Cyclic Hypoxia Conditioning Alters the Content of Myoblast-Derived Extracellular Vesicles and Enhances Their Cell-Protective Functions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cyclic Hypoxia-Reoxygenation (HR) Treatment
2.2. QRT-PCR for Cells
2.3. EV Isolation
2.4. Nanoparticle Tracking Analysis (NTA) and Protein Measurements
2.5. Transmission Electron Microscopy (TEM)
2.6. Western Blot
2.7. Small RNA Library Preparation and Sequencing
2.8. Small RNA Sequencing Data Analysis
2.9. Taqman miRNA Assay for EVs
2.10. Mass Spectrometry (MS) Sample Preparation
2.11. MS Data Analysis
2.12. Cell Viability Test
2.13. Macrophage Activation
2.14. Angiogenesis Assay
2.15. Transient Middle Cerebral Artery Occlusion Model (tMCAO) and Injection of EVs
2.16. Statistical Analysis
3. Results
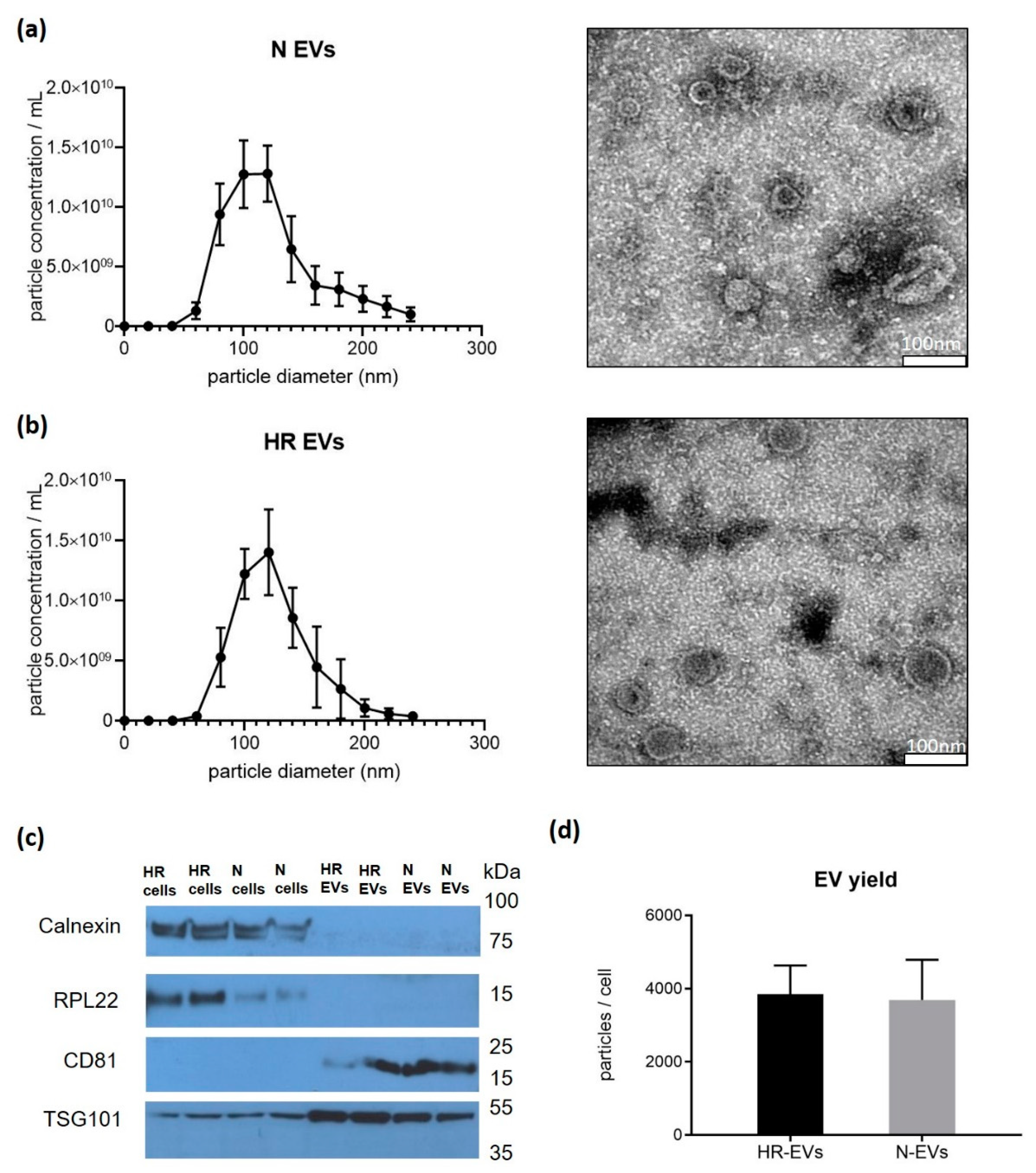
3.1. Cyclic Hypoxia-Reoxygenation (HR) Treatment Has No Effect on EV Size and Quantity
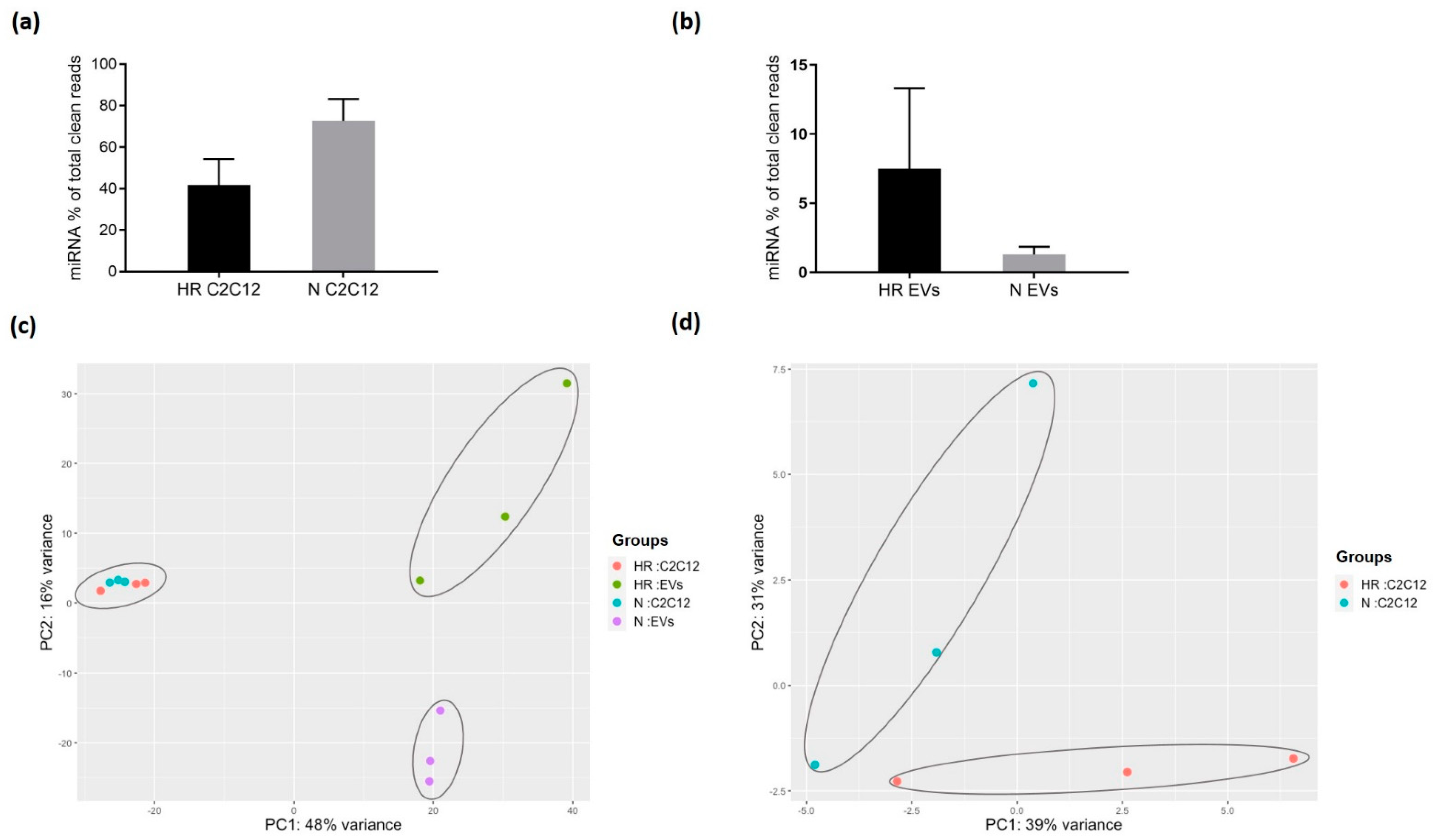
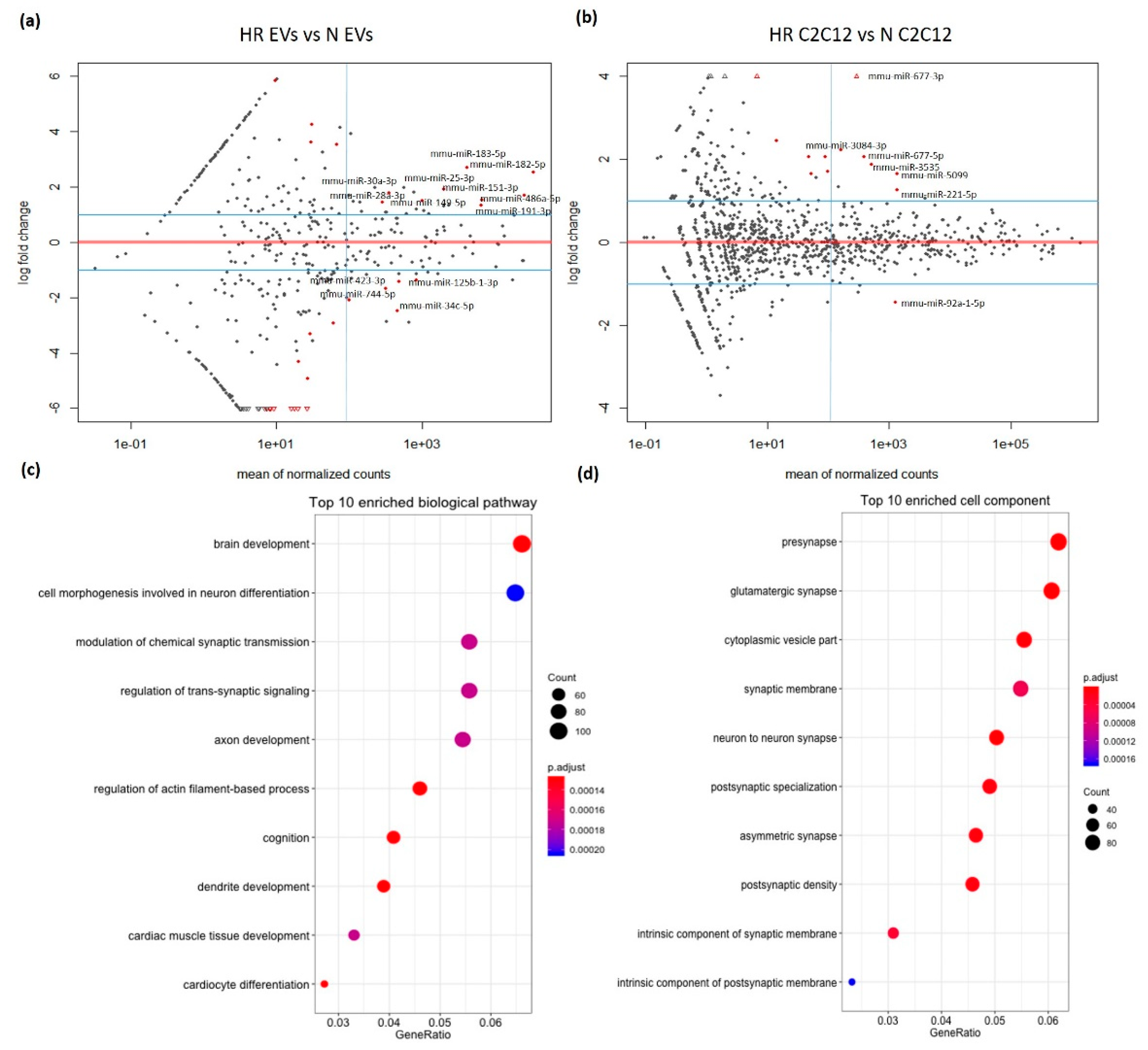
3.2. HR-Treatment Alters the miRNA Profile of EVs Secreted from C2C12 Cells
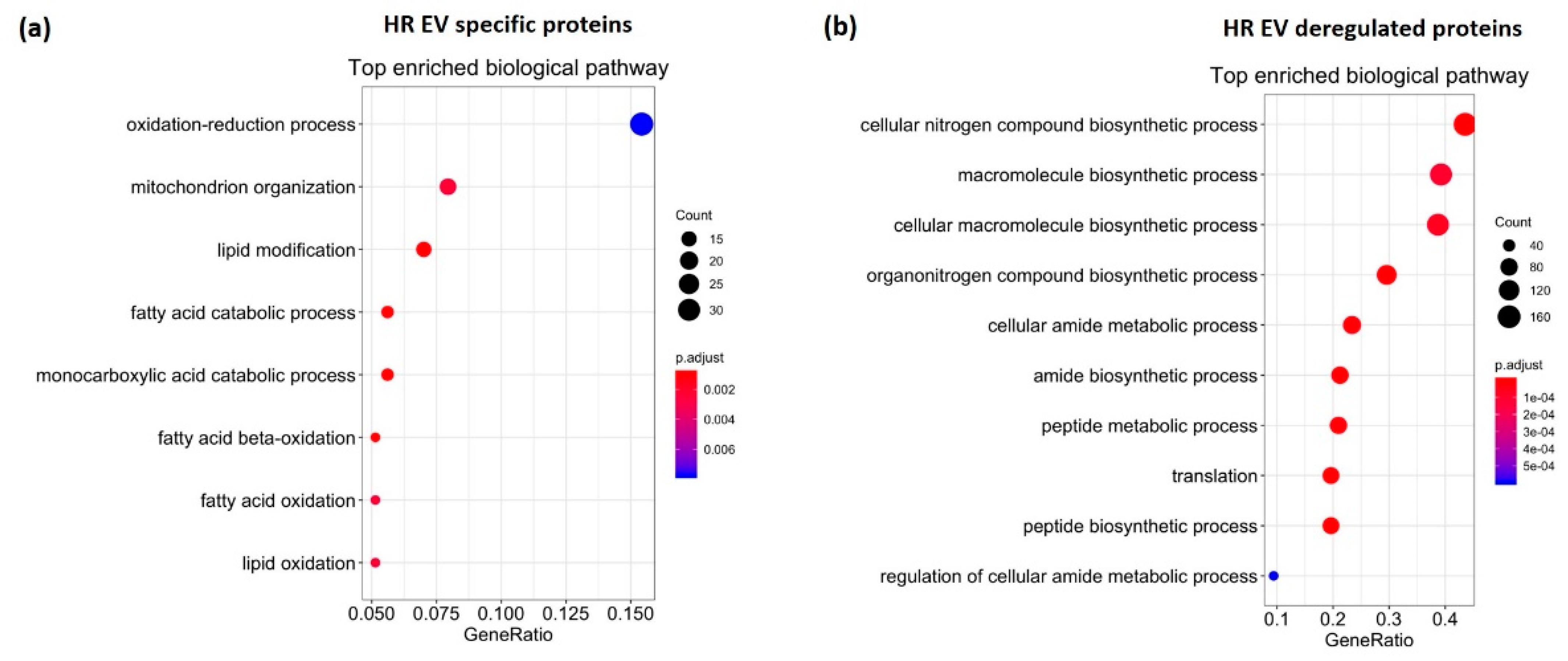
3.3. HR Treatment Alters the Protein Profile of EVs Secreted from C2C12 Cells
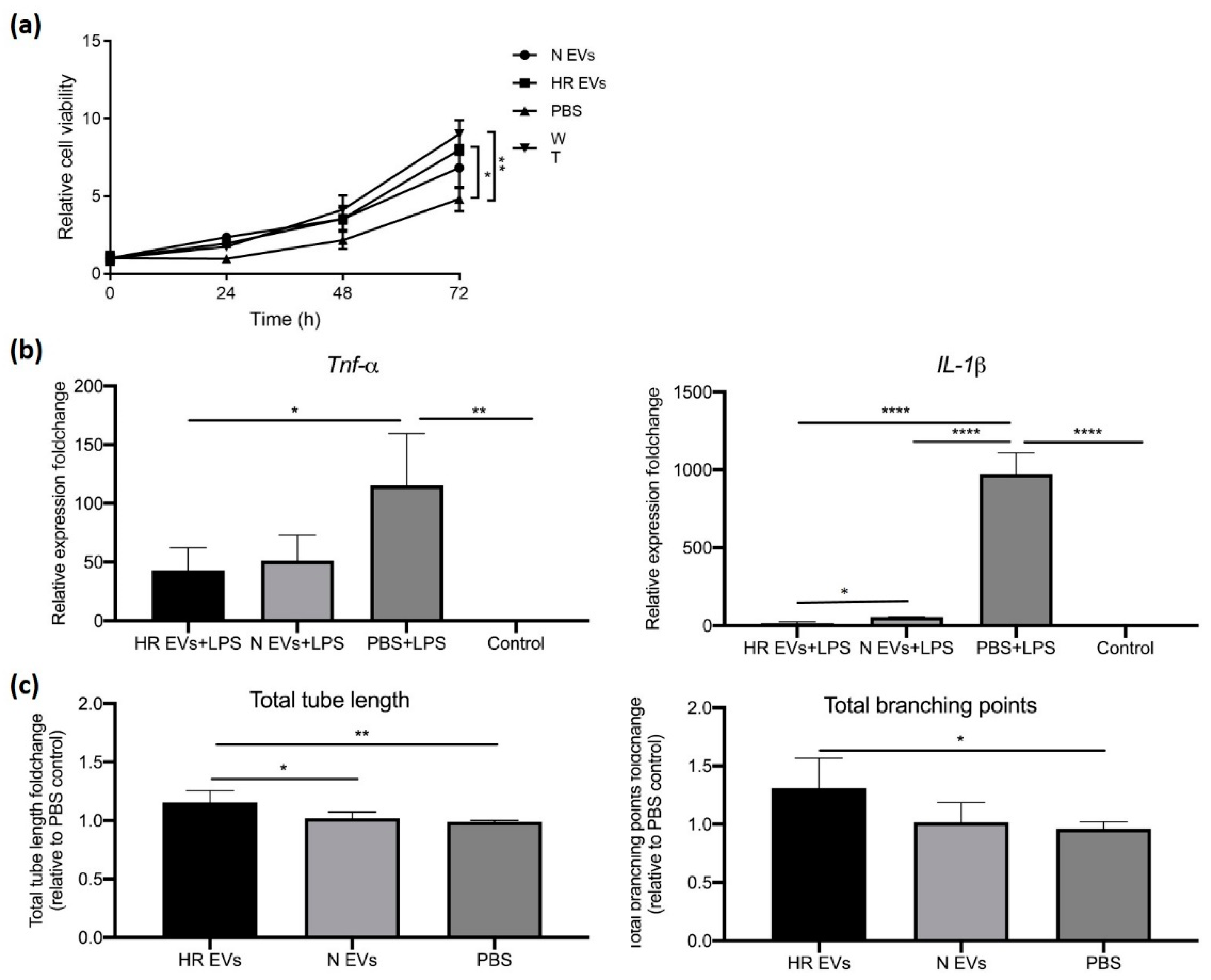
3.4. HR EVs Restore Cell Viability of Hypoxia-Treated C2C12 Cells
3.5. HR EVs Protect Macrophages from LPS-Induced Inflammation
3.6. HR EVs Enhance Angiogenesis
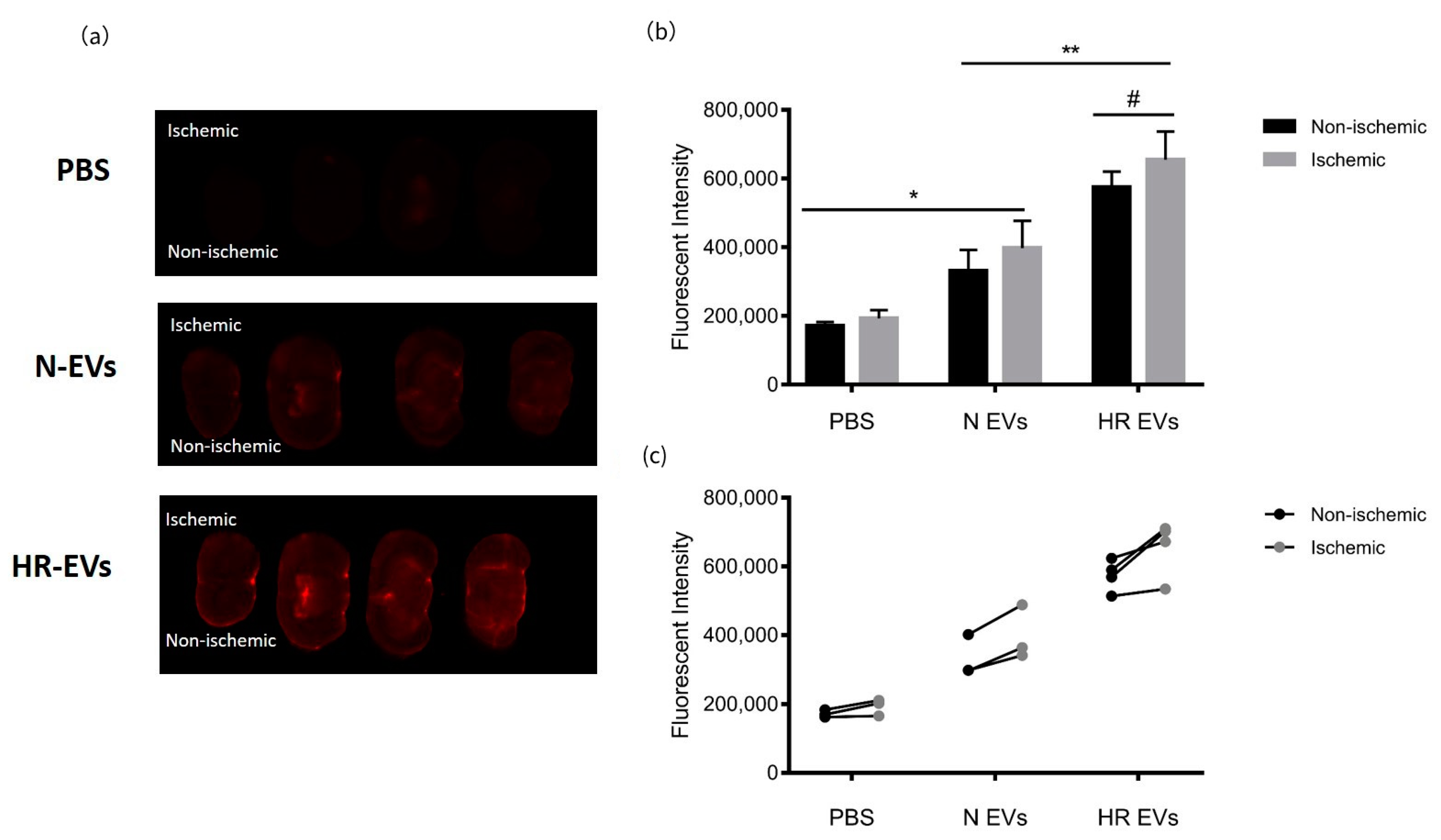
3.7. HR EVs Accumulate in the Ischemic Hemisphere
