Abstract
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder. The neuropathological hallmark of the disease is the loss of dopamine neurons of the substantia nigra pars compacta. The clinical manifestations of PD are bradykinesia, rigidity, resting tremors and postural instability. PD patients often display non-motor symptoms such as depression, anxiety, weakness, sleep disturbances and cognitive disorders. Although, in 90% of cases, PD has a sporadic onset of unknown etiology, highly penetrant rare genetic mutations in many genes have been linked with typical familial PD. Understanding the mechanisms behind the DA neuron death in these Mendelian forms may help to illuminate the pathogenesis of DA neuron degeneration in the more common forms of PD. A key step in the identification of the molecular pathways underlying DA neuron death, and in the development of therapeutic strategies, is the creation and characterization of animal models that faithfully recapitulate the human disease. In this review, we outline the current status of PD modeling using mouse, rat and non-mammalian models, focusing on animal models for autosomal recessive PD.
1. Introduction
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder, affecting more than 6.1 million people worldwide [1]. It is characterized by progressive dysfunction and the death of dopamine (DA) neurons of the substantia nigra pars compacta (SNc). The ensuing DA depletion in the striatum causes motor symptoms (e.g., bradykinesia, rigidity, resting tremor, postural instability) and non-motor symptoms (e.g., depression, anxiety, weakness, sleep disturbances, cognitive disorders) [2,3]. Most often (90% of the cases), PD has a sporadic onset of unknown etiology possibly caused by the association of genetic and environmental risk factors. In 5–10% of cases, however, PD is monogenic with Mendelian inheritance [4]. At least 19 genetic loci for Parkinsonism have been identified to date, ten of which are autosomal dominant genes: SNCA (PARK1/PARK4), PARK3, UCHL1 (PARK5), LRRK2 (PARK8), GIGYF2 (PARK11), HTRA2 (PARK13), VPS35 (PARK17), EIF4G1 (PARK18), TMEM230 (PARK21), and CHCHD2 (PARK22). Autosomal-recessive (AR) homozygous or compound heterozygous mutations have been identified in nine genes: PARKIN (PARK2), PINK1 (PARK6), DJ-1 (PARK7), ATP13A2 (PARK9), PLA2G6 (PARK14), FBXO7 (PARK15), DNAJC6 (PARK19), SYNJ1 (PARK20) and VPS13C (PARK23) [4] (Figure 1). Understanding the mechanisms behind DA neuron death in each of these Mendelian forms may help to illuminate the pathogenesis of DA neuron degeneration in the more common forms of PD.
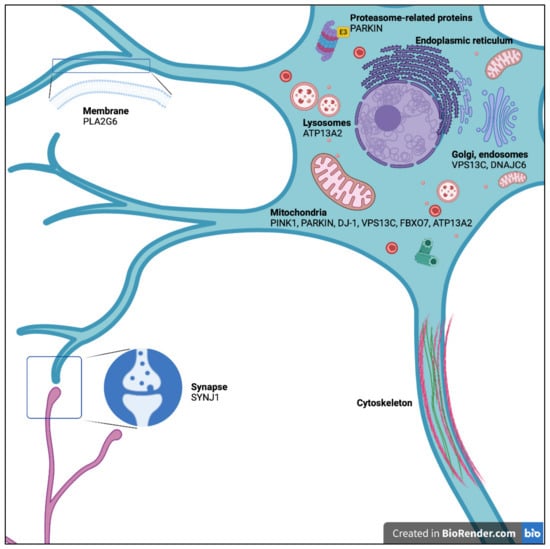
Figure 1.
Subcellular localization of proteins encoded by genes involved in autosomal recessive Parkinson’s disease.
