In Vitro Miniaturized Tuberculosis Spheroid Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. Host Cell Culture
2.2. Bacterial Strains
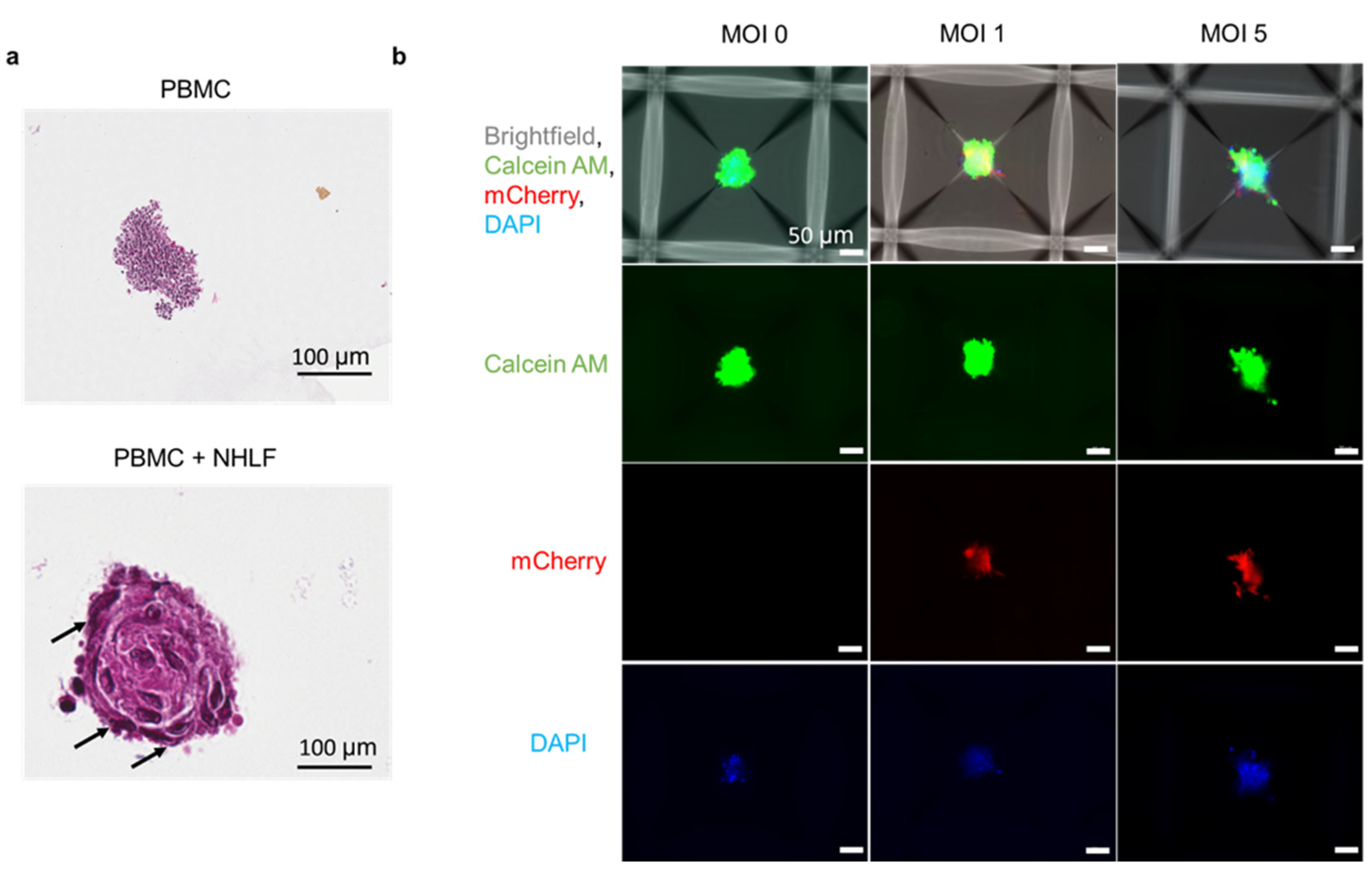
2.3. Three-Dimensional TB Spheroid Formation
2.4. Three-Dimensional PBMC-Stroma Co-Cultures
2.5. Staining and Fluorescent Microscopy
2.5.1. Live Dead Staining
2.5.2. Hypoxia Staining
2.5.3. Nile Red Staining
2.6. Generation of Spheroid Sections and Staining
2.7. Multiplex Cytokine Analysis
2.8. Mass Spectrometry Imaging
2.9. MALDI-MSI Data Analysis
2.10. Transmission Electron Microscopy
2.11. Drug Treatment
2.12. Bacterial Colony Formation (CFU Assay)
3. Results
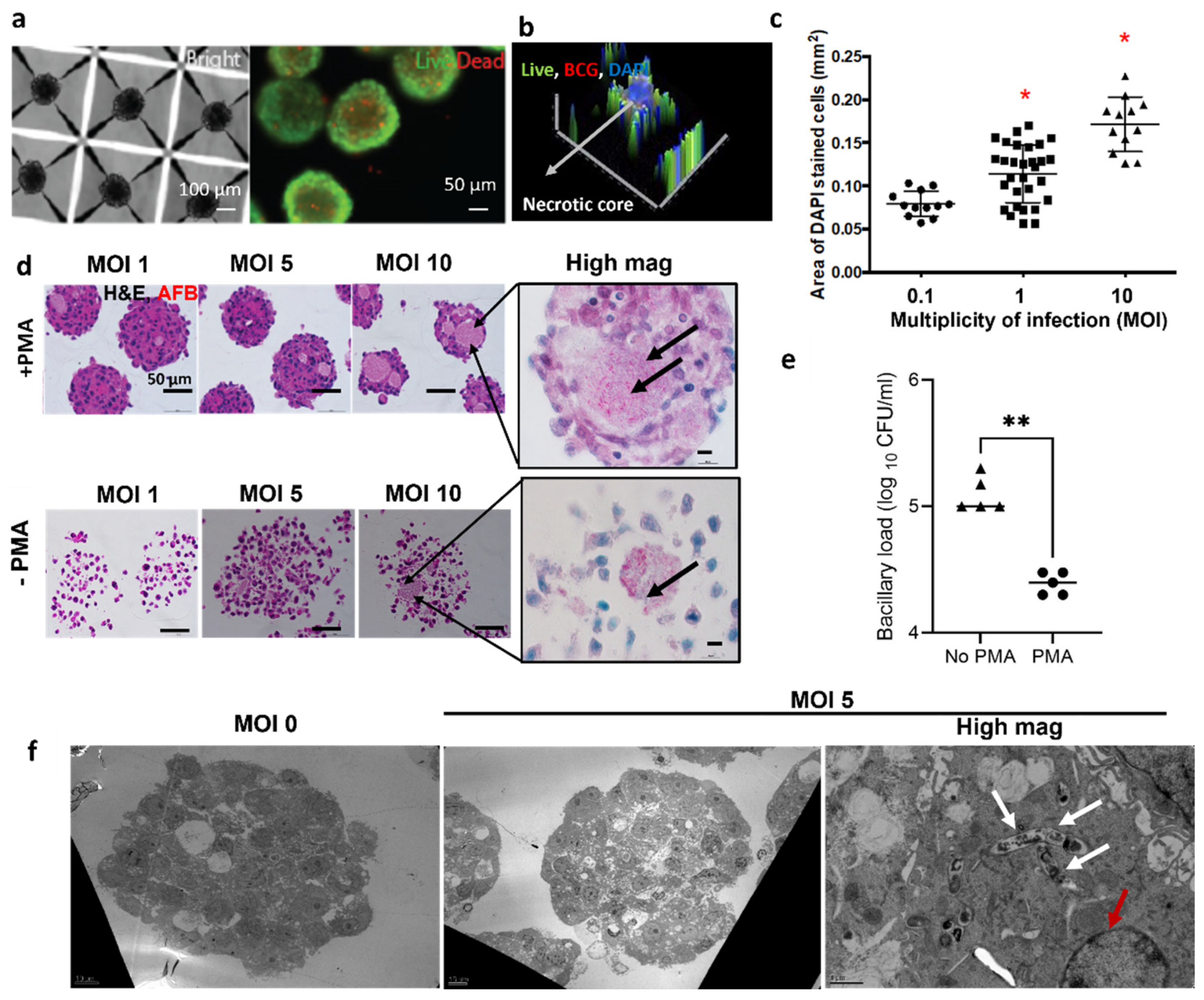
3.1. Modelling the TB Spheroid Infection In Vitro
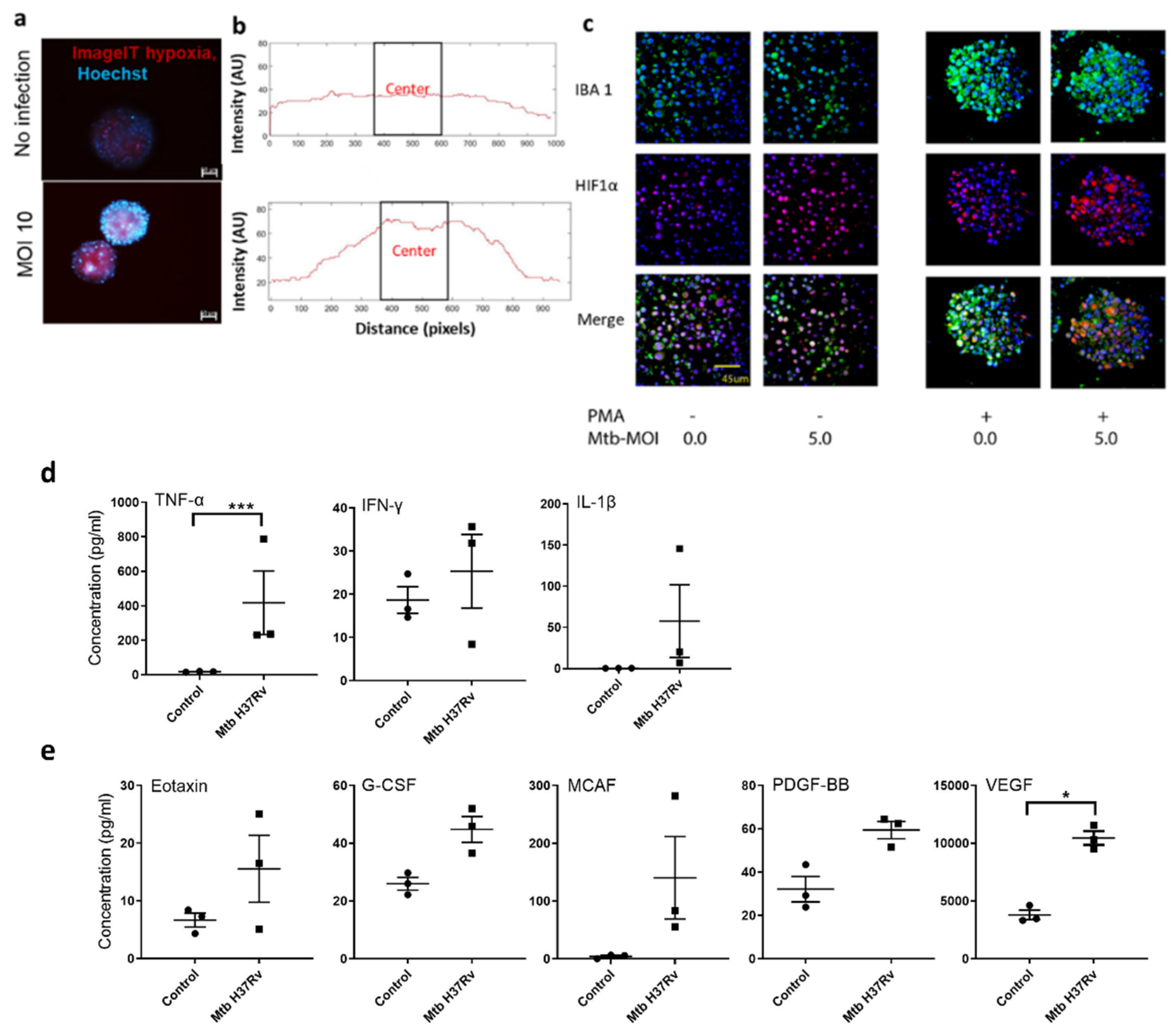
3.2. TB Spheroid Are Hypoxic and Promote Proinflammatory Milieu
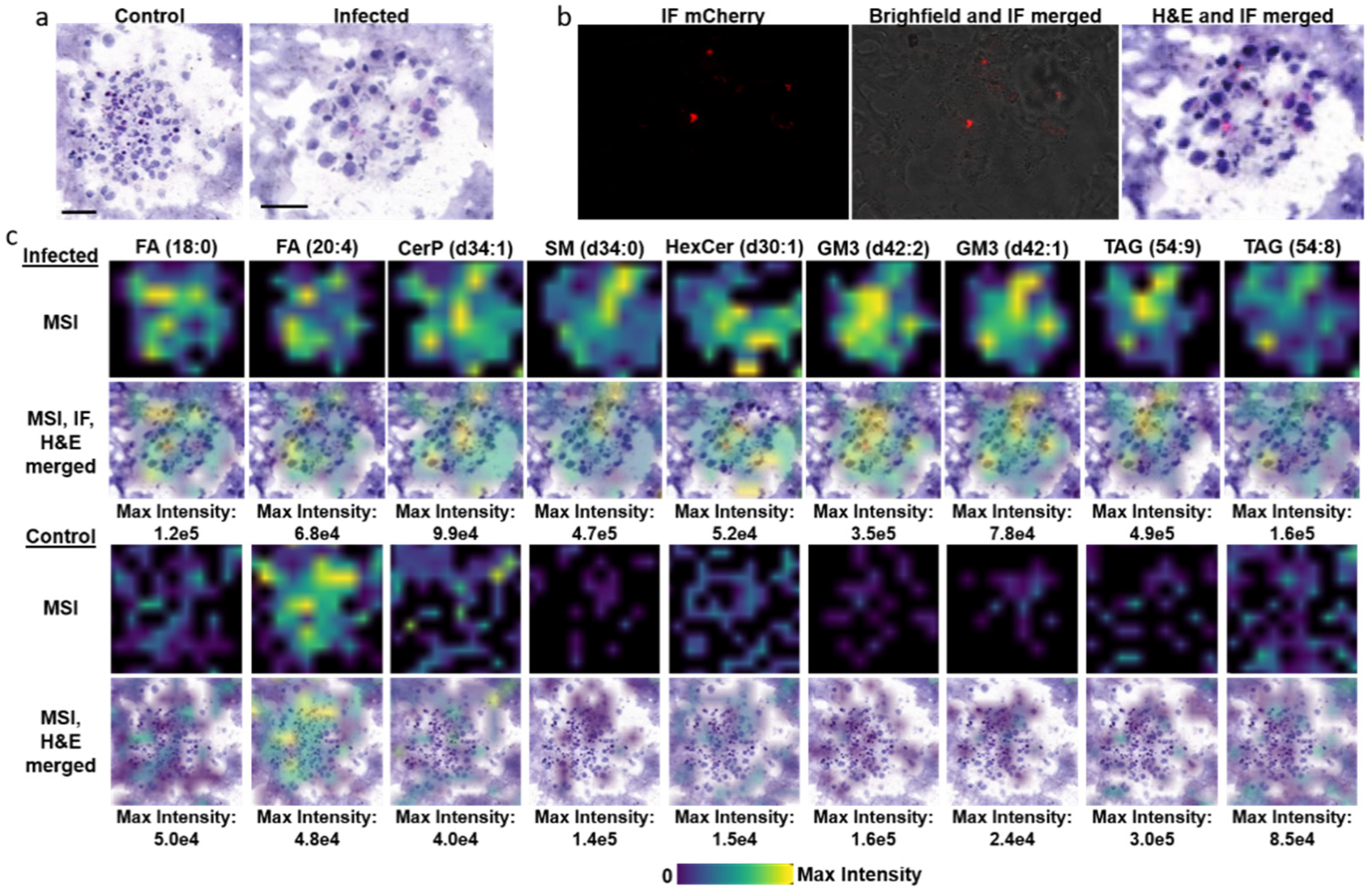
3.3. TB Spheroids Display Extensive Lipid Deposition
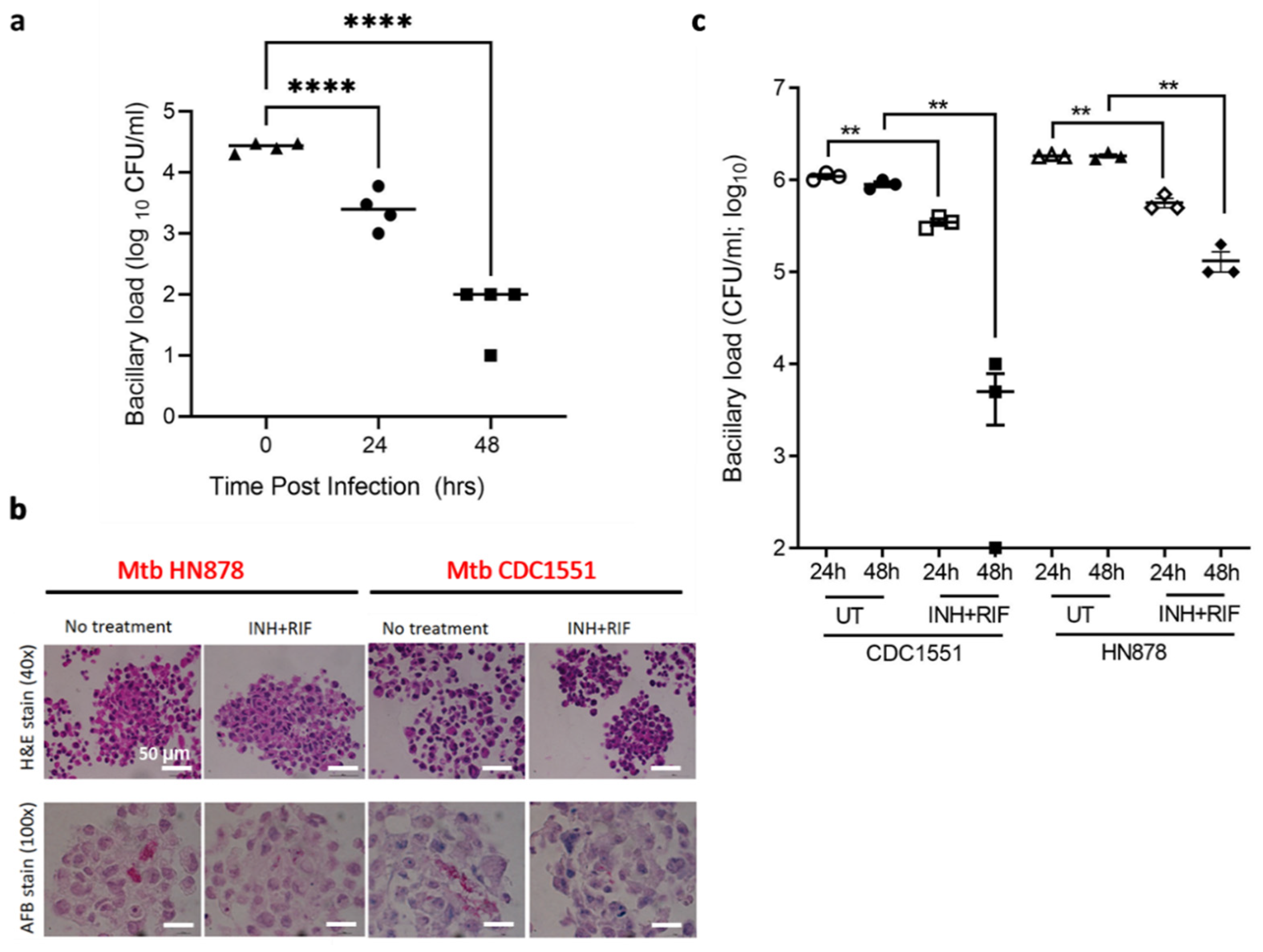
3.4. TB Spheroids Display Bacterial Strain Dependent Response to Anti-TB Drugs
3.5. Formation of PBMC-NHLF Co-Culture TB Spheroids
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Appendix A
- MATLAB Code for surface intensity plot (used in Figure 2b)
- Image = imread(‘Image.tiff’);
- x = [0 size(Image,2)];
- y = [size(Image,1)/2 size(Image,1)/2];
- s = improfile(I,x,y);
- figure
- subplot (2,1,1)
- imshow (Image)
- hold on
- plot(x,y,‘r’)
- subplot (2,1,2)
- plot(s(:,1,1),‘r’)
- axis ([0 1000 0 80])
References
- Birkness, K.A.; Guarner, J.; Sable, S.B.; Tripp, R.A.; Kellar, K.L.; Bartlett, J.; Quinn, F.D. An in vitro model of the leukocyte interactions associated with granuloma formation in Mycobacterium tuberculosis infection. Immunol. Cell Biol. 2007, 85, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.H.; Warit, S.; Wan, B.; Hwang, C.H.; Pauli, G.F.; Franzblau, S.G. Low-oxygen-recovery assay for high-throughput screening of compounds against nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2007, 51, 1380–1385. [Google Scholar] [CrossRef] [Green Version]
- Rustad, T.R.; Harrell, M.I.; Liao, R.; Sherman, D.R. The Enduring Hypoxic Response of Mycobacterium tuberculosis. PLoS ONE 2008, 3, e1502. [Google Scholar] [CrossRef] [PubMed]
- Voskuil, M.I.; Schnappinger, D.; Visconti, K.C.; Harrell, M.I.; Dolganov, G.M.; Sherman, D.R.; Schoolnik, G.K. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 2003, 198, 705–713. [Google Scholar] [CrossRef] [Green Version]
- Sarathy, J.; Dartois, V.; Dick, T.; Gengenbacher, M. Reduced drug uptake in phenotypically resistant nutrient-starved nonreplicating Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 2013, 57, 1648–1653. [Google Scholar] [CrossRef] [Green Version]
- Manabe, Y.C.; Bishai, W.R. Latent Mycobacterium tuberculosis–persistence, patience, and winning by waiting. Nat. Med. 2000, 6, 1327–1329. [Google Scholar] [CrossRef] [PubMed]
- Ollinger, J.; Bailey, M.A.; Moraski, G.C.; Casey, A.; Florio, S.; Alling, T.; Miller, M.J.; Parish, T. A Dual Read-Out Assay to Evaluate the Potency of Compounds Active against Mycobacterium tuberculosis. PLoS ONE 2013, 8, e60531. [Google Scholar] [CrossRef] [Green Version]