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Heusch, G.; Bøtker, H.E.; Przyklenk, K.; Redington, A.; Yellon, D. Remote ischemic conditioning. J. Am. Coll. Cardiol. 2015, 65, 177–195. [Google Scholar] [CrossRef] [Green Version]
- Kanoria, S.; Jalan, R.; Seifalian, A.M.; Williams, R.; Davidson, B.R. Protocols and mechanisms for remote ischemic preconditioning: A novel method for reducing ischemia reperfusion injury. Transplantation 2007, 84, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Hausenloy, D.J.; Yellon, D.M. Remote ischaemic preconditioning: Underlying mechanisms and clinical application. Cardiovasc. Res. 2008, 79, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Hausenloy, D.J.; Lim, S.Y. Remote ischemic conditioning: From bench to bedside. Front. Physiol. 2012, 3, 27. [Google Scholar]
- Silachev, D.N.; Isaev, N.K.; Pevzner, I.B.; Zorova, L.D.; Stelmashook, E.V.; Novikova, S.V.; Plotnikov, E.Y.; Skulachev, V.P.; Zorov, D.B. The mitochondria-targeted antioxidants and remote kidney preconditioning ameliorate brain damage through kidney-to-brain cross-talk. PLoS ONE 2012, 7, e51553. [Google Scholar] [CrossRef] [Green Version]
- Pan, J.; Li, X.; Peng, Y. Remote ischemic conditioning for acute ischemic stroke: Dawn in the darkness. Rev. Neurosci. 2016, 27, 501–510. [Google Scholar] [CrossRef]
- Mastitskaya, S.; Marina, N.; Gourine, A.; Gilbey, M.P.; Spyer, K.M.; Teschemacher, A.G.; Kasparov, S.; Trapp, S.; Ackland, G.L.; Gourine, A.V. Cardioprotection evoked by remote ischaemic preconditioning is critically dependent on the activity of vagal pre-ganglionic neurones. Cardiovasc. Res. 2012, 95, 487–494. [Google Scholar] [CrossRef]