A key step in the identification of the molecular pathways underlying DA neuron death, and in the development of therapeutic strategies, is the creation and characterization of animal models that faithfully recapitulate human disease. Ideally, an animal model for PD will display a progressive loss of SNc DA neurons and a major reduction of DA in the striatum. The models should also manifest motor symptoms (e.g., bradykinesia, rigidity, postural instability, resting tremor) and responsivity to L-DOPA. Widely employed in biomedical research because of their many advantages, mice and rats share biological similarity with humans (Chia 2020). Rodent models of PD can be broadly divided into environmental models, such as the widely used 1-methyl-4-phenyl-1,2,3,6-tetrapyridine (MPTP) mouse and the 6-hydroxydopamine (6-OHDA) rat model, and genetic models with knock-in (KI) and knock-out (KO) rodents based on known PD-associated genes [5]. Each model has its own strengths and limitations which determine the suitability of the model for a specific experiment [6].
Non-mammalian models are also extensively used to model PD. Among teleost fishes, zebrafish (Danio rerio) provides an interesting vertebrate model for the study of movement disorders because the neuronal circuitries involved in movement in zebrafish are well characterized, and most of the associated molecular mechanisms are highly conserved [7], demonstrating similarities with the mammalian central nervous system (CNS). Additionally, the DA neurons (14 in total) in the posterior tuberculum of the zebrafish (homologous to the substantia nigra in humans) are well characterized [7]. Clusters of DA neurons in the ventral diencephalon have ascending projections to the subpallium of the telencephalon, suggesting that these neurons are homologous to the ascending midbrain DA neurons of the mammalian nigrostriatal pathway [8]. Their relatively small size, optical transparency, rapid lifecycle and genetic similarity to humans make zebrafish a simple model for the evaluation of the pathological mechanisms in PD [7]. Several transgenic, knock-down (KD) and mutant zebrafish models of PD have been generated and characterized, advancing our understanding of the role of several genes implicated in the disease. Furthermore, the zebrafish vertebrate model is particularly suited for large-scale drug screening [6,7,9].
Although the zebrafish is the most widely used fish model, the Japanese Medaka teleost fish, Oryzias latipes, is also extensively used to model PD. Similarly to the zebrafish, Medaka fish are relatively transparent, produce large numbers of progeny per generation, are easy to maintain, and there are well-established techniques for the manipulation of their genomes [10].
Furthermore, Drosophila melanogaster provides a simple, yet powerful in vivo system to model PD pathobiology [11]. Clusters of dopaminergic neurons are detectable in the developing and adult fly, and metabolic pathways for DA synthesis are conserved between Drosophila and humans [9]. In the adult brain, Drosophila has distinct DA neuronal clusters, including about 200 DA neurons, and displays complicated behaviors mimicking some DA-dependent human behaviors [11]. Drosophila are cheap and easy to maintain in the laboratory; they have a rather short life span (40–120 days), and a variety of techniques and tools to manipulate their gene expression are currently available [11].
Other small organisms, such as Caenorhabditis (C.) elegans, are good experimental models to study PD because of their easy genetic manipulation and rapid reproduction and growth rate. C. elegans is a 959-cell nematomorph with a well-characterized genome. It exists primarily as a self-fertilizing hermaphrodite, in which the progeny are genetically identical. Males exist as a small fraction of the population (<0.1%) [12]. C. elegans has a well-defined, simple nervous system comprising 302 neurons and a DA network formed by eight neurons exactly: six anterior (four cephalic sesilla (CEP) neurons, two anterior deirids (ADE) neurons) and two posterior deirids (PDE) neurons. Male C. elegans possess six additional DA neurons in the tail ray [12]. Unlike other species in which staining for DA neurons is possible only in fixed tissues, neuronal survival in C. elegans can be assessed in live animals by expressing a fluorescent protein specifically in DA neurons, thanks to the existence of a transparent cuticle [12]. The quantification of more subtle phenotypes besides the loss of neuron cell bodies can be achieved by documenting the disappearance of axons, broken neurites, the retreat of dendritic terminals, and axonal and dendritic blebbing [12]. Here, we review the current state of animal models for autosomal recessive PD (ARPD) and discuss their limitations and utility.