- Abramovitch, R.B. Mycobacterium tuberculosis reporter strains as tools for drug discovery and development. IUBMB Life 2018, 70, 818–825. [Google Scholar] [CrossRef]
- Bashiri, G.; Rehan, A.M.; Greenwood, D.R.; Dickson, J.M.; Baker, E.N. Metabolic engineering of cofactor F 420 production in Mycobacterium smegmatis. PLoS ONE 2010, 5, e15803. [Google Scholar] [CrossRef] [Green Version]
- Choudhary, E.; Thakur, P.; Pareek, M.; Agarwal, N. Gene silencing by CRISPR interference in mycobacteria. Nat. Commun. 2015, 6, 6267. [Google Scholar] [CrossRef] [Green Version]
- Gibson, S.E.R.; Harrison, J.; Cox, J.A.G. Modelling a Silent Epidemic: A Review of the In Vitro Models of Latent Tuberculosis. Pathogens 2018, 7, 88. [Google Scholar] [CrossRef] [PubMed]
- Santucci, P.; Bouzid, F.; Smichi, N.; Poncin, I.; Kremer, L.; De Chastellier, C.; Drancourt, M.; Canaan, S. Experimental Models of Foamy Macrophages and Approaches for Dissecting the Mechanisms of Lipid Accumulation and Consumption during Dormancy and Reactivation of Tuberculosis. Front. Cell. Infect. Microbiol. 2016, 6, 122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Je, S.; Quan, H.; Na, Y.; Cho, S.-N.; Kim, B.-J.; Seok, S.H. An in vitro model of granuloma-like cell aggregates substantiates early host immune responses against Mycobacterium massiliense infection. Biol. Open 2016, 5, 1118–1127. [Google Scholar] [CrossRef] [Green Version]
- Bhavanam, S.; Rayat, G.R.; Keelan, M.; Kunimoto, D.; Drews, S.J. Characterization of immune responses of human PBMCs infected with Mycobacterium tuberculosis H37Ra: Impact of donor declared BCG vaccination history on immune responses and M. tuberculosis growth. PLoS ONE 2018, 13, e0203822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lay, G.; Poquet, Y.; Salek-Peyron, P.; Puissegur, M.P.; Botanch, C.; Bon, H.; Levillain, F.; Duteyrat, J.L.; Emile, J.F.; Altare, F. Langhans giant cells from M. tuberculosis-induced human granulomas cannot mediate mycobacterial uptake. J. Pathol. 2007, 211, 76–85. [Google Scholar] [CrossRef]
- Al Shammari, B.; Shiomi, T.; Tezera, L.; Bielecka, M.K.; Workman, V.; Sathyamoorthy, T.; Mauri, F.; Jayasinghe, S.N.; Robertson, B.D.; D’Armiento, J.; et al. The Extracellular Matrix Regulates Granuloma Necrosis in Tuberculosis. J. Infect. Dis. 2015, 212, 463–473. [Google Scholar] [CrossRef] [Green Version]
- Guirado, E.; Mbawuike, U.; Keiser, T.L.; Arcos, J.; Azad, A.K.; Wang, S.H.; Schlesinger, L.S. Characterization of host and microbial determinants in individuals with latent tuberculosis infection using a human granuloma model. mBio 2015, 6, e02537-14. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, N.; Pawar, S.; Sirakova, T.D.; Deb, C.; Warren, W.L.; Kolattukudy, P.E. Human granuloma in vitro model, for TB dormancy and resuscitation. PLoS ONE 2013, 8, e53657. [Google Scholar] [CrossRef] [Green Version]
- Puissegur, M.-P.; Botanch, C.; Duteyrat, J.-L.; Delsol, G.; Caratero, C.; Altare, F. An in vitro dual model of mycobacterial granulomas to investigate the molecular interactions between mycobacteria and human host cells. Cell. Microbiol. 2004, 6, 423–433. [Google Scholar] [CrossRef]
- Tezera, L.B.; Bielecka, M.K.; Chancellor, A.; Reichmann, M.T.; Shammari, B.A.; Brace, P.; Batty, A.; Tocheva, A.; Jogai, S.; Marshall, B.G.; et al. Dissection of the host-pathogen interaction in human tuberculosis using a bioengineered 3-dimensional model. eLife 2017, 6, e21283. [Google Scholar] [CrossRef] [PubMed]
- Workman, V.L.; Tezera, L.B.; Elkington, P.T.; Jayasinghe, S.N. Controlled Generation of Microspheres Incorporating Extracellular Matrix Fibrils for Three-Dimensional Cell Culture. Adv. Funct. Mater. 2014, 24, 2648–2657. [Google Scholar] [CrossRef]
- Fonseca, K.L.; Rodrigues, P.N.S.; Olsson, I.A.S.; Saraiva, M. Experimental study of tuberculosis: From animal models to complex cell systems and organoids. PLoS Pathog. 2017, 13, e1006421. [Google Scholar] [CrossRef] [Green Version]
- Votanopoulos, K.I.; Forsythe, S.; Sivakumar, H.; Mazzocchi, A.; Aleman, J.; Miller, L.; Levine, E.; Triozzi, P.; Skardal, A. Model of Patient-Specific Immune-Enhanced Organoids for Immunotherapy Screening: Feasibility Study. Ann. Surg. Oncol. 2020, 27, 1956–1967. [Google Scholar] [CrossRef]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Nadauld, L.; Ootani, A.; Corney, D.C.; Pai, R.K.; Gevaert, O.; Cantrell, M.A.; Rack, P.G.; Neal, J.T.; Chan, C.W. Oncogenic transformation of diverse gastrointestinal tissues in primary organoid culture. Nat. Med. 2014, 20, 769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purwada, A.; Shah, S.B.; Beguelin, W.; Melnick, A.M.; Singh, A. Modular Immune Organoids with Integrin Ligand Specificity Differentially Regulate Ex Vivo B Cell Activation. ACS Biomater. Sci. Eng. 2017, 3, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Purwada, A.; Jaiswal, M.K.; Ahn, H.; Nojima, T.; Kitamura, D.; Gaharwar, A.K.; Cerchietti, L.; Singh, A. Ex vivo engineered immune organoids for controlled germinal center reactions. Biomaterials 2015, 63, 24–34. [Google Scholar] [CrossRef] [Green Version]
- Huh, D.; Matthews, B.D.; Mammoto, A.; Montoya-Zavala, M.; Hsin, H.Y.; Ingber, D.E. Reconstituting Organ-Level Lung Functions on a Chip. Science 2010, 328, 1662–1668. [Google Scholar] [CrossRef] [Green Version]
- Benam, K.H.; Villenave, R.; Lucchesi, C.; Varone, A.; Hubeau, C.; Lee, H.-H.; Alves, S.E.; Salmon, M.; Ferrante, T.C.; Weaver, J.C.; et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat. Methods 2016, 13, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Braian, C.; Svensson, M.; Brighenti, S.; Lerm, M.; Parasa, V.R. A 3D Human Lung Tissue Model for Functional Studies on Mycobacterium tuberculosis Infection. J. Vis. Exp. 2015, e53084. [Google Scholar] [CrossRef] [Green Version]
- Harimoto, T.; Singer, Z.S.; Velazquez, O.S.; Zhang, J.; Castro, S.; Hinchliffe, T.E.; Mather, W.; Danino, T. Rapid screening of engineered microbial therapies in a 3D multicellular model. Proc. Natl. Acad. Sci. USA 2019, 116, 9002–9007. [Google Scholar] [CrossRef] [Green Version]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology 2015, 148, 126–136.e6. [Google Scholar] [CrossRef] [Green Version]
- Singh, M.; Close, D.A.; Mukundan, S.; Johnston, P.A.; Sant, S. Production of Uniform 3D Microtumors in Hydrogel Microwell Arrays for Measurement of Viability, Morphology, and Signaling Pathway Activation. ASSAY Drug Dev. Technol. 2015, 13, 570–583. [Google Scholar] [CrossRef] [PubMed]