- Liu, Z.-J.; Chen, C.; Li, X.-R.; Ran, Y.-Y.; Xu, T.; Zhang, Y.; Geng, X.-K.; Zhang, Y.; Du, H.-S.; Leak, R.K.; et al. Remote ischemic preconditioning-mediated neuroprotection against stroke is associated with significant alterations in peripheral immune responses. CNS Neurosci. Ther. 2016, 22, 43–52. [Google Scholar] [CrossRef]
- Schoemaker, R.G.; van Heijningen, C.L. Bradykinin mediates cardiac preconditioning at a distance. Am. J. Physiol. Heart Circ. Physiol. 2000, 278, H1571–H1576. [Google Scholar] [CrossRef] [Green Version]
- Davidson, S.M.; Selvaraj, P.; He, D.; Boi-Doku, C.; Yellon, R.L.; Vicencio, J.M.; Yellon, D.M. Remote ischaemic preconditioning involves signalling through the SDF-1α/CXCR4 signalling axis. Basic Res. Cardiol. 2013, 108, 377. [Google Scholar] [CrossRef]
- Leung, C.H.; Wang, L.; Nielsen, J.M.; Tropak, M.B.; Fu, Y.Y.; Kato, H.; Callahan, J.; Redington, A.N.; Caldarone, C.A. Remote cardioprotection by transfer of coronary effluent from ischemic preconditioned rabbit heart preserves mitochondrial integrity and function via adenosine receptor activation. Cardiovasc. Drugs Ther. 2014, 28, 7–17. [Google Scholar] [CrossRef]
- Giricz, Z.; Varga, Z.V.; Baranyai, T.; Sipos, P.; Pálóczi, K.; Kittel, Á.; Buzás, E.I.; Ferdinandy, P. Cardioprotection by remote ischemic preconditioning of the rat heart is mediated by extracellular vesicles. J. Mol. Cell. Cardiol. 