2. PARK2: The Parkin RBR E3 Ubiquitin Protein Ligase Gene (PARKIN)
PARK2-Parkin (PRKN, OMIM 600116) was the first gene to be discovered to be linked to genetic Parkinsonism with an AR mode of inheritance [13,14]. Mutations in the PARK2 gene are the most common known factor responsible for autosomal recessive juvenile Parkinsonism (ARJP) (10–20%) [15]. PARK2 spans a large genomic interval (1.4 Mb) and is located in a region of genomic instability. The majority of parkin-proven PD cases result from large genomic alterations (deletion, duplication or inversion) that affect one or more exons. There are several other mutations, including missense and stop point mutations, but at a lower frequency than deletion [16,17]. PARK2 encodes the protein PARKIN, a ubiquitin E3 ligase that, at the intracellular level, catalyzes the transfer of ubiquitin to various protein substrates. Through ubiquitination, PARKIN regulates the turnover and localization of many proteins, thus modulating a variety of cellular processes, including mitochondrial turnover [18,19,20]. Several transgenic overexpression and KO models of PARK2 have been generated to gain insight into wild type (WT) PARKIN function and the mechanism of the disease [21] (Table 1).

Table 1.
Observed phenotype in animal models of Parkin-Linked Parkinson’s disease (PARK2).
2.1. Mouse Model
Several PARKIN mouse models have been generated, seven of which were Parkin KO models. In 2003, Itier and coauthors created the first Parkin KO mouse model by replacing murine Park2 exon 3 with the PGK-NeoR cassette (B6;129S2-Park2tm1Roo) [22]. While these mutant mice were viable and fertile, with normal brain morphology, weight and size, their bodyweight and body temperature were characteristically low. No overt behavioral change was observed up to 24 months of age. Furthermore, histological sections of the brain at the level of the striatum, hippocampus, brain stem and cerebellum revealed no differences versus WT animals, nor were differences in tyrosine hydroxylase (TH) staining observed in the SNc and the striatum. The immunohistochemical examination of the dopamine transporter (DAT) showed no significant differences between the WT and mutant mice. However, the levels of DAT and vesicular monoamine transporter type-2 (VMAT2) were significantly reduced in the striatum of the Parkin KO mice at 15 months of age. Behavioral tests showed that the Parkin KO mice displayed a deficit in learning and memory. Hence, this model did not display a clear PD phenotype. This led to the hypothesis that the nigrostriatal DA system in mice is less vulnerable to the neuronal damage induced by PARKIN absence than the human brain [22].
That same year, Goldberg and colleagues developed another Parkin KO mouse model carrying an exon 3 deletion (B6;129S4-Park2tm1Shn) [23]. They reported that the Parkin KO mice were viable, fertile, and exhibited normal brain morphology. Quantitative in vivo microdialysis revealed an increase in the extracellular DA concentration in the striatum. Intracellular recordings of medium-sized striatal spiny neurons showed a reduction in their synaptic excitability; in addition, behavioral tasks disclosed poorer performance on tests sensitive to nigrostriatal dysfunction [23]. Palacino and coauthors used this model to investigate whether the loss of PARKIN function resulted in an abnormal abundance and/or modification of proteins in the ventral midbrain [24]. Two-dimensional gel electrophoresis followed by mass spectrometry revealed a decrease in several proteins involved in mitochondrial function or oxidative stress [24].
A further exon 3-deleted Parkin mutant was generated by Stichel et al. in 2007 following the complete replacement of exon 3 with a NeoR cassette [25]. They observed severe mitochondria changes in the neuronal somata in the SNc. The mitochondria showed electron-dense inclusion bodies and dilated, disorganized cristae. These alterations coincided with a reduced complex I capacity in the SNc. Nonetheless, neither SNc DA neuron neurodegeneration nor motor disabilities were observed [25].