- Tung, Y.-C.; Hsiao, A.Y.; Allen, S.G.; Torisawa, Y.-S.; Ho, M.; Takayama, S. High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array. Analyst 2011, 136, 473–478. [Google Scholar] [CrossRef]
- Wallberg, F.; Tenev, T.; Meier, P. Analysis of Apoptosis and Necroptosis by Fluorescence-Activated Cell Sorting. Cold Spring Harb. Protoc. 2016, 2016. [Google Scholar] [CrossRef] [Green Version]
- Jurchen, J.C.; Rubakhin, S.S.; Sweedler, J.V. MALDI-MS imaging of features smaller than the size of the laser beam. J. Am. Soc. Mass. Spectrom. 2005, 16, 1654–1659. [Google Scholar] [CrossRef] [Green Version]
- Rohde, K.H.; Veiga, D.F.T.; Caldwell, S.; Balázsi, G.; Russell, D.G. Linking the Transcriptional Profiles and the Physiological States of Mycobacterium tuberculosis during an Extended Intracellular Infection. PLoS Pathog. 2012, 8, e1002769. [Google Scholar] [CrossRef]
- Cummings, B.S.; Schnellmann, R.G. Measurement of cell death in mammalian cells. Curr. Protoc. Pharm. 2004, 25, 12.8.1–12.8.22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belton, M.; Brilha, S.; Manavaki, R.; Mauri, F.; Nijran, K.; Hong, Y.T.; Patel, N.H.; Dembek, M.; Tezera, L.; Green, J.; et al. Hypoxia and tissue destruction in pulmonary TB. Thorax 2016, 71, 1145–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Via, L.E.; Lin, P.L.; Ray, S.M.; Carrillo, J.; Allen, S.S.; Eum, S.Y.; Taylor, K.; Klein, E.; Manjunatha, U.; Gonzales, J.; et al. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 2008, 76, 2333–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peyssonnaux, C.; Datta, V.; Cramer, T.; Doedens, A.; Theodorakis, E.A.; Gallo, R.L.; Hurtado-Ziola, N.; Nizet, V.; Johnson, R.S. HIF-1alpha expression regulates the bactericidal capacity of phagocytes. J. Clin. Investig. 2005, 115, 1806–1815. [Google Scholar] [CrossRef] [Green Version]
- Russell, D.G.; Cardona, P.J.; Kim, M.J.; Allain, S.; Altare, F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat. Immunol. 2009, 10, 943–948. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.J.; Wainwright, H.C.; Locketz, M.; Bekker, L.G.; Walther, G.B.; Dittrich, C.; Visser, A.; Wang, W.; Hsu, F.F.; Wiehart, U.; et al. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol. Med. 2010, 2, 258–274. [Google Scholar] [CrossRef]
- Lee, W.; VanderVen, B.C.; Fahey, R.J.; Russell, D.G. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J. Biol. Chem. 2013, 288, 6788–6800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmins, G.S.; Deretic, V. Mechanisms of action of isoniazid. Mol. Microbiol. 2006, 62, 1220–1227. [Google Scholar] [CrossRef] [PubMed]
- Iacobino, A.; Piccaro, G.; Giannoni, F.; Mustazzolu, A.; Fattorini, L. Mycobacterium tuberculosis Is Selectively Killed by Rifampin and Rifapentine in Hypoxia at Neutral pH. Antimicrob. Agents Chemother. 2017, 61, e02296-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterling, T.R.; Villarino, M.E.; Borisov, A.S.; Shang, N.; Gordin, F.; Bliven-Sizemore, E.; Hackman, J.; Hamilton, C.D.; Menzies, D.; Kerrigan, A.; et al. Three Months of Rifapentine and Isoniazid for Latent Tuberculosis Infection. N. Engl. J. Med. 2011, 365, 2155–2166. [Google Scholar] [CrossRef] [Green Version]