2014, 68, 75–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Billah, M.; Ridiandries, A.; Allahwala, U.; Mudaliar, H.; Dona, A.; Hunyor, S.; Khachigian, L.M.; Bhindi, R. Circulating mediators of remote ischemic preconditioning: Search for the missing link between non-lethal ischemia and cardioprotection. Oncotarget 2019, 10, 216. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moreno-Gonzalo, O.; Fernandez-Delgado, I.; Sanchez-Madrid, F. Post-translational add-ons mark the path in exosomal protein sorting. Cell. Mol. Life Sci. 2018, 75, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Vallabhaneni, K.C.; Penfornis, P.; Dhule, S.; Guillonneau, F.; Adams, K.V.; Yuan Mo, Y.; Xu, R.; Liu, Y.; Watabe, K.; Vemuri, M.C.; et al. Extracellular vesicles from bone marrow mesenchymal stem/stromal cells transport tumor regulatory microRNA, proteins, and metabolites. Oncotarget 2015, 6, 4953. [Google Scholar] [CrossRef] [Green Version]
- Van Niel, G.; d’Angelo, G.; Raposo, G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol. 2018, 19, 213–228. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kogure, T.; Lin, W.L.; Yan, I.K.; Braconi, C.; Patel, T. Intercellular nanovesicle-mediated microRNA transfer: A mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology 2011, 54, 1237–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minghua, W.; Zhijian, G.; Chahua, H.; Qiang, L.; Minxuan, X.; luqiao, W.; Weifang, Z.; Peng, L.; Biming, Z.; Lingling, Y.; et al. Plasma exosomes induced by remote ischaemic preconditioning attenuate myocardial ischaemia/reperfusion injury by transferring miR-24. Cell Death Dis. 2018, 9, 1–14. [Google Scholar] [CrossRef]