In 2005, Perez and Palmiter created a new Park2 KO model involving the complete replacement of exon 2 with the Polr2a-NeoR cassette (B6;129S4-Park2tm1Rpa) [26]. The mice displayed no differences in their behavioral phenotype or in their catecholamine levels, suggesting that the nigrostriatal and the noradrenergic system were conserved [26]. Sato et al. created another exon 2-deleted Park2 KO mouse model via the partial replacement of exon 2 with the MC1-NeoR cassette, followed by a frame insertion of tau-green fluorescent protein (tau-GFP) fusion protein [27]. This Park2 KO mouse showed no obvious behavioral or locomotor activity deficits, no change in TH-positive nigra neurons, and no major decrease in DAT. The abnormally low levels of DA release were interpreted by the authors as a predictive factor for neuronal death [27]. Von Coelln and coauthors created another Park2 KO model by deleting Parkin exon 7 (B6;129S7/S4-Park2tm1Tmd) [28]. The germline exon 7-deleted mice showed the early loss of catecholaminergic neurons in the locus coeruleus (LC) that mirrored the loss of LC neurons, as occurs in patients with PARK2 mutations [47]. In addition to the impairment of the central noradrenergic system, there was a dramatic reduction in the norepinephrine-dependent acoustic startle response. A lower concentration of norepinephrine was also found in the olfactory bulb and in the spinal cord, two major target regions of projecting axons from the LC. There was no impairment of the nigrostriatal dopaminergic system [28]. The reasons behind the absence of SNc neuron degeneration in these Park2 KO models remain to be elucidated. One plausible explanation is that genetic compensation occurs in response to the gene knockout [48].
In order to overcome any potential compensatory mechanisms that may underlie the absence of SNc DA neuron degeneration, Shin et al. followed a different strategy [29]. Because previous studies showed that the embryonic deletion of key factors often has no deleterious effects while the conditional KO of the same factor in adult animals leads to mice with profound neuron degeneration [49], Shin et al. generated a new conditional Parkin exon 7 KO mouse using the lentiviral delivery of GFP-tagged Cre-recombinase delivered stereotaxically to the midbrain of adult mice. Following this strategy, a progressive loss of DA neurons could be observed. Increased levels of PARIS (parkin interacting substrate) were also reported, as observed in both ARJP and sporadic PD human SNc [29]. Furthermore, Stevens and coauthors showed that the deletion of PARKIN in adult mice led to a decrease in mitochondrial size, mitochondria number, and protein markers in the ventral midbrain, consistent with a defect in mitochondrial biogenesis [30]. Hence, this conditional KO strategy seems to be promising.
A completely different model is the spontaneous quakingviable mouse mutant. The genetic lesion in this model is a large deletion of approximately one megabase on chromosome 17 [31]. This deletion includes the GkI gene, which encodes the RNA-binding proteins of the STAR family [32]. The lack of GKL protein expression in oligodendrocytes leads to the improper myelination of the CNS in animals homozygous for the deletion. The quakingviable mouse has been extensively studied for the dysmyelination of the CNS. The analysis of the sequence of the deleted region revealed that because the mouse homolog of the human gene PARKIN is partially contained in this interval, this deletion eliminates the expression of the mouse Parkin gene product and generates a mouse Park2 KO [31]. Quakingviable mice displayed normal cellular conformation in the grey matter. Although they have not been extensively studied, alterations in the DA metabolism have been reported in the quakingviable mouse. Quakingviable homozygotes showed reduced locomotor/exploratory activity, particularly in the open field test, and tremors in the caudal part of the trunk and proximal portions of the hind extremities. However, because this model did not show the loss of DA neurons in the SNc, the quakingviable mouse cannot be considered a reliable model of PD [31]. The predominant dysmyelination phenotype further complicates behavioral, neuropathological and biochemical analysis [32].
In 2010, Ramsey and Giasson first observed a novel spontaneous missense mutation in the C3H mouse strain, resulting in PARKIN amino acid substitution E398Q [50]. Results from in vitro studies suggested that the novel E398Q Parkin mutant had reduced solubility and functional impairment [50]. This mutation is equivalent to an E399Q mutation in human PARKIN, because mouse Parkin has one residue fewer (Gly 139) than human PARKIN [51]. Because the E399Q variant has never been found in human subjects, it is not clear whether this variant can be considered a benign variant or a pathogenic mutation.