- Subbian, S.; Tsenova, L.; O’Brien, P.; Yang, G.; Kushner, N.L.; Parsons, S.; Peixoto, B.; Fallows, D.; Kaplan, G. Spontaneous latency in a rabbit model of pulmonary tuberculosis. Am. J. Pathol. 2012, 181, 1711–1724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Subbian, S.; Tsenova, L.; Yang, G.; O’Brien, P.; Parsons, S.; Peixoto, B.; Taylor, L.; Fallows, D.; Kaplan, G. Chronic pulmonary cavitary tuberculosis in rabbits: A failed host immune response. Open Biol. 2011, 1, 110016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koo, M.S.; Subbian, S.; Kaplan, G. Strain specific transcriptional response in Mycobacterium tuberculosis infected macrophages. Cell Commun. Signal. 2012, 10, 2. [Google Scholar] [CrossRef] [Green Version]
- Leemans, J.C.; Thepen, T.; Weijer, S.; Florquin, S.; van Rooijen, N.; van de Winkel, J.G.; van der Poll, T. Macrophages Play a Dual Role during Pulmonary Tuberculosis in Mice. J. Infect. Dis. 2005, 191, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Peng, C.-C.; Kuo, C.W.; Chueh, D.-Y.; Wu, H.-M.; Liu, Y.-H.; Chen, P.; Tung, Y.-C. Study of oxygen tension variation within live tumor spheroids using microfluidic devices and multi-photon laser scanning microscopy. RSC Adv. 2018, 8, 30320–30329. [Google Scholar] [CrossRef] [Green Version]
- Voissiere, A.; Jouberton, E.; Maubert, E.; Degoul, F.; Peyrode, C.; Chezal, J.-M.; Miot-Noirault, É. Development and characterization of a human three-dimensional chondrosarcoma culture for in vitro drug testing. PLoS ONE 2017, 12, e0181340. [Google Scholar] [CrossRef]
- Singh, M.; Mukundan, S.; Jaramillo, M.; Oesterreich, S.; Sant, S. Three-Dimensional Breast Cancer Models Mimic Hallmarks of Size-Induced Tumor Progression. Cancer Res. 2016, 76, 3732–3743. [Google Scholar] [CrossRef] [Green Version]
- Yoshihara, T.; Hirakawa, Y.; Hosaka, M.; Nangaku, M.; Tobita, S. Oxygen imaging of living cells and tissues using luminescent molecular probes. J. Photochem. Photobiol. C Photochem. Rev. 2017, 30, 71–95. [Google Scholar] [CrossRef]
- Gilmore, A.C.; Flaherty, S.J.; Somasundaram, V.; Scheiblin, D.A.; Lockett, S.J.; Wink, D.A.; Heinz, W.F. An in vitro tumorigenesis model based on live-cell-generated oxygen and nutrient gradients. Commun. Biol. 2021, 4, 477. [Google Scholar] [CrossRef]
- Jankovic, B.; Aquino-Parsons, C.; Raleigh, J.A.; Stanbridge, E.J.; Durand, R.E.; Banath, J.P.; MacPhail, S.H.; Olive, P.L. Comparison between pimonidazole binding, oxygen electrode measurements, and expression of endogenous hypoxia markers in cancer of the uterine cervix. Cytom. Part B Clin. Cytom. 2006, 70B, 45–55. [Google Scholar] [CrossRef]
- Sobhanifar, S.; Aquino-Parsons, C.; Stanbridge, E.J.; Olive, P. Reduced expression of hypoxia-inducible factor-1alpha in perinecrotic regions of solid tumors. Cancer Res 2005, 65, 7259–7266. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wakeham, J.; Harkness, R.; Xing, Z. Macrophages are a significant source of type 1 cytokines during mycobacterial infection. J. Clin. Investig. 1999, 103, 1023–1029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desmedt, M.; Rottiers, P.; Dooms, H.; Fiers, W.; Grooten, J. Macrophages Induce Cellular Immunity by Activating Th1 Cell Responses and Suppressing Th2 Cell Responses. J. Immunol. 1998, 160, 5300–5308. [Google Scholar] [PubMed]