- Sohel, M.H. Extracellular/circulating microRNAs: Release mechanisms, functions and challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Vicencio, J.M.; Yellon, D.M.; Sivaraman, V.; Das, D.; Boi-Doku, C.; Arjun, S.; Zheng, Y.; Riquelme, J.A.; Kearney, J.; Sharma, V. Plasma exosomes protect the myocardium from ischemia-reperfusion injury. J. Am. Coll. Cardiol. 2015, 65, 1525–1536. [Google Scholar] [CrossRef] [Green Version]
- Davidson, S.M.; Riquelme, J.A.; Zheng, Y.; Vicencio, J.M.; Lavandero, S.; Yellon, D.M. Endothelial cells release cardioprotective exosomes that may contribute to ischaemic preconditioning. Sci. Rep. 2018, 8, 1–9. [Google Scholar]
- Li, J.; Rohailla, S.; Gelber, N.; Rutka, J.; Sabah, N.; Gladstone, R.A.; Wei, C.; Hu, P.; Kharbanda, R.K.; Redington, A.N. MicroRNA-144 is a circulating effector of remote ischemic preconditioning. Basic Res. Cardiol. 2014, 109, 423. [Google Scholar] [CrossRef]
- Just, J.; Yan, Y.; Farup, J.; Sieljacks, P.; Sloth, M.; Venø, M.; Gu, T.; de Paoli, F.V.; Nyengaard, J.R.; Bæk, R.; et al. Blood flow-restricted resistance exercise alters the surface profile, miRNA cargo and functional impact of circulating extracellular vesicles. Sci. Rep. 2020, 10, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Xu, Q.; Cui, Y.; Luan, J.; Zhou, X.; Li, H.; Han, J. Exosomes from C2C12 myoblasts enhance osteogenic differentiation of MC3T3-E1 pre-osteoblasts by delivering miR-27a-3p. Biochem. Biophys. Res. Commun. 2018, 498, 32–37. [Google Scholar] [CrossRef]
