Lactococcus lactis Delivery of Surface Layer Protein A Protects Mice from Colitis by Re-Setting Host Immune Repertoire
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of R110
2.2. Growth and Maintenance of R110
2.3. Mice
2.4. Ethical Animal Care
2.5. T Cell Colitis Induction
2.6. Fecal Occult Blood Determination
2.7. FITC Dextran Assay
2.8. Fecal Albumin Assay
2.9. Isolation of Colonic Lamina Propria Cells
2.10. Flow Cytometry Analysis of Isolated Cells
2.11. Microbial Composition Analysis
2.12. Quantitative Real-Time PCR
2.13. Murine Dendritic Cells Experiment
2.14. Human Dendritic Cells Experiment
2.15. RNAseq Analysis
2.16. Statistics
3. Results
3.1. Generation of Lactococcus Lactis Expressing SlpA, a Novel Clinical Candidate for Treating IBD
3.2. R110 Protects Mice from T Cell-Mediated Colitis
3.3. R110 Induces Positive Changes in the Transcriptome of Colon Tissue
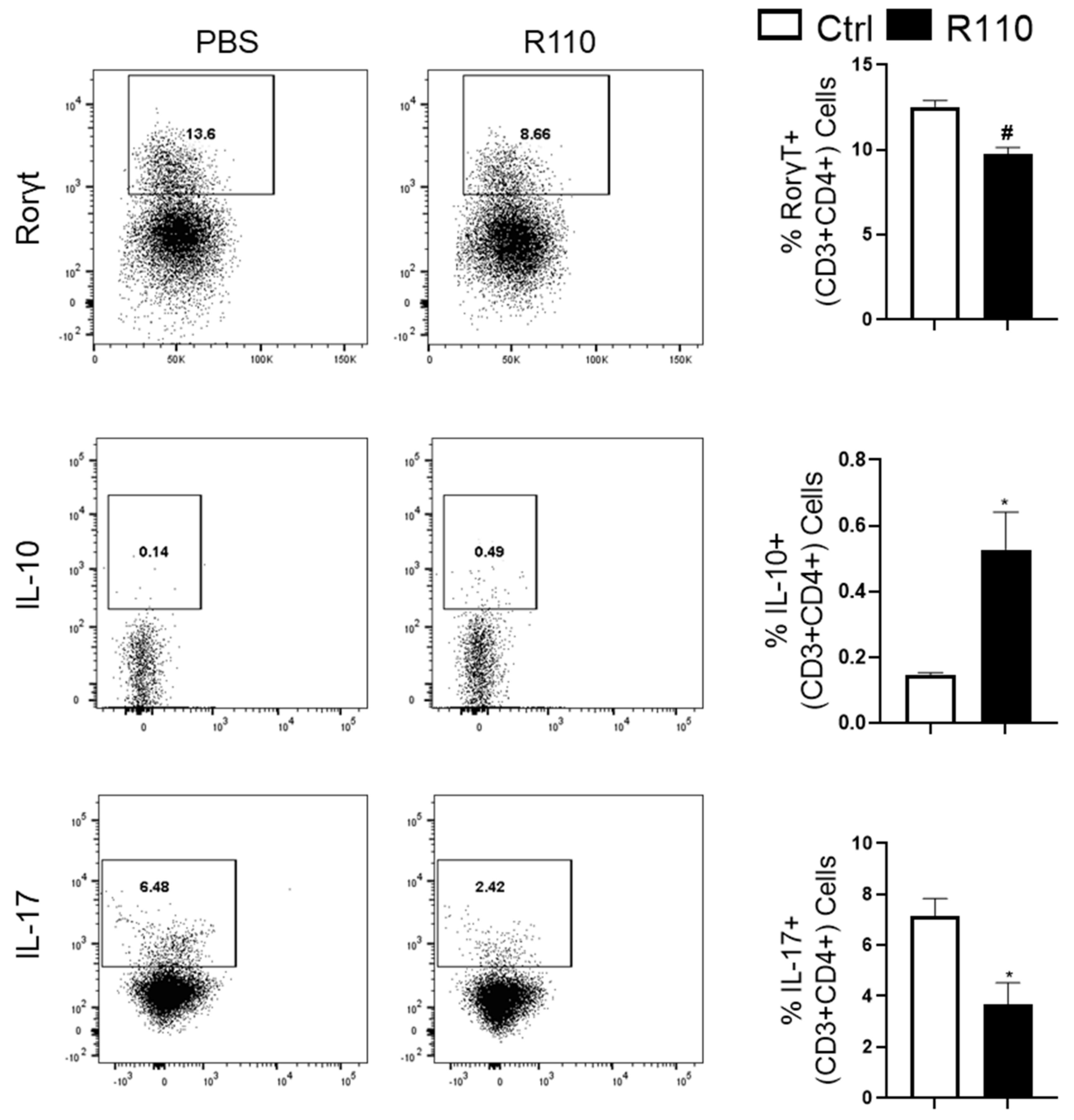
3.4. R110 Induces IL-10 and Reduces Rorγt and IL-17 Production
3.5. R110 Reduces Critical Inflammatory Cytokines in the Sera of Diseased Mice
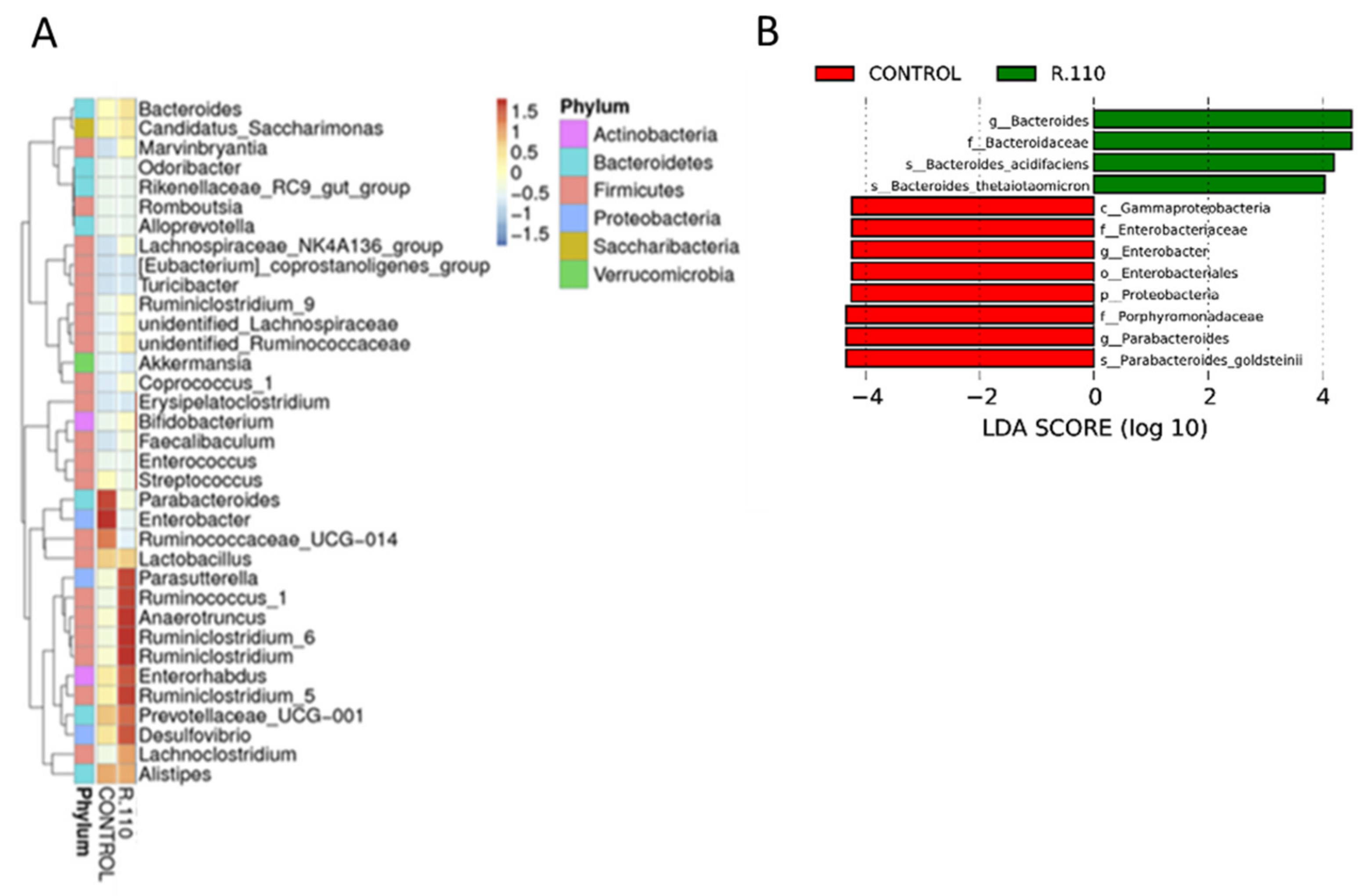
3.6. R110 Protects the Gut Microbiota
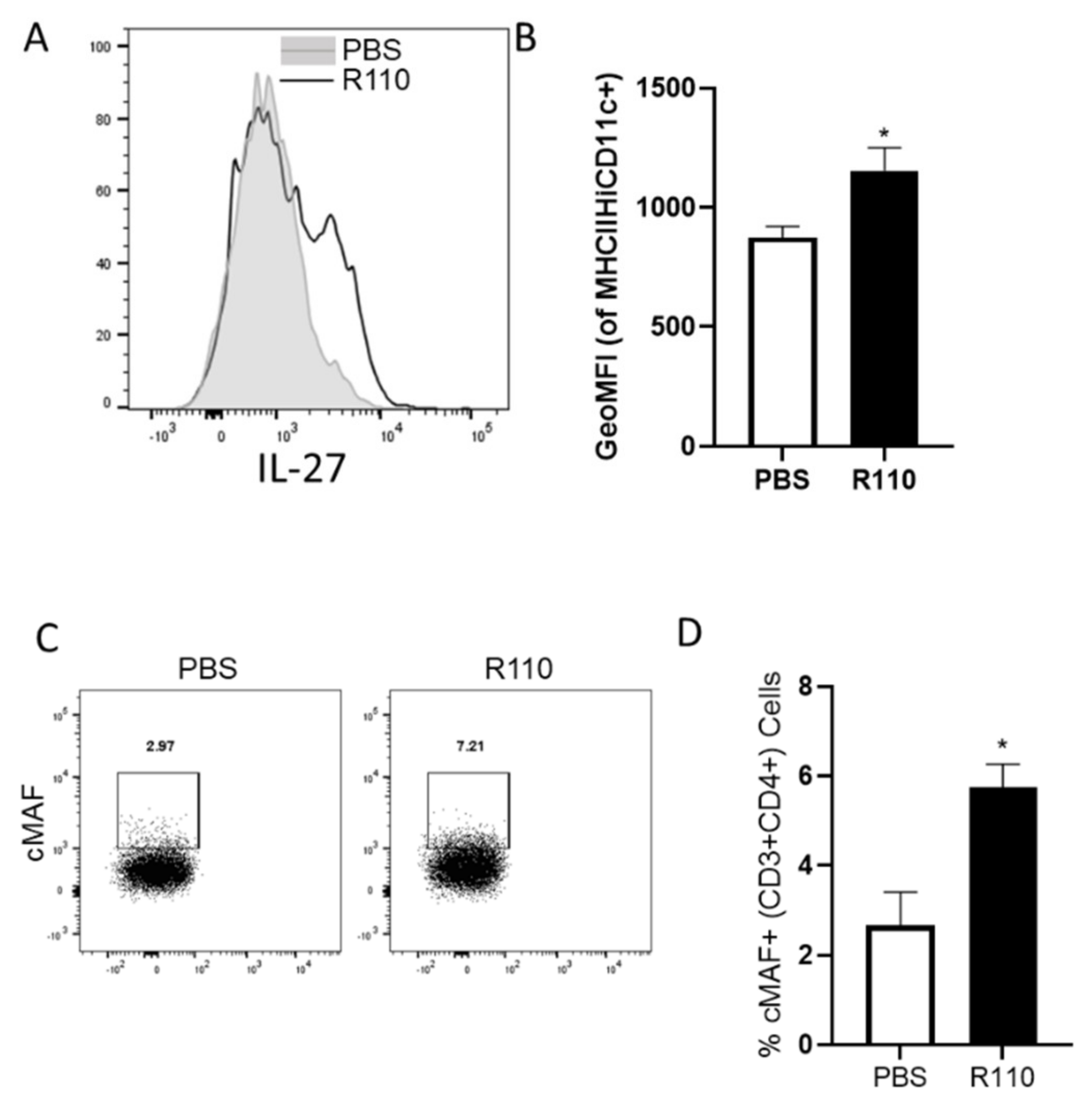
3.7. R110 Induces Regulatory Cytokines in Human Dendritic Cells
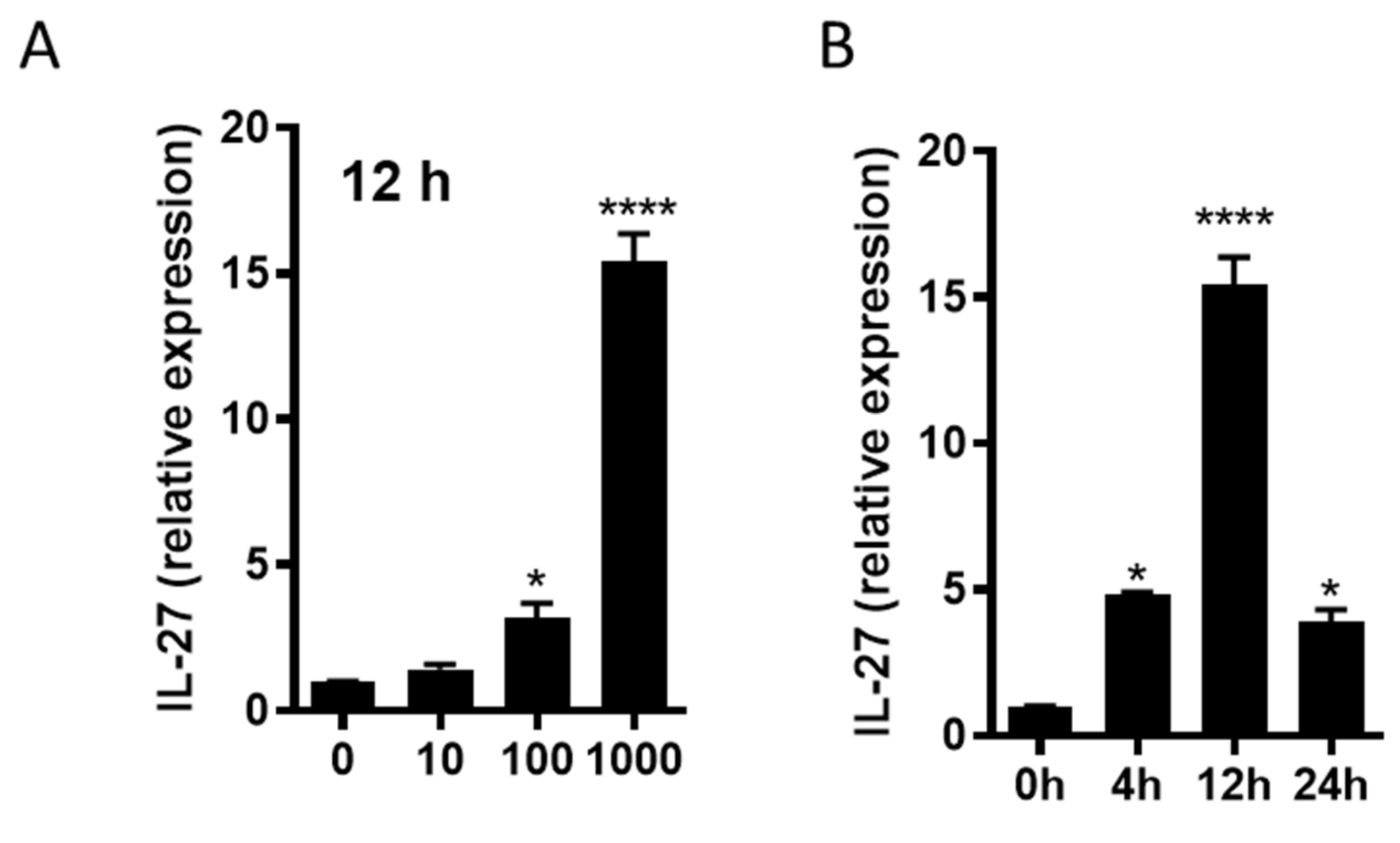
3.8. R110 Induces IL-27 in Murine Dendritic Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alatab, S.; Sepanlou, S.G.; Ikuta, K.; Vahedi, H.; Bisignano, C.; Safiri, S.; Sadeghi, A.; Nixon, M.R.; Abdoli, A.; Abolhassani, H.; et al. The global, regional, and national burden of inflammatory bowel disease in 195 countries and territories, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2020, 5, 17–30. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Manne, S.; Treem, W.R.; Bennett, D. Prevalence of Inflammatory Bowel Disease in Pediatric and Adult Populations: Recent Estimates From Large National Databases in the United States, 2007–2016. Inflamm. Bowel Dis. 2019, 26, 619–625. [Google Scholar] [CrossRef]
- Leirisalo-Repo, M.; Turunen, U.; Stenman, S.; Helenius, P.; Seppälä, K. High Frequency of Silent Inflammatory Bowel Disease in Spondylarthropathy. Arthritis Rheum. 1994, 37, 23–31. [Google Scholar] [CrossRef]
- Di Jiang, C.; Raine, T. IBD considerations in spondyloarthritis. Ther. Adv. Musculoskelet. Dis. 2020, 12. [Google Scholar] [CrossRef]
- Fragoulis, G.E.; Liava, C.; Daoussis, D.; Akriviadis, E.; Garyfallos, A.; Dimitroulas, T. Inflammatory bowel diseases and spondyloarthropathies: From pathogenesis to treatment. World J. Gastroenterol. 2019, 25, 2162–2176. [Google Scholar] [CrossRef] [PubMed]
- Orchard, T.R.; Wordsworth, B.P.; Jewell, D.P. Peripheral arthropathies in inflammatory bowel disease: Their articular distribution and natural history. Gut 1998, 42, 387–391. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, C.N.; Benchimol, E.I.; Bitton, A.; Murthy, S.K.; Nguyen, G.C.; Lee, K.; Cooke-Lauder, J.; Kaplan, G.G. The Impact of Inflammatory Bowel Disease in Canada 2018: Extra-intestinal Diseases in IBD. J. Can. Assoc. Gastroenterol. 2019, 2, S73–S80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pouillon, L.; Bossuyt, P.; Vanderstukken, J.; Moulin, D.; Netter, P.; Danese, S.; Jouzeau, J.-Y.; Loeuille, D.; Peyrin-Biroulet, L. Management of patients with inflammatory bowel disease and spondyloarthritis. Expert Rev. Clin. Pharmacol. 