- Kumar, N.P.; Moideen, K.; Banurekha, V.V.; Nair, D.; Babu, S. Plasma Proinflammatory Cytokines Are Markers of Disease Severity and Bacterial Burden in Pulmonary Tuberculosis. Open Forum Infect. Dis. 2019, 6, ofz257. [Google Scholar] [CrossRef] [PubMed]
- Sahiratmadja, E.; Alisjahbana, B.; de Boer, T.; Adnan, I.; Maya, A.; Danusantoso, H.; Nelwan, R.H.H.; Marzuki, S.; van der Meer, J.W.M.; van Crevel, R.; et al. Dynamic Changes in Pro- and Anti-Inflammatory Cytokine Profiles and Gamma Interferon Receptor Signaling Integrity Correlate with Tuberculosis Disease Activity and Response to Curative Treatment. Infect. Immun. 2007, 75, 820–829. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Tan, S.; Huang, L.; Abramovitch, R.B.; Rohde, K.H.; Zimmerman, M.D.; Chen, C.; Dartois, V.; VanderVen, B.C.; Russell, D.G. Immune activation of the host cell induces drug tolerance in Mycobacterium tuberculosis both in vitro and in vivo. J. Exp. Med. 2016, 213, 809–825. [Google Scholar] [CrossRef] [Green Version]
- Guerrini, V.; Prideaux, B.; Blanc, L.; Bruiners, N.; Arrigucci, R.; Singh, S.; Ho-Liang, H.P.; Salamon, H.; Chen, P.Y.; Lakehal, K.; et al. Storage lipid studies in tuberculosis reveal that foam cell biogenesis is disease-specific. PLoS Pathog. 2018, 14, e1007223. [Google Scholar] [CrossRef] [Green Version]
- Daniel, J.; Maamar, H.; Deb, C.; Sirakova, T.D.; Kolattukudy, P.E. Mycobacterium tuberculosis uses host triacylglycerol to accumulate lipid droplets and acquires a dormancy-like phenotype in lipid-loaded macrophages. PLoS Pathog. 2011, 7, e1002093. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yañez, M.J.; Marín, T.; Balboa, E.; Klein, A.D.; Alvarez, A.R.; Zanlungo, S. Finding pathogenic commonalities between Niemann-Pick type C and other lysosomal storage disorders: Opportunities for shared therapeutic interventions. Biochim. Biophys. Acta Mol. Basis Dis. 2020, 1866, 165875. [Google Scholar] [CrossRef]
- Schulze, H.; Sandhoff, K. Lysosomal lipid storage diseases. Cold Spring Harb Perspect. Biol. 2011, 3, a004804. [Google Scholar] [CrossRef]
- Fineran, P.; Lloyd-Evans, E.; Lack, N.A.; Platt, N.; Davis, L.C.; Morgan, A.J.; Höglinger, D.; Tatituri, R.V.V.; Clark, S.; Williams, I.M.; et al. Pathogenic mycobacteria achieve cellular persistence by inhibiting the Niemann-Pick Type C disease cellular pathway. Wellcome Open Res. 2016, 1, 18. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, L.A.; Pearce, E.J. Immunometabolism governs dendritic cell and macrophage function. J. Exp. Med. 2016, 213, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Russell, D.G.; Huang, L.; VanderVen, B.C. Immunometabolism at the interface between macrophages and pathogens. Nat. Rev. Immunol. 2019, 19, 291–304. [Google Scholar] [CrossRef]
- Huang, L.; Nazarova, E.V.; Tan, S.; Liu, Y.; Russell, D.G. Growth of Mycobacterium tuberculosis in vivo segregates with host macrophage metabolism and ontogeny. J. Exp. Med. 2018, 215, 1135–1152. [Google Scholar] [CrossRef] [Green Version]