- Cai, Z.; Luo, W.; Zhan, H.; Semenza, G.L. Hypoxia-inducible factor 1 is required for remote ischemic preconditioning of the heart. Proc. Natl. Acad. Sci. USA 2013, 110, 17462–17467. [Google Scholar] [CrossRef] [Green Version]
- Bøtker, H.E.; Kharbanda, R.; Schmidt, M.R.; Bøttcher, M.; Kaltoft, A.K.; Terkelsen, C.J.; Munk, K.; Andersen, N.H.; Hansen, T.M.; Trautner, S.; et al. Remote ischaemic conditioning before hospital admission, as a complement to angioplasty, and effect on myocardial salvage in patients with acute myocardial infarction: A randomised trial. Lancet 2010, 375, 727–734. [Google Scholar] [CrossRef]
- Johnsen, J.; Pryds, K.; Salman, R.; Løfgren, B.; Kristiansen, S.B.; Bøtker, H.E. The remote ischemic preconditioning algorithm: Effect of number of cycles, cycle duration and effector organ mass on efficacy of protection. Basic Res. Cardiol. 2016, 111, 10. [Google Scholar] [CrossRef]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Gordon, A.; Hannon, G. FASTX-Toolkit. FASTQ/A Short-Reads Pre-Processing Tools (version 0.0. 13). 2010. Available online: http://hannonlab.cshl.edu/fastx_toolkit/download.html (accessed on 7 September 2021).
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [Green Version]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pajak, M.; Simpson, T. miRNAtap: miRNAtap: microRNA Targets—Aggregated Predictions. R package version. 2016. Available online: https://bioconductor.org/packages/release/bioc/manuals/miRNAtap/man/miRNAtap.pdf (accessed on 7 September 2021).