2017, 10, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- McDermott, A.J.; Huffnagle, G.B. The microbiome and regulation of mucosal immunity. Immunology 2014, 142, 24–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glassner, K.L.; Abraham, B.P.; Quigley, E.M. The microbiome and inflammatory bowel disease. J. Allergy Clin. Immunol. 2020, 145, 16–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeFilipp, Z.; Bloom, P.P.; Soto, M.T.; Mansour, M.K.; Sater, M.; Huntley, M.H.; Turbett, S.; Chung, R.T.; Chen, Y.-B.; Hohmann, E.L. Drug-Resistant E. coli Bacteremia Transmitted by Fecal Microbiota Transplant. N. Engl. J. Med. 2019, 381, 2043–2050. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; De Groot, P.F.; Geerlings, S.E.; Hodiamont, C.J.; Belzer, C.; Berge, I.J.M.T.; De Vos, W.M.; Bemelman, F.J.; Nieuwdorp, M. Fecal microbiota transplantation against intestinal colonization by extended spectrum beta-lactamase producing Enterobacteriaceae: A proof of principle study. BMC Res. Notes 2018, 11, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Basso, P.J.; Câmara, N.O.S.; Sales-Campos, H. Microbial-Based Therapies in the Treatment of Inflammatory Bowel Disease—An Overview of Human Studies. Front. Pharmacol. 2019, 9, 1571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangan, K.J.; Pedicord, V.A.; Wang, Y.-C.; Kim, B.; Lu, Y.; Shaham, S.; Mucida, D.; Hang, H.C. A secreted bacterial peptidoglycan hydrolase enhances tolerance to enteric pathogens. Science 2016, 353, 1434–1437. [Google Scholar] [CrossRef] [Green Version]
- Lightfoot, Y.L.; Selle, K.; Yang, T.; Goh, Y.J.; Sahay, B.; Zadeh, M.; Owen, J.L.; Colliou, N.; Li, E.; Johannssen, T.; et al. SIGNR 3-dependent immune regulation by Lactobacillus acidophilus surface layer protein A in colitis. EMBO J. 2015, 34, 881–895. [Google Scholar] [CrossRef] [Green Version]
- Konstantinov, S.R.; Smidt, H.; de Vos, W.M.; Bruijns, S.C.M.; Singh, S.K.; Valence, F.; Molle, D.; Lortal, S.; Altermann, E.; Klaenhammer, T.R.; et al. S layer protein A of Lactobacillus acidophilus NCFM regulates immature dendritic cell and T cell functions. Proc. Natl. Acad. Sci. USA 2008, 105, 19474–19479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sahay, B.; Ge, Y.; Colliou, N.; Zadeh, M.; Weiner, C.; Mila, A.; Owen, J.L.; Mohamadzadeh, M. Advancing the use of Lactobacillus acidophilus surface layer protein A for the treatment of intestinal disorders in humans. Gut Microbes 2015, 6, 392–397. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.; Pollard, J.W.; Friesen, J.D.; Stanners, C.P. Stuttering: High-level mistranslation in animal and bacterial cells. Proc. Natl. Acad. Sci. USA 1978, 75, 1091–1095. [Google Scholar] [CrossRef] [Green Version]
- Leenhouts, K.J.; Kok, J.; Venema, G. Replacement recombination in Lactococcus lactis. J. Bacteriol. 1991, 173, 4794–4798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steidler, L. Gene exchange of thyA for interleukin-10 secures live GMO bacterial therapeutics. Discov. Med. 2003, 3, 49–51. [Google Scholar] [PubMed]
- Simons, G.; Nijhuis, M.; de Vos, W.M. Integration and gene replacement in the Lactococcus lactis lac operon: Induction of a cryptic phospho-beta-glucosidase in LacG-deficient strains. J. Bacteriol. 1993, 175, 5168–5175. [Google Scholar] [CrossRef] [Green Version]