In order to test the hypothesis that Parkin mutants may have toxic effects on DA neurons, Lu and colleagues generated a bacterial artificial chromosome (BAC) transgenic mouse expressing the truncated human parkinQ311X mutation in DA neurons, driven by a DAT promoter [33]. The private mutation Q311X is a C-to-T transition, replacing a Gln codon (CAG) with a stop codon (TAG) at amino acid position 311 in exon 8 [14]. ParkinQ311X mice have normal WT Parkin alleles on both chromosomes, and they express WT endogenous Parkin in addition to the exogenous human parkinQ311X variant ectopically expressed from the BAC chromosome. ParkinQ311X mice exhibit multiple late-onset and progressive hypokinetic motor deficits. Stereological analyses revealed that the mutant mice develop age-dependent DA neuron degeneration in the SNc accompanied by a marked loss of DA neuron terminals in the striatum. Neurochemical analyses showed a reduction in the striatal DA in mutant mice, which correlated with their hypokinetic motor deficits. Finally, mutant parkinQ311X mice exhibited the age-dependent accumulation of proteinase K-resistant endogenous α-syn in the SNc and increased levels of nitrotyrosine, a marker for oxidative protein damage [33]. This model was recently used to test a pharmacological neuroprotective strategy [52]. Although the genetics of this model do not exactly recapitulate AR transmission, it can be a useful model to study the deleterious effects of PARKIN mutants.
2.2. Rat Model
Recent advances in technology for mammalian genome engineering and the optimization of viral expression vectors have increased the use of genetic rat models of PD [53]. Compared to mice, some genetic rat models of PD better reproduce key aspects of PD; the reasons for the different phenotypes between mice and rats are not clear. Many behavioral, physiological and biochemical differences exist between mice and rats, in addition to the substantial differences in the gene sequence and expression [53]. Compared to mice, the rat neuronal circuitry more closely resembles that of humans; furthermore, rats are less prone to anxiety, which affords a major advantage for behavioral evaluation [9].
The Parkin KO rat model was generated using the zinc finger nuclease (ZFN)-mediated targeted disruption of exon 4 of the Parkin gene [34]. The rats displayed unaltered core body temperature and increased body weight, normal behavior, and no neurochemical changes [34]. A further characterization by Gemechu et al. showed that 2-month-old Parkin KO rats displayed lower monoamine oxidase (MAO) enzyme activity and higher levels of ß-phenylenthylamine (ß-PEA) than their WT counterparts [35]. Parkin KO rats displayed normal locomotor activity but a decreased locomotor response when tested with a low dose of psychostimulant methamphetamine, suggesting altered DA neurotransmission in the striatum when challenged with an indirect agonist [35]. Stauch and coauthors isolated non-synaptic mitochondria from the striatum of Parkin KO rats and analyzed the mitochondrial proteome: 15 proteins exhibited differential expression in Parkin KO rats [36]. This result suggested a pathological alteration; however, no mitochondrial dysfunction was reported [36]. As regards neuroanatomical changes in the DA neuron number, Parkin KO rats did not differ from WT rats, except for a small and not statistically significant 20% reduction in the DA neuron number at 8 months of age [34]. However, the Parkin rats were not analyzed at an older age. Given this small reduction in DA neurons, it is possible that in Parkin KO rats a potential PD-like phenotype is delayed, warranting further characterization in older rats [34,36]. Overall, this model can be useful to recapitulate some aspects of ARJP, but more data are necessary.
Building on previous work that suggested a dominant negative effect of PARKIN mutants [54], Van Rompuy and coauthors used a recombinant type 2 adeno-associated viral vector pseudotyped with type 8 capsid (rAAV2/8) to develop a novel rat model that overexpressed the human PARKIN-T240R mutation in the DA neurons of adult animals [37]. Surprisingly, the overexpression of PARKIN-T240R and WT PARKIN induced progressive DA neuron death in rats, starting at 8 weeks after viral injection [37]. The degeneration was specific for PARKIN because the similar overexpression of GFP did not lead to nigral degeneration. Nonspecific cell death induced by an inflammatory response to the vector preparations was excluded by immunohistochemical staining specific for activated and phagocytic microglia. This result contrasts with the many studies describing the neuroprotective capacity of PARKIN. The authors speculated that there might be a small window of physiologic PARKIN levels in DA neurons. Further data are needed to better characterize this model.