- Pisu, D.; Huang, L.; Grenier, J.K.; Russell, D.G. Dual RNA-Seq of Mtb-Infected Macrophages In Vivo Reveals Ontologically Distinct Host-Pathogen Interactions. Cell Rep 2020, 30, 335–350.e4. [Google Scholar] [CrossRef]
- VanderVen, B.C.; Fahey, R.J.; Lee, W.; Liu, Y.; Abramovitch, R.B.; Memmott, C.; Crowe, A.M.; Eltis, L.D.; Perola, E.; Deininger, D.D.; et al. Novel inhibitors of cholesterol degradation in Mycobacterium tuberculosis reveal how the bacterium’s metabolism is constrained by the intracellular environment. PLoS Pathog. 2015, 11, e1004679. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.; Schmidt, S.V.; Sander, J.; Draffehn, A.; Krebs, W.; Quester, I.; De Nardo, D.; Gohel, T.D.; Emde, M.; Schmidleithner, L.; et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity 2014, 40, 274–288. [Google Scholar] [CrossRef] [Green Version]
- Lachmandas, E.; Beigier-Bompadre, M.; Cheng, S.C.; Kumar, V.; van Laarhoven, A.; Wang, X.; Ammerdorffer, A.; Boutens, L.; de Jong, D.; Kanneganti, T.D.; et al. Rewiring cellular metabolism via the AKT/mTOR pathway contributes to host defence against Mycobacterium tuberculosis in human and murine cells. Eur. J. Immunol. 2016, 46, 2574–2586. [Google Scholar] [CrossRef]
- Koeken, V.; Lachmandas, E.; Riza, A.; Matzaraki, V.; Li, Y.; Kumar, V.; Oosting, M.; Joosten, L.A.B.; Netea, M.G.; van Crevel, R. Role of Glutamine Metabolism in Host Defense Against Mycobacterium tuberculosis Infection. J. Infect. Dis. 2019, 219, 1662–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, S.L.; Lamprecht, D.A.; Mandizvo, T.; Jones, T.T.; Naidoo, V.; Addicott, K.W.; Moodley, C.; Ngcobo, B.; Crossman, D.K.; Wells, G.; et al. Compromised Metabolic Reprogramming Is an Early Indicator of CD8(+) T Cell Dysfunction during Chronic Mycobacterium tuberculosis Infection. Cell Rep. 2019, 29, 3564–3579.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hackett, E.E.; Charles-Messance, H.; O’Leary, S.M.; Gleeson, L.E.; Muñoz-Wolf, N.; Case, S.; Wedderburn, A.; Johnston, D.G.W.; Williams, M.A.; Smyth, A.; et al. Mycobacterium tuberculosis Limits Host Glycolysis and IL-1β by Restriction of PFK-M via MicroRNA-21. Cell Rep. 2020, 30, 124–136.e4. [Google Scholar] [CrossRef]
- Gleeson, L.E.; Sheedy, F.J.; Palsson-McDermott, E.M.; Triglia, D.; O’Leary, S.M.; O’Sullivan, M.P.; O’Neill, L.A.; Keane, J. Cutting Edge: Mycobacterium tuberculosis Induces Aerobic Glycolysis in Human Alveolar Macrophages That Is Required for Control of Intracellular Bacillary Replication. J. Immunol. 2016, 196, 2444–2449. [Google Scholar] [CrossRef] [Green Version]
- Sreevatsan, S.; Pan, X.; Stockbauer, K.E.; Connell, N.D.; Kreiswirth, B.N.; Whittam, T.S.; Musser, J.M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc. Natl. Acad. Sci. USA 1997, 94, 9869–9874. [Google Scholar] [CrossRef] [Green Version]
- Gopal, R.; Monin, L.; Slight, S.; Uche, U.; Blanchard, E.; Fallert Junecko, B.A.; Ramos-Payan, R.; Stallings, C.L.; Reinhart, T.A.; Kolls, J.K.; et al. Unexpected Role for IL-17 in Protective Immunity against Hypervirulent Mycobacterium tuberculosis HN878 Infection. PLoS Pathog. 2014, 10, e1004099. [Google Scholar] [CrossRef]