- Steidler, L. Genetically engineered probiotics. Best Prac. Res. Clin. Gastroenterol. 2003, 17, 861–876. [Google Scholar] [CrossRef]
- McNealy, T.; Steidler, A.; Schaaf, A.; Alken, P.; Michel, M.S. Entwicklung und Evaluation eines neuartigen 3D-Blasenmatrixmodelles zur standardisierten Untersuchung verschiedener Transfektionsmethoden. Aktuel Urol. 2003, 34, 172–175. [Google Scholar] [CrossRef]
- Sasaki, Y.; Ito, Y.; Sasaki, T. thyA as a Selection Marker in Construction of Food-Grade Host-Vector and Integration Systems for Streptococcus thermophilus. Appl. Environ. Microbiol. 2004, 70, 1858–1864. [Google Scholar] [CrossRef] [Green Version]
- Wong, Q.N.Y. Efficient and seamless DNA recombineering using a thymidylate synthase A selection system in Escherichia coli. Nucleic Acids Res. 2005, 33, e59. [Google Scholar] [CrossRef]
- Weiss, A.A.; Babyatsky, M.W.; Ogata, S.; Chen, A.; Itzkowitz, S.H. Expression of MUC2 and MUC3 mRNA in human normal, malignant, and inflammatory intestinal tissues. J. Histochem. Cytochem. 1996, 44, 1161–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bergstrom, K.S.B.; Kissoon-Singh, V.; Gibson, D.L.; Ma, C.; Montero, M.; Sham, H.P.; Ryz, N.; Huang, T.; Velcich, A.; Finlay, B.B.; et al. Muc2 Protects against Lethal Infectious Colitis by Disassociating Pathogenic and Commensal Bacteria from the Colonic Mucosa. PLoS Pathog. 2010, 6, e1000902. [Google Scholar] [CrossRef] [PubMed]
- Van der Sluis, M.; De Koning, B.A.; De Bruijn, A.C.; Velcich, A.; Meijerink, J.; van Goudoever, J.; Büller, H.A.; Dekker, J.; VAN Seuningen, I.; Renes, I.B.; et al. Muc2-Deficient Mice Spontaneously Develop Colitis, Indicating That MUC2 Is Critical for Colonic Protection. Gastroenterology 2006, 131, 117–129. [Google Scholar] [CrossRef]
- Liu, B.; Yang, L.; Cui, Z.; Zheng, J.; Huang, J.; Zhao, Q.; Su, Z.; Wang, M.; Zhang, W.; Liu, J.; et al. Anti-TNF-α therapy alters the gut microbiota in proteoglycan-induced ankylosing spondylitis in mice. Microbiologyopen 2019, 8, e927. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, N.A.; Heap, G.; Green, H.; Hamilton, B.; Bewshea, C.; Walker, G.J.; Thomas, A.; Nice, R.; Perry, M.H.; Bouri, S.; et al. Predictors of anti-TNF treatment failure in anti-TNF-naive patients with active luminal Crohn’s disease: A prospective, multicentre, cohort study. Lancet Gastroenterol. Hepatol. 2019, 4, 341–353. [Google Scholar] [CrossRef] [Green Version]
- Lin, I.Y.C.; Van, T.T.H.; Smooker, P.M. Live-Attenuated Bacterial Vectors: Tools for Vaccine and Therapeutic Agent Delivery. Vaccines 2015, 3, 940–972. [Google Scholar] [CrossRef] [Green Version]
- Steidler, L.; Neirynck, S.; Huyghebaert, N.; Snoeck, V.; Vermeire, A.; Goddeeris, B.; Cox, E.; Remon, J.P.; Remaut, E. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nat. Biotechnol. 2003, 21, 785–789. [Google Scholar] [CrossRef] [PubMed]
- Dorofeyev, A.E.; Vasilenko, I.V.; Rassokhina, O.A.; Kondratiuk, R.B. Mucosal Barrier in Ulcerative Colitis and Crohn’s Disease. Gastroenterol. Res. Pract. 2013, 2013, 431231. [Google Scholar] [CrossRef]
- Carrato, C.; Balague, C.; De Bolos, C.; Gonzalez, E.; Gambus, G.; Planas, J.; Perini, J.M.; Andreu, D.; Real, F.X. Differential apomucin expression in normal and neoplastic human gastrointestinal tissues. Gastroenterology 1994, 107, 160–172. [Google Scholar] [CrossRef]