2.3. Zebrafish (Danio rerio)
Zebrafish Parkin protein is 62% identical to its human counterpart, with 78% identity in functionally relevant regions. The park2 gene is expressed throughout zebrafish development and ubiquitously in adult zebrafish tissues [55]. Flinn et al. generated a zebrafish model of parkin KO using anti-sense oligonucleotides [38]. Parkin silencing led to a marked decrease in the number of ascending DA neurons in the posterior tuberculum (homologous to the substantia nigra in humans), which was enhanced by exposure to the toxin 1-methyl-4-phenylpyridinium (MPP+). Other neuronal populations were spared. Neither serotonergic nor motor neurons were affected; therefore, the toxic effect of parkin silencing was specific for DA neurons. Notably, parkin silencing impaired the mitochondrial function. Because this vertebrate model shared pathogenic mechanisms (reactive oxygen species (ROS), complex I deficiency) and a pathological hallmark (DA cell loss) with human PARKIN-mutated patients, the authors concluded that it mirrors the human disease better than any other currently available animal model for PARKIN disease [38]. Taniguchi et al. identified, in a library of fishes mutagenized by N-ethyl-N-nitrosourea (ENU), a nonsense mutation that resulted in a truncated Parkin protein at Tyr314. No further information is available regarding this model [56].
2.4. Drosophila melanogaster
A Drosophila model of ARJP was first created using a mutational approach to inactivate the highly conserved parkin ortholog. Flies bearing null alleles of parkin were viable but exhibited a reduced lifespan and male sterility [39]. The histological analysis of the major flight muscles revealed the severe disruption of muscle integrity, which correlated with locomotion defects [40]. Progressive mitochondrial pathology preceded myofibril degeneration; the muscle cells showed many swollen mitochondria with degenerated cristae. The standard histologic analysis of the brain revealed the appropriate development of the major brain centers in young and old mutant flies. DA neurons of the dorsomedial (DM), dorsolateral (DL) and anteromedial (AM) clusters of the medulla were analyzed but no clear neuronal loss was observed in any of these cell groups [40]. Lacking DA neurodegeneration, this model failed to recapitulate the prominent hallmark of PD.
The observed mitochondrial dysfunction was then investigated in order to understand the mitochondrial impairment occurring in DA neurons. One year later, Pesah et al. used P-element mutagenesis to generate another parkin KO mutant, and they confirmed previous findings by Greene et al. (2003). They observed female infertility and increased sensitivity to chemical and environmental stress in mutant animals compared to controls [41]. Again, no major loss of DA neurons in the parkin mutant was observed. In order to better understand the effects of parkin mutations in vivo, Wang and colleagues generated transgenic Drosophila overexpressing the human PARKIN mutant R275W associated with ARJP [42]. Transgenic flies overexpressing R275W displayed an age-dependent degeneration of specific DA neuronal clusters and concomitant locomotor deficits that accelerated with age or in response to rotenone treatment. The R275W mutant flies exhibited prominent mitochondrial abnormalities in their flight muscles. These defects were caused by the expression of human parkinR275W and were highly similar to those observed in parkin null flies.
These findings raised the interesting hypothesis that selected parkin mutations may directly exert neurotoxicity in vivo [42]. Cha et al. generated two other transgenic lines in Drosophila expressing an N-terminal deleted parkin (containing only 108–482 aa) and parkinK71P, a point mutant similar to R42P found in ARJP patients [43]. Both parkin mutant flies showed reduced longevity, a drooped wing phenotype, locomotor dysfunction and muscle degeneration accompanied by apoptosis. Among the histopathological hallmarks, there was a severe loss of DA neurons in the parkin mutant flies and a shrunken morphology of DA neurons [43]. These phenotypes were completely restored by the overexpression of WT parkin. In order to further explore whether the parkin mutations identified in familial PD can exert cell-specific toxic effects in vivo, Sang and colleagues generated Drosophila lines expressing parkinQ311X or T240R mutants under the control of a ddc-GAL4 driver to lead specific Parkin expression in DA neurons and serotonin (5-HT) neurons [44]. Flies expressing parkinT240R or parkinQ311X showed a dramatic decline in their climbing ability compared to the controls. Motor tests revealed severe deficits with onset 2–3 weeks after eclosion, which suggested a degenerative, rather than a developmentally induced, defect. A histopathological hallmark was age-dependent selective DA neuron degeneration [44]. The model provided a robust behavioral and neuropathological phenotype.