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Akimov, V.; Barrio-Hernandez, I.; Hansen, S.V.; Hallenborg, P.; Pedersen, A.-K.; Bekker-Jensen, D.B.; Puglia, M.; Christensen, S.D.; Vanselow, J.T.; Nielsen, M.M. UbiSite approach for comprehensive mapping of lysine and N-terminal ubiquitination sites. Nat. Struct. Mol. Biol. 2018, 25, 631–640. [Google Scholar] [CrossRef] [PubMed]
- Ægidius, H.M.; Veidal, S.S.; Feigh, M.; Hallenborg, P.; Puglia, M.; Pers, T.H.; Vrang, N.; Jelsing, J.; Kornum, B.R.; Blagoev, B. Multi-omics characterization of a diet-induced obese model of non-alcoholic steatohepatitis. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9. 1: Protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2012, 41, D808–D815. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef] [Green Version]
- Engel, O.; Kolodziej, S.; Dirnagl, U.; Prinz, V. Modeling stroke in mice-middle cerebral artery occlusion with the filament model. J. Vis. Exp. 2011, 47, e2423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Oksvold, M.P.; Kullmann, A.; Forfang, L.; Kierulf, B.; Li, M.; Brech, A.; Vlassov, A.V.; Smeland, E.B.; Neurauter, A.; Pedersen, K.W. Expression of B-cell surface antigens in subpopulations of exosomes released from B-cell lymphoma cells. Clin. Ther. 2014, 36, 847–862. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Q.; Younus, M.; Thompson, J.; Hu, Y.; Hollander, J.M.; Lesnefsky, E.J. Intermediary metabolism and fatty acid oxidation: Novel targets of electron transport chain-driven injury during ischemia and reperfusion. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H787–H795. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, Å.B.; Gottlieb, R.A. Heart mitochondria: Gates of life and death. Cardiovasc. Res. 2008, 77, 334–343. [Google Scholar] [CrossRef]
- Kalogeris, T.; Baines, C.P.; Krenz, M.; Korthuis, R.J. Cell biology of ischemia/reperfusion injury. In International Review of Cell and Molecular Biology; Elsevier: Amsterdam, The Netherlands, 2012; pp. 229–317. [Google Scholar]
- Nour, M.; Scalzo, F.; Liebeskind, D.S. Ischemia-reperfusion injury in stroke. Interv. Neurol. 2012, 1, 185–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slegtenhorst, B.R.; Dor, F.J.; Rodriguez, H.; Voskuil, F.J.; Tullius, S.G. Ischemia/reperfusion injury and its consequences on immunity and inflammation. Curr. Transplant. Rep. 2014, 1, 147–154. [Google Scholar] [CrossRef] [Green Version]
- Konstantinov, I.E.; Arab, S.; Li, J.; Coles, J.G.; Boscarino, C.; Mori, A.; Cukerman, E.; Dawood, F.; Cheung, M.M.; Shimizu, M. Remote ischemic preconditioning stimulus modifies gene expression in mouse myocardium. J. Thorac. Cardiovasc. Surg. 2005, 130, 1326–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peralta, C.; Fernández, L.; Panés, J.; Prats, N.; Sans, M.; Piqué, J.M.; Gelpıí, E.; Roselló-Catafau, J. Preconditioning protects against systemic disorders associated with hepatic ischemia-reperfusion through blockade of tumor necrosis factor–induced P-selectin up-regulation in the rat. Hepatology 2001, 33, 100–113. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.-H.; Yoon, D.-W.; Kim, J.-H.; Lee, J.-H.; Lim, C.-H. Effect of remote ischemic post-conditioning on systemic inflammatory response and survival rate in lipopolysaccharide-induced systemic inflammation model. J. Inflamm. 2014, 11, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pryds, K.; Rahbek Schmidt, M.; Bjerre, M.; Thiel, S.; Refsgaard, J.; Bøtker, H.E.; Drage Østgård, R.; Ranghøj Nielsen, R. Effect of long-term remote ischemic conditioning on inflammation and cardiac remodeling. Scand. Cardiovasc. J. 2019, 53, 183–191. [Google Scholar] [CrossRef]
- Carmeliet, P. Angiogenesis in health and disease. Nat. Med. 2003, 9, 653–660. [Google Scholar] [CrossRef]
- Beck, H.; Plate, K.H. Angiogenesis after cerebral ischemia. Acta Neuropathol. 2009, 117, 481–496. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, D.A. Cerebral angiogenesis: A realistic therapy for ischemic disease? In Cerebral Angiogenesis; Springer: Berlin/Heidelberg, Germany, 2014; pp. 21–24. [Google Scholar]
- Liu, J.; Wang, Y.; Akamatsu, Y.; Lee, C.C.; Stetler, R.A.; Lawton, M.T.; Yang, G.-Y. Vascular remodeling after ischemic stroke: Mechanisms and therapeutic potentials. Prog. Neurobiol. 2014, 115, 138–156. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; Xia, F.C.; Wagner, P.D.; Breen, E.C. Exercise-induced VEGF transcriptional activation in brain, lung and skeletal muscle. Respir. Physiol. Neurobiol. 2010, 170, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Ren, C.; Li, N.; Li, S.; Han, R.; Huang, Q.; Hu, J.; Jin, K.; Ji, X. Limb ischemic conditioning improved cognitive deficits via eNOS-dependent augmentation of angiogenesis after chronic cerebral hypoperfusion in rats. Aging Dis. 2018, 9, 869. [Google Scholar] [CrossRef] [Green Version]
- Yoshimura, A.; Ito, M. Resolution of inflammation and repair after ischemic brain injury. Neuroimmunol. Neuroinflammation 2020, 7, 264–276. [Google Scholar] [CrossRef]
- Webb, R.L.; Kaiser, E.E.; Scoville, S.L.; Thompson, T.A.; Fatima, S.; Pandya, C.; Sriram, K.; Swetenburg, R.L.; Vaibhav, K.; Arbab, A.S. Human neural stem cell extracellular vesicles improve tissue and functional recovery in the murine thromboembolic stroke model. Transl. Stroke Res. 2018, 9, 530–539. [Google Scholar] [CrossRef] [Green Version]