- Ahrne, S.; Hagslatt, M.-L.J. Effect of Lactobacilli on Paracellular Permeability in the Gut. Nutrients 2011, 3, 104–117. [Google Scholar] [CrossRef]
- Yoshida, H.; Hunter, C.A. The Immunobiology of Interleukin-27. Annu. Rev. Immunol. 2015, 33, 417–443. [Google Scholar] [CrossRef] [PubMed]
- Wheaton, J.D.; Yeh, C.-H.; Ciofani, M. Cutting Edge: C-Maf Is Required for Regulatory T Cells To Adopt RORγt+ and Follicular Phenotypes. J. Immunol. 2017, 199, 3931–3936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neumann, C.; Blume, J.; Roy, U.; Teh, P.P.; Vasanthakumar, A.; Beller, A.; Liao, Y.; Heinrich, F.; Arenzana, T.L.; Hackney, J.A.; et al. c-Maf-dependent Treg cell control of intestinal TH17 cells and IgA establishes host–microbiota homeostasis. Nat. Immunol. 2019, 20, 471–481. [Google Scholar] [CrossRef]
- Apetoh, L.; Quintana, F.J.; Pot, C.; Joller, N.; Xiao, S.; Kumar, D.; Burns, E.J.; Sherr, D.; Weiner, H.L.; Kuchroo, V.K. The aryl hydrocarbon receptor interacts with c-Maf to promote the differentiation of type 1 regulatory T cells induced by IL-27. Nat. Immunol. 2010, 11, 854–861. [Google Scholar] [CrossRef] [Green Version]
- Carrier, Y.; Whitters, M.J.; Miyashiro, J.S.; Labranche, T.P.; Ramon, H.E.; Benoit, S.E.; Ryan, M.S.; Keegan, S.P.; Guay, H.; Douhan, J.; et al. Enhanced GITR/GITRL interactions augment IL-27 expression and induce IL-10-producing Tr-1 like cells. Eur. J. Immunol. 2012, 42, 1393–1404. [Google Scholar] [CrossRef] [Green Version]
- Pot, C.; Jin, H.; Awasthi, A.; Liu, S.M.; Lai, C.-Y.; Madan, R.; Sharpe, A.H.; Karp, C.L.; Miaw, S.-C.; Ho, I.-C.; et al. Cutting Edge: IL-27 Induces the Transcription Factor c-Maf, Cytokine IL-21, and the Costimulatory Receptor ICOS that Coordinately Act Together to Promote Differentiation of IL-10-Producing Tr1 Cells. J. Immunol. 2009, 183, 797–801. [Google Scholar] [CrossRef] [Green Version]
- VasanthaKumar, A.; Kallies, A. IL-27 paves different roads to Tr1. Eur. J. Immunol. 2013, 43, 882–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, A.M.; Khader, S.A. IL-12p40: An inherently agonistic cytokine. Trends Immunol. 2007, 28, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Mitsialis, V.; Wall, S.; Liu, P.; Ordovas-Montanes, J.; Parmet, T.; Vukovic, M.; Spencer, D.; Field, M.; McCourt, C.; Toothaker, J.; et al. Single-Cell Analyses of Colon and Blood Reveal Distinct Immune Cell Signatures of Ulcerative Colitis and Crohn’s Disease. Gastroenterology 2020, 159, 591–608.e10. [Google Scholar] [CrossRef] [PubMed]
- Ansari, N.; Abdulla, J.; Zayyani, N.; Brahmi, U.; Taha, S.; Satir, A.A. Comparison of RANTES expression in Crohn’s disease and ulcerative colitis: An aid in the differential diagnosis? J. Clin. Pathol. 2006, 59, 1066–1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, D.Y.; Clark, E.A. The role of CD40 and CD154/CD40L in dendritic cells. Semin. Immunol. 2009, 21, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Martin, S.; Agarwal, R.; Murugaiyan, G.; Saha, B. CD40 Expression Levels Modulate Regulatory T Cells inLeishmania donovaniInfection. J. Immunol. 2010, 185, 551–559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guiducci, C.; Valzasina, B.; Dislich, H.; Colombo, M.P. CD40/CD40L interaction regulates CD4+CD25+ T reg homeostasis through dendritic cell-produced IL-2. Eur. J. Immunol. 2005, 35, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Manzotti, C.; Liu, M.; Burke, F.; Mead, K.I.; Sansom, D. CD86 and CD80 Differentially Modulate the Suppressive Function of Human Regulatory T Cells. J. Immunol. 2004, 172, 2778–2784. [Google Scholar] [CrossRef] [PubMed]