2.5. Caenorhabditis elegans
C. elegans has orthologs to many of the genes implicated in PD (e.g., LRRK2/lrk-1, PINK1/pink-1, PARKIN/pdr-1, DJ-1/djr-1.1/djr-1.2, ATP13A2/catp-6). This phylogenetic similarity to humans makes C. elegans a suitable PD model [12]. Ved and colleagues generated a parkin KO C. elegans line [45] that displayed normal development but had a shorter lifespan than the WT. The strain was also more vulnerable to several mitochondrial complex I inhibitors than nontransgenic nematodes [45]. Bornhorst et al. used a PARK2-pdr1(gk448) III (CGC) C. elegans strain to investigate manganese (Mn)-induced toxicity in dopaminergic neurotoxicity. The mutants exhibited hypersensitivity to Mn-induced lethality compared to WT worms and a time-dependent increase in Mn-induced reactive oxygen and nitrogen species (RONS). However, when the authors used worms expressing GFP under the control of a promoter for the dopamine re-uptake transporter 1 (C. elegans ortholog for vertebrate DAT), they observed in these mutants no Mn-induced degeneration of the CEP dopaminergic neurons [46].
3. PARK6: PTEN-Induced Putative Kinase 1 (PINK1)
Identified in 2004, mutations in the PARK6 gene (PINK1, OMIM 605909) are the second most common genetic cause of autosomal recessive early-onset PD (AR EOPD) [57]. PARK6 encodes a 581-amino-acid protein named (PTEN)-induced kinase 1 (PINK1) with an N-terminal mitochondrial targeting sequence (MTS) and a short transmembrane domain (TMD), followed by a C-terminal serine/threonine kinase catalytic domain [58]. PINK1 is imported into the mitochondria through the translocase of the outer (TOM) and inner (TIM23) membrane complexes; its MTS is cleaved off by the mitochondrial processing peptidase (MPP) located in the matrix [58]. The Pink1 TMD acts as a stop-transfer sequence leading to integration into the inner membrane, where PINK1 exposes its kinase domain to the mitochondrial intermembrane space (IMS) [59]. Subsequently, the inner mitochondrial membrane protease presenilin-associated rhomboid-like protease (PARL) cleaves PINK1 into a short soluble form which is then released into the cytosol [58,60]. The disruption of the mitochondrial membrane potential interferes with PINK1 mitochondrial import through the TIM23 complex and induces PINK1 retention and accumulation on the outer mitochondrial membrane. Here, PINK1 participates in inducing mitochondrial turnover [61,62,63].
PINK1 has been suggested to provide protection against oxidative stress by assisting in condemning damaged mitochondria to degradation/mitophagy and maintaining mitochondrial homeostasis [64]. PINK1 deficiency in mammalian neurons alters the mitochondrial buffering capacity, increases ROS and impairs respiration [65], suggesting a mechanism by which the loss of PINK1 function would confer vulnerability to cell death. Most PD-linked mutations are located within the kinase domain and result in impaired or the loss of function of kinase activity and the decreased neuroprotective function of PINK1 [66]. In order to investigate the physiological function of PINK1 and its potential neuroprotective role, several PINK1-deficient animal models have been generated (Table 2).

Table 2.
Observed phenotype in animal models of PTEN-induced kinase 1-linked Parkinson’s disease (PARK6).
