Years) | Gender (Male/ Female) | Individual Code | Age (Years) | Gender (Male/ Female) | Individual Code | Age (Years) | Gender (Male/ Female) |
| A1 | 34 | M | B1 | 36 | F | C1 | 48 | M |
| A2 | 31 | M | B2 | 23 | M | C2 | 55 | M |
| A3 | 27 | M | B3 | 31 | M | C3 | 60 | M |
| A4 | 23 | M | B4 | 26 | M | C4 | 50 | F |
| A5 | 32 | M | B5 | 33 | M | C5 | 63 | M |
| A6 | 37 | M | B6 | 34 | F | C6 | 45 | M |
| A7 | 33 | M | B7 | 25 | F | C7 | 45 | M |
| A8 | 22 | M | B8 | 30 | M | C8 | 55 | M |
| A9 | 36 | M | B9 | 35 | M | C9 | 45 | F |
| A10 | 17 | M | B10 | 22 | M | C10 | 60 | F |
| A11 | 37 | M | B11 | 35 | M | C11 | 66 | M |
| A12 | 20 | M | B12 | 34 | M | C12 | 63 | M |
| A13 | 26 | M | B13 | 29 | M | C13 | 44 | F |
| C14 | 49 | M | ||||||
| C15 | 45 | F | ||||||
| C16 | 60 | M | ||||||
| C17 | 38 | F | ||||||
| C18 | 40 | M | ||||||
| Group median age | 31 ± 6.79 | Group median age | 31 ± 4.85 | Group median age | 49.5 ± 8.66 | |||
| Primary Antibody * | Antibody Characteristics ** | Clone | Dilution | Manufacturer |
|---|---|---|---|---|
| CD68 | Monoclonal mouse AB against human AG | Kp-1 | 1:200 | Cell Marque (USA) |
| CD11b | Monoclonal rabbit AB against human AG | EP45 | 1:100 | Epitomics (USA) |
| Iba1 | Monoclonal rabbit AB against human AG | EP289 | 1:150 | Epitomics (USA) |
| HHV-6 (20) | Monoclonal mouse AB against viral lysate | - | 1:200 | Santa Cruz (USA) |
| Primer | Primer Sequence |
|---|---|
| Outer primer O1 | 5′-AGTCATCACGATCGGCGTGCTATC-3′ |
| Outer primer O2 | 5′-TATCTAGCGCAATCGCTATGTCG-3′ |
| Inner primer I3 | 5′-TCGACTCTCACCCTACTGAACGAG-3′ |
| Inner primer I4 | 5′-TGACTAGAGAGCGACAAATTGGAG-3′ |
| Region | Group A | Group B | Group C | p Value † |
|---|---|---|---|---|
| Pars Compacta (SNpc) | ||||
| Gray matter | 6.50 ± 0.27 | 6.32 ± 0.20 | 6.22 ± 0.15 | 0.908 |
| White matter | 6.55 ± 0.25 | 6.07 ± 0.18 | 5.97 ± 0.20 | 0.360 |
| p value ‡ | 0.749 | 0.145 | 0.519 | - |
| Pars Reticulata (SNpr) | ||||
| Gray matter | 6.50 ± 0.25 | 6.34 ± 0.22 | 5.61 ± 0.25 | 0.031 ** |
| White matter | 7.40 ± 0.29 | 6.75 ± 0.25 | 5.86 ± 0.19 | <0.001 ** |
| p value ‡ | 0.001 ** | 0.002 ** | 0.020 ** | - |
| Region | Group A | Group B | Group C | p Value † |
|---|---|---|---|---|
| Pars Compacta (SNpc) | ||||
| Gray matter | 2.97 ± 0.15 | 4.41 ± 0.16 | 4.72 ± 0.17 | <0.001 ** |
| White matter | 3.50 ± 0.17 | 4.57 ± 0.14 | 4.80 ± 0.17 | <0.001 ** |
| p value ‡ | <0.001 ** | <0.001 ** | 0.232 | - |
| Pars Reticulata (SNpr) | ||||
| Gray matter | 1.97 ± 0.13 | 2.20 ± 0.15 | 3.27 ± 0.14 | <0.001 ** |
| White matter | 2.30 ± 0.16 | 2.45 ± 0.16 | 3.31 ± 0.13 | <0.001 ** |
| p value ‡ | <0.001 ** | 0.342 | 0.752 | - |
| Region | Group A | Group B | Group C | p Value † |
|---|---|---|---|---|
| Pars Compacta (SNpc) | ||||
| Gray matter | 1.02 ± 0.07 | 1.47 ± 0.07 | 1.58 ± 0.09 | <0.001 ** |
| White matter | 1.34 ± 0.09 | 1.60 ± 0.07 | 1.73 ± 0.09 | 0.005 ** |
| p value ‡ | 0.002 ** | 0.191 | 0.165 | - |
| Pars Reticulata (SNpr) | ||||
| Gray matter | 0.72 ± 0.08 | 0.83 ± 0.08 | 0.88 ± 0.07 | 0.143 |
| White matter | 0.79 ± 0.09 | 0.83 ± 0.07 | 0.95 ± 0.09 | 0.724 |
| p value ‡ | 0.815 | 0.270 | 0.603 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jain, N.; Smirnovs, M.; Strojeva, S.; Murovska, M.; Skuja, S. Chronic Alcoholism and HHV-6 Infection Synergistically Promote Neuroinflammatory Microglial Phenotypes in the Substantia Nigra of the Adult Human Brain. Biomedicines 2021, 9, 1216. https://doi.org/10.3390/biomedicines9091216
Jain N, Smirnovs M, Strojeva S, Murovska M, Skuja S. Chronic Alcoholism and HHV-6 Infection Synergistically Promote Neuroinflammatory Microglial Phenotypes in the Substantia Nigra of the Adult Human Brain. Biomedicines. 2021; 9(9):1216. https://doi.org/10.3390/biomedicines9091216
Chicago/Turabian StyleJain, Nityanand, Marks Smirnovs, Samanta Strojeva, Modra Murovska, and Sandra Skuja. 2021. "Chronic Alcoholism and HHV-6 Infection Synergistically Promote Neuroinflammatory Microglial Phenotypes in the Substantia Nigra of the Adult Human Brain" Biomedicines 9, no. 9: 1216. https://doi.org/10.3390/biomedicines9091216
APA StyleJain, N., Smirnovs, M., Strojeva, S., Murovska, M., & Skuja, S. (2021). Chronic Alcoholism and HHV-6 Infection Synergistically Promote Neuroinflammatory Microglial Phenotypes in the Substantia Nigra of the Adult Human Brain. Biomedicines, 9(9), 1216. https://doi.org/10.3390/biomedicines9091216