- Su-Ling, F.; Du, Y.-M.; Yan, Z.-D.; Yan, J.; Zhuansun, Y.-X.; Chen, R.; Zhang, W.; Feng, S.-L.; Ran, P.-X. CD80 and CD86 knockdown in dendritic cells regulates Th1/Th2 cytokine production in asthmatic mice. Exp. Ther. Med. 2016, 11, 878–884. [Google Scholar] [CrossRef] [Green Version]
- Brckalo, T.; Calzetti, F.; Pérez-Cabezas, B.; Borras, F.E.; Cassatella, M.A.; López-Botet, M. Functional analysis of the CD300e receptor in human monocytes and myeloid dendritic cells. Eur. J. Immunol. 2010, 40, 722–732. [Google Scholar] [CrossRef]
- Aarhus, R.; Graeff, R.M.; Dickey, D.M.; Walseth, T.F.; Hon, C.L. ADP-ribosyl Cyclase and CD38 Catalyze the Synthesis of a Calcium-mobilizing Metabolite from NADP+. J. Biol. Chem. 1995, 270, 30327–30333. [Google Scholar] [CrossRef] [Green Version]
- Lischke, T.; Heesch, K.; Schumacher, V.; Schneider, M.; Haag, F.; Koch-Nolte, F.; Mittrücker, H.-W. CD38 Controls the Innate Immune Response against Listeria monocytogenes. Infect. Immun. 2013, 81, 4091–4099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viegas, M.S.; Carmo, A.; Silva, T.; Seco, F.; Serra, V.; Lacerda, M.; Martins, T. CD38 plays a role in effective containment of mycobacteria within granulomata and polarization of Th1 immune responses against Mycobacterium avium. Microbes Infect. 2007, 9, 847–854. [Google Scholar] [CrossRef] [PubMed]
- Partida-Sánchez, S.; Cockayne, D.A.; Monard, S.; Jacobson, E.L.; Oppenheimer, N.; Garvy, B.; Kusser, K.; Goodrich, S.; Howard, M.; Harmsen, A.; et al. Cyclic ADP-ribose production by CD38 regulates intracellular calcium release, extracellular calcium influx and chemotaxis in neutrophils and is required for bacterial clearance in vivo. Nat. Med. 2001, 7, 1209–1216. [Google Scholar] [CrossRef] [PubMed]
- Jablonski, K.A.; Amici, S.A.; Webb, L.M.; Ruiz-Rosado, J.D.D.; Popovich, P.G.; Partida-Sanchez, S.; Guerau-De-Arellano, M. Novel Markers to Delineate Murine M1 and M2 Macrophages. PLoS ONE 2015, 10, e0145342. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Zuo, H.; Xiong, H.; Kolar, M.; Chu, Q.; Saghatelian, A.; Siegwart, D.J.; Wan, Y. Gpr132 sensing of lactate mediates tumor–macrophage interplay to promote breast cancer metastasis. Proc. Natl. Acad. Sci. USA 2017, 114, 580–585. [Google Scholar] [CrossRef] [Green Version]
- Kern, K.; Schäfer, S.M.G.; Cohnen, J.; Pierre, S.; Osthues, T.; Tarighi, N.; Hohmann, S.; Ferreiros, N.; Brüne, B.; Weigert, A.; et al. The G2A Receptor Controls Polarization of Macrophage by Determining Their Localization Within the Inflamed Tissue. Front. Immunol. 2018, 9, 2261. [Google Scholar] [CrossRef]
- Osthues, T.; Zimmer, B.; Rimola, V.; Klann, K.; Schilling, K.; Mathoor, P.; Angioni, C.; Weigert, A.; Geisslinger, G.; Münch, C.; et al. The Lipid Receptor G2A (GPR132) Mediates Macrophage Migration in Nerve Injury-Induced Neuropathic Pain. Cells 2020, 9, 1740. [Google Scholar] [CrossRef] [PubMed]