3.1. Mouse Model
In 2007, Zhou et al. used a transgenic RNA interference (RNAi) approach to generate a conditional Pink1-silenced mouse. In these transgenic mice, PINK1 expression was regulated by the Cre-LoxP system; the activation of the RNAi transgene resulted in the silencing of the Pink1 gene to less than 5% of normal levels in the CNS [67]. The silencing of the Pink1 gene expression did not cause a loss of the DA neurons in the SNc, an alteration in the DA level in the striatum, or abnormalities in motor function [67]. In addition, Pink1 KO mouse models were generated and characterized [68,69]. The Pink1 KO mice generated by germline deletions of exons 4–7 did not exhibit major abnormalities or PD-associated pathology in the brain. No changes in the number of DA neurons or striatal DA levels were observed, and no alterations in DA synthesis or DA receptor levels were found [69]. Nevertheless, the absence of PINK1 caused a marked decrease in the evoked DA release in striatal slices and a reduction in the quantal size and release frequency of catecholamines in dissociated chromaffin cells [69]. The impairment of DA release in Pink1 KO mice compromised striatal synaptic plasticity, with a reduction in corticostriatal long-term potentiation (LTP) and long-term depression (LTD), which could be restored by either DA receptor agonists or agents that increase DA release, such as amphetamine or L-DOPA [69].
Further studies with the same mouse model showed that the loss of PINK1 also resulted in mitochondrial dysfunction [68]. While the absence of PINK1 did not cause gross ultrastructural alterations or changes in the total number of mitochondria, an increased number of larger mitochondria was observed in the striatum of Pink1 KO mice at 3–4 and 24 months. Mitochondrial respiration was also impaired in young Pink1 KO mice, and the activity of complex I, complex II and aconitase was reduced in the striatum but not in the cerebral cortex. At 2 years of age, mitochondrial respiration was also impaired in the cerebral cortex, indicating an age-dependent impairment of mitochondrial function [68]. The mitochondria in the cerebral cortex were found to be more sensitive to oxidative stress [68].
Glasl and colleagues generated another Pink1-deficient mouse by deleting exons 2 and 3; this excision generates a shift of the reading frame that results in a stop codon in exon 4 [70]. As observed in other Pink1 KO mice [67,68,69], these mice did not show morphological alterations in the DA system. No differences in the number of TH-positive neurons in the SNc or in the striatal DA content were observed at 6 or 19 months of age. The neurons in the Pink1 KO mice showed less fragmented mitochondria. Behavioral analysis in the open field showed that the mice, at the ages of 3 and 24 months, did not manifest a strong impairment in their spontaneous locomotor activity in a novel environment. The mice demonstrated normal motor coordination and balance [70]. Nonetheless, at 26 months of age the Pink1-deficient mice displayed symptoms resembling the early phases of PD, symptoms that in humans are known to precede the severe motor-phase of the disease: gait alterations, especially in the hind paws, and olfactory dysfunction. The density of the serotonergic fibers in the glomerular layer of the olfactory bulb was considerably reduced and the aged male Pink1 KO mice showed a marked deficit in their fine olfactory discrimination and smell sensitivity, which are two non-motor symptoms of PD [70].
In line with these findings, Kelm-Nelson and colleagues used the Pink1 KO mouse (Pink1tm1Shn) to investigate early-onset locomotor and sensorimotor deficits, including deficits in the limbs, and impaired vocalization, articulation and swallowing [71]. The Pink1 KO mice exhibited impaired vocalization: they produced simple calls at a lower intensity compared to WT. The assessment of their spontaneous motor activity showed a severe impairment of limb motor skills, with fewer hindlimb and forelimb steps and reduced rearing and landing on the cylinder test compared to WT. Additionally, the Pink1 KO mice took longer to turn and traverse during the pole test. These motor deficits occurred in the absence of DA neuron loss in the SNc or a reduction in TH optical density in the striatum [71]. While Pink1 KO mice do not recapitulate DA neuron loss, they could be useful to study early dysfunctions associated with PINK1-related PD.
Gispert and colleagues generated a transgenic mouse bearing the human pathogenic G309D-PINK1 missense mutation inserted by homologous recombination into the orthologous mouse Pink1 locus. The G309D mutation is a loss-of-function mutation that induces Pink1 mRNA instability and dampens PINK1 protein expression [72]. An analysis of Pink1 mRNA showed a 97% reduction in the brain of homozygous mice. There was an age-dependent reduction in DA and a decrease in locomotor activity, with selectivity for spontaneous movements [72], along with a progressive reduction in body weight starting from middle age. No Lewy body formation or nigrostriatal degeneration was observed up to 18 months of age; however, impaired