- Betts, J.C.; Dodson, P.; Quan, S.; Lewis, A.P.; Thomas, P.J.; Duncan, K.; McAdam, R.A. Comparison of the proteome of Mycobacterium tuberculosis strain H37Rv with clinical isolate CDC 1551. Microbiology 2000, 146, 3205–3216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manca, C.; Tsenova, L.; Barry, C.E.; Bergtold, A.; Freeman, S.; Haslett, P.A.J.; Musser, J.M.; Freedman, V.H.; Kaplan, G. Mycobacterium tuberculosis CDC1551 Induces a More Vigorous Host Response In Vivo and In Vitro, but Is Not More Virulent Than Other Clinical Isolates. J. Immunol. 1999, 162, 6740–6746. [Google Scholar]
- DiFazio, R.M.; Mattila, J.T.; Klein, E.C.; Cirrincione, L.R.; Howard, M.; Wong, E.A.; Flynn, J.L. Active transforming growth factor-β is associated with phenotypic changes in granulomas after drug treatment in pulmonary tuberculosis. Fibrogenes. Tissue Repair 2016, 9, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warsinske, H.C.; DiFazio, R.M.; Linderman, J.J.; Flynn, J.L.; Kirschner, D.E. Identifying mechanisms driving formation of granuloma-associated fibrosis during Mycobacterium tuberculosis infection. J. Theor. Biol. 2017, 429, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Marrazzo, P.; Maccari, S.; Taddei, A.; Bevan, L.; Telford, J.; Soriani, M.; Pezzicoli, A. 3D Reconstruction of the Human Airway Mucosa In Vitro as an Experimental Model to Study NTHi Infections. PLoS ONE 2016, 11, e0153985. [Google Scholar] [CrossRef]
- Kuo, C.-T.; Chiang, C.-L.; Huang, R.; Lee, H.; Wo, A. Configurable 2D and 3D spheroid tissue cultures on bioengineered surfaces with acquisition of epithelial–mesenchymal transition characteristics. NPG Asia Mater. 2012, 4, e27. [Google Scholar] [CrossRef] [Green Version]
- Grist, S.M.; Nasseri, S.S.; Laplatine, L.; Schmok, J.C.; Yao, D.; Hua, J.; Chrostowski, L.; Cheung, K.C. Long-term monitoring in a microfluidic system to study tumour spheroid response to chronic and cycling hypoxia. Sci. Rep. 2019, 9, 17782. [Google Scholar] [CrossRef] [Green Version]
- Choi, J.H.; Loarca, L.; Hoyos-Vega, J.M.D.; Dadgar, N.; Loutherback, K.; Shah, V.H.; Stybayeva, G.; Revzin, A. Microfluidic confinement enhances phenotype and function of hepatocyte spheroids. Am. J. Physiol.-Cell Physiol. 2020, 319, C552–C560. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukundan, S.; Singh, P.; Shah, A.; Kumar, R.; O’Neill, K.C.; Carter, C.L.; Russell, D.G.; Subbian, S.; Parekkadan, B. In Vitro Miniaturized Tuberculosis Spheroid Model. Biomedicines 2021, 9, 1209. https://doi.org/10.3390/biomedicines9091209
Mukundan S, Singh P, Shah A, Kumar R, O’Neill KC, Carter CL, Russell DG, Subbian S, Parekkadan B. In Vitro Miniaturized Tuberculosis Spheroid Model. Biomedicines. 2021; 9(9):1209. https://doi.org/10.3390/biomedicines9091209
Chicago/Turabian StyleMukundan, Shilpaa, Pooja Singh, Aditi Shah, Ranjeet Kumar, Kelly C. O’Neill, Claire L. Carter, David G. Russell, Selvakumar Subbian, and Biju Parekkadan. 2021. "In Vitro Miniaturized Tuberculosis Spheroid Model" Biomedicines 9, no. 9: 1209. https://doi.org/10.3390/biomedicines9091209