- Park, A.J.; Agak, G.W.; Qin, M.; Hisaw, L.D.; Pirouz, A.; Kao, S.; Marinelli, L.J.; Garbán, H.J.; Thiboutot, D.; Liu, P.T.; et al. G2A AttenuatesPropionibacterium acnesInduction of Inflammatory Cytokines in Human Monocytes. Ann. Dermatol. 2017, 29, 688–698. [Google Scholar] [CrossRef] [Green Version]
- Xiu, W.; Chen, Q.; Wang, Z.; Wang, J.; Zhou, Z. Microbiota-derived short chain fatty acid promotion of Amphiregulin expression by dendritic cells is regulated by GPR43 and Blimp-1. Biochem. Biophys. Res. Commun. 2020, 533, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Sun, M.; Chen, F.; Cao, A.T.; Liu, H.; Zhao, Y.; Huang, X.; Xiao, Y.; Yao, S.; Zhao, Q.; et al. Microbiota metabolite short-chain fatty acid acetate promotes intestinal IgA response to microbiota which is mediated by GPR43. Mucosal Immunol. 2017, 10, 946–956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavoie, S.; Chun, E.; Bae, S.; Brennan, C.A.; Comeau, C.A.G.; Lang, J.K.; Michaud, M.; Hoveyda, H.R.; Fraser, G.L.; Fuller, M.H.; et al. Expression of Free Fatty Acid Receptor 2 by Dendritic Cells Prevents Their Expression of Interleukin 27 and Is Required for Maintenance of Mucosal Barrier and Immune Response Against Colorectal Tumors in Mice. Gastroenterology 2020, 158, 1359–1372.e9. [Google Scholar] [CrossRef] [PubMed]
- Osaka, T.; Moriyama, E.; Arai, S.; Date, Y.; Yagi, J.; Kikuchi, J.; Tsuneda, S. Meta-Analysis of Fecal Microbiota and Metabolites in Experimental Colitic Mice during the Inflammatory and Healing Phases. Nutrients 2017, 9, 1329. [Google Scholar] [CrossRef] [Green Version]
- Togo, A.; Diop, A.; Dubourg, G.; Khelaifia, S.; Richez, M.; Armstrong, N.; Maraninchi, M.; Fournier, P.-E.; Raoult, D.; Million, M. Anaerotruncus massiliensis sp. nov., a succinate-producing bacterium isolated from human stool from an obese patient after bariatric surgery. New Microbes New Infect. 2019, 29, 100508. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Sonnenburg, J.L. Eating For Two: How Metabolism Establishes Interspecies Interactions in the Gut. Cell Host Microbe 2011, 10, 336–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, P.; Flint, H.J. Formation of propionate and butyrate by the human colonic microbiota. Environ. Microbiol. 2017, 19, 29–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banerjee, A.; Herring, C.A.; Chen, B.; Kim, H.; Simmons, A.J.; Southard-Smith, A.; Allaman, M.M.; White, J.R.; Macedonia, M.C.; Mckinley, E.T.; et al. Succinate Produced by Intestinal Microbes Promotes Specification of Tuft Cells to Suppress Ileal Inflammation. Gastroenterology 2020, 159, 2101–2115.e5. [Google Scholar] [CrossRef]
- Han, R.; Ma, Y.; Xiao, J.; You, L.; Pedisić, S.; Liao, L. The possible mechanism of the protective effect of a sulfated polysaccharide from Gracilaria Lemaneiformis against colitis induced by dextran sulfate sodium in mice. Food Chem. Toxicol. 2021, 149, 112001. [Google Scholar] [CrossRef] [PubMed]
- Antharam, V.C.; Li, E.C.; Ishmael, A.; Sharma, A.; Mai, V.; Rand, K.H.; Wang, G.P. Intestinal Dysbiosis and Depletion of Butyrogenic Bacteria in Clostridium difficile Infection and Nosocomial Diarrhea. J. Clin. Microbiol. 2013, 51, 2884–2892. [Google Scholar] [CrossRef] [Green Version]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; White, M.B.; Monteith, P.; Noble, N.A.; Unser, A.B.; Daita, K.; Fisher, A.R.; et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 2014, 60, 940–947. [Google Scholar] [CrossRef] [Green Version]