- Jeanneteau, J.; Hibert, P.; Martinez, M.C.; Tual-Chalot, S.; Tamareille, S.; Furber, A.; Andriantsitohaina, R.; Prunier, F. Microparticle release in remote ischemic conditioning mechanism. Am. J. Physiol. Heart Circ. Physiol. 2012, 303, H871–H877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, C.; Weng, X.; Hu, W.; Lv, Z.; Xiao, H.; Ding, C.; Gyabaah, O.-A.K.; Xie, H.; Zhou, L.; Wu, J.; et al. Hypoxia-inducible MiR-182 promotes angiogenesis by targeting RASA1 in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2015, 34, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Roser, A.-E.; Gomes, L.C.; Halder, R.; Jain, G.; Maass, F.; Tönges, L.; Tatenhorst, L.; Bähr, M.; Fischer, A.; Lingor, P. miR-182-5p and miR-183-5p act as GDNF mimics in dopaminergic midbrain neurons. Mol. Ther. Nucleic Acids 2018, 11, 9–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viñas, J.L.; Burger, D.; Zimpelmann, J.; Haneef, R.; Knoll, W.; Campbell, P.; Gutsol, A.; Carter, A.; Allan, D.S.; Burns, K.D. Transfer of microRNA-486-5p from human endothelial colony forming cell–derived exosomes reduces ischemic kidney injury. Kidney Int. 2016, 90, 1238–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aspuria, P.-J.; Tamanoi, F. The Rheb family of GTP-binding proteins. Cell. Signal. 2004, 16, 1105–1112. [Google Scholar] [CrossRef] [PubMed]
- Hoda, M.N.; Siddiqui, S.; Herberg, S.; Periyasamy-Thandavan, S.; Bhatia, K.; Hafez, S.S.; Johnson, M.H.; Hill, W.D.; Ergul, A.; Fagan, S.C.; et al. Remote ischemic perconditioning is effective alone and in combination with intravenous tissue-type plasminogen activator in murine model of embolic stroke. Stroke 2012, 43, 2794–2799. [Google Scholar] [CrossRef] [PubMed]
- Wei, M.; Huo, K.; Liu, R.; Yang, J.; Cheng, Y.; Chang, S.; Ren, D.; Luo, G. The design and rationale of a clinical trial evaluating limb postconditioning in young patients with intracranial arterial stenosis. J. Stroke Cerebrovasc. Dis. 2016, 25, 2506–2512. [Google Scholar] [CrossRef]
- Venkat, P.; Chopp, M.; Chen, J. Blood–brain barrier disruption, vascular impairment, and ischemia/reperfusion damage in diabetic stroke. J. Am. Heart Assoc. 2017, 6, e005819. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Ségaliny, A.; et al. Elucidation of exosome migration across the blood–brain barrier model in vitro. Cell. Mol. Bioeng. 2016, 9, 509–529. [Google Scholar] [CrossRef] [PubMed]
- Gupta, D.; Liang, X.; Pavlova, S.; Wiklander, O.P.B.; Corso, G.; Zhao, Y.; Saher, O.; Bost, J.; Zickler, A.M.; Piffko, A.; et al. Quantification of extracellular vesicles in vitro and in vivo using sensitive bioluminescence imaging. J. Extracell. Vesicles 2020, 9, 1800222. [Google Scholar] [CrossRef]
- Toribio, V.; Morales, S.; López-Martín, S.; Cardeñes, B.; Cabañas, C.; Yáñez-Mó, M. Development of a quantitative method to measure EV uptake. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Kolluri, K.K.; Gowers, K.H.; Janes, S.M. TRAIL delivery by MSC-derived extracellular vesicles is an effective anticancer therapy. J. Extracell. Vesicles 2017, 6, 1265291. [Google Scholar] [CrossRef] [PubMed]
- Vajen, T.; Benedikter, B.J.; Heinzmann, A.C.A.; Vasina, E.M.; Henskens, Y.; Parsons, M.; Maguire, P.B.; Stassen, F.R.; Heemskerk, J.W.M.; Schurgers, L.J.; et al. Platelet extracellular vesicles induce a pro-inflammatory smooth muscle cell phenotype. J. Extracell. Vesicles 2017, 6, 1322454. [Google Scholar] [CrossRef]
- Nakazaki, M.; Morita, T.; Lankford, K.L.; Askenase, P.W.; Kocsis, J.D. Small extracellular vesicles released by infused mesenchymal stromal cells target M2 macrophages and promote TGF-β upregulation, microvascular stabilization and functional recovery in a rodent model of severe spinal cord injury. J. Extracell. Vesicles 2021, 10, e12137. [Google Scholar] [CrossRef]
- Zakeri, A.; Whitehead, B.J.; Stensballe, A.; de Korne, C.; Williams, A.R.; Everts, B.; Nejsum, P. Parasite worm antigens instruct macrophages to release immunoregulatory extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12131. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, Y.; Gu, T.; Christensen, S.D.K.; Su, J.; Lassen, T.R.; Hjortbak, M.V.; Lo, I.; Venø, S.T.; Tóth, A.E.; Song, P.; et al. Cyclic Hypoxia Conditioning Alters the Content of Myoblast-Derived Extracellular Vesicles and Enhances Their Cell-Protective Functions. Biomedicines 2021, 9, 1211. https://doi.org/10.3390/biomedicines9091211
Yan Y, Gu T, Christensen SDK, Su J, Lassen TR, Hjortbak MV, Lo I, Venø ST, Tóth AE, Song P, et al. Cyclic Hypoxia Conditioning Alters the Content of Myoblast-Derived Extracellular Vesicles and Enhances Their Cell-Protective Functions. Biomedicines. 2021; 9(9):1211. https://doi.org/10.3390/biomedicines9091211
Chicago/Turabian StyleYan, Yan, Tingting Gu, Stine Duelund Kaas Christensen, Junyi Su, Thomas Ravn Lassen, Marie Vognstoft Hjortbak, IJu Lo, Susanne Trillingsgaard Venø, Andrea Erzsebet Tóth, Ping Song, and et al. 2021. "Cyclic Hypoxia Conditioning Alters the Content of Myoblast-Derived Extracellular Vesicles and Enhances Their Cell-Protective Functions" Biomedicines 9, no. 9: 1211. https://doi.org/10.3390/biomedicines9091211
APA StyleYan, Y., Gu, T., Christensen, S. D. K., Su, J., Lassen, T. R., Hjortbak, M. V., Lo, I., Venø, S. T., Tóth, A. E., Song, P., Nielsen, M. S., Bøtker, H. E., Blagoev, B., Drasbek, K. R., & Kjems, J. (2021). Cyclic Hypoxia Conditioning Alters the Content of Myoblast-Derived Extracellular Vesicles and Enhances Their Cell-Protective Functions. Biomedicines, 9(9), 1211. https://doi.org/10.3390/biomedicines9091211