- Saresella, M.; Mendozzi, L.; Rossi, V.; Mazzali, F.; Piancone, F.; La Rosa, F.; Marventano, I.; Caputo, D.; Felis, G.E.; Clerici, M. Immunological and Clinical Effect of Diet Modulation of the Gut Microbiome in Multiple Sclerosis Patients: A Pilot Study. Front. Immunol. 2017, 8, 1391. [Google Scholar] [CrossRef] [PubMed]
- Pasolli, E.; Truong, D.T.; Malik, F.; Waldron, L.; Segata, N. Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS Comput. Biol. 2016, 12, e1004977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allegretti, J.R.; Kearney, S.M.; Li, N.; Bogart, E.; Bullock, K.; Gerber, G.K.; Bry, L.; Clish, C.; Alm, E.J.; Korzenik, J. RecurrentClostridium difficileinfection associates with distinct bile acid and microbiome profiles. Aliment. Pharmacol. Ther. 2016, 43, 1142–1153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stappenbeck, T.S.; Hooper, L.V.; Gordon, J.I. Nonlinear partial differential equations and applications: Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 2002, 99, 15451–15455. [Google Scholar] [CrossRef] [Green Version]
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [Green Version]
- Cash, H.L.; Whitham, C.V.; Behrendt, C.L.; Hooper, L.V. Symbiotic Bacteria Direct Expression of an Intestinal Bactericidal Lectin. Science 2006, 313, 1126–1130. [Google Scholar] [CrossRef] [Green Version]
- Kamada, N.; Kim, Y.-G.; Sham, H.P.; Vallance, B.; Puente, J.L.; Martens, E.C.; Núñez, G. Regulated Virulence Controls the Ability of a Pathogen to Compete with the Gut Microbiota. Science 2012, 336, 1325–1329. [Google Scholar] [CrossRef] [Green Version]
- Zhu, W.; Yan, J.; Zhi, C.; Zhou, Q.; Yuan, X. 1,25(OH)2D3 deficiency-induced gut microbial dysbiosis degrades the colonic mucus barrier in Cyp27b1 knockout mouse model. Gut Pathog. 2019, 11, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Fukatsu, S.; Horinouchi, H.; Nagata, S.; Kamei, R.; Tanaka, D.; Hong, W.; Kazami, Y.; Fujimori, M.; Itoh, K.; Momose, Y.; et al. Post-translational suppression of the high affinity IgE receptor expression on mast cells by an intestinal bacterium. Immunobiology 2021, 226, 152056. [Google Scholar] [CrossRef]
- Yanagibashi, T.; Hosono, A.; Oyama, A.; Tsuda, M.; Suzuki, A.; Hachimura, S.; Takahashi, Y.; Momose, Y.; Itoh, K.; Hirayama, K.; et al. IgA production in the large intestine is modulated by a different mechanism than in the small intestine: Bacteroides acidifaciens promotes IgA production in the large intestine by inducing germinal center formation and increasing the number of IgA+ B cells. Immunobiology 2013, 218, 645–651. [Google Scholar] [CrossRef] [PubMed]
- Then, C.K.; Paillas, S.; Wang, X.; Hampson, A.; Kiltie, A.E. Association of Bacteroides acidifaciens relative abundance with high-fibre diet-associated radiosensitisation. BMC Biol. 2020, 18, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Miki, T.; Goto, R.; Fujimoto, M.; Okada, N.; Hardt, W.-D. The Bactericidal Lectin RegIIIβ Prolongs Gut Colonization and Enteropathy in the Streptomycin Mouse Model for Salmonella Diarrhea. Cell Host Microbe 2017, 21, 195–207. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Yan, Q.; Chen, F.; Cao, C.; Wang, S. Bupleuri radix extract ameliorates impaired lipid metabolism in high-fat diet-induced obese mice via gut microbia-mediated regulation of FGF21 signaling pathway. Biomed. Pharmacother. 2021, 135, 111187. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, X.; Wang, S.; Li, H.; Lu, Z.; Shi, J.; Xu, Z. Mannan-oligosaccharide modulates the obesity and gut microbiota in high-fat diet-fed mice. Food Funct. 2018, 9, 3916–3929. [Google Scholar] [CrossRef]
- Yang, J.-Y.; Lee, Y.-S.; Kim, Y.; Lee, S.-H.; Ryu, S.; Fukuda, S.; Hase, K.; Yang, C.-S.; Lim, H.S.; Kim, M.-S.; et al. Gut commensal Bacteroides acidifaciens prevents obesity and improves insulin sensitivity in mice. Mucosal Immunol. 2017, 10, 104–116. [Google Scholar] [CrossRef] [Green Version]
- Scott, S.A.; Fu, J.; Chang, P.V. Microbial tryptophan metabolites regulate gut barrier function via the aryl hydrocarbon receptor. Proc. Natl. Acad. Sci. USA 2020, 117, 19376–19387. [Google Scholar] [CrossRef] [PubMed]
- Mazmanian, S.K.; Liu, C.H.; Tzianabos, A.O.; Kasper, D.L. An Immunomodulatory Molecule of Symbiotic Bacteria Directs Maturation of the Host Immune System. Cell 2005, 122, 107–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rüter, C.; Buss, C.; Scharnert, J.; Heusipp, G.; Schmidt, M.A. A newly identified bacterial cell-penetrating peptide that reduces the transcription of pro-inflammatory cytokines. J. Cell Sci. 2010, 123, 2190–2198. [Google Scholar] [CrossRef] [Green Version]
- Kochetkova, I.; Thornburg, T.; Callis, G.; Holderness, K.; Maddaloni, M.; Pascual, D.W. OralEscherichia coliColonization Factor Antigen I Fimbriae Ameliorate Arthritis via IL-35, Not IL-27. J. Immunol. 2014, 192, 804–816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vignali, D.A.; Kuchroo, V.K. IL-12 family cytokines: Immunological playmakers. Nat. Immunol. 2012, 13, 722–728. [Google Scholar] [CrossRef] [PubMed] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arukha, A.P.; Freguia, C.F.; Mishra, M.; Jha, J.K.; Kariyawasam, S.; Fanger, N.A.; Zimmermann, E.M.; Fanger, G.R.; Sahay, B. Lactococcus lactis Delivery of Surface Layer Protein A Protects Mice from Colitis by Re-Setting Host Immune Repertoire. Biomedicines 2021, 9, 1098. https://doi.org/10.3390/biomedicines9091098
Arukha AP, Freguia CF, Mishra M, Jha JK, Kariyawasam S, Fanger NA, Zimmermann EM, Fanger GR, Sahay B. Lactococcus lactis Delivery of Surface Layer Protein A Protects Mice from Colitis by Re-Setting Host Immune Repertoire. Biomedicines. 2021; 9(9):1098. https://doi.org/10.3390/biomedicines9091098
Chicago/Turabian StyleArukha, Ananta Prasad, Christian Furlan Freguia, Meerambika Mishra, Jyoti K. Jha, Subhashinie Kariyawasam, Neil A. Fanger, Ellen M. Zimmermann, Gary R. Fanger, and Bikash Sahay. 2021. "Lactococcus lactis Delivery of Surface Layer Protein A Protects Mice from Colitis by Re-Setting Host Immune Repertoire" Biomedicines 9, no. 9: 1098. https://doi.org/10.3390/biomedicines9091098
APA StyleArukha, A. P., Freguia, C. F., Mishra, M., Jha, J. K., Kariyawasam, S., Fanger, N. A., Zimmermann, E. M., Fanger, G. R., & Sahay, B. (2021). Lactococcus lactis Delivery of Surface Layer Protein A Protects Mice from Colitis by Re-Setting Host Immune Repertoire. Biomedicines, 9(9), 1098. https://doi.org/10.3390/biomedicines9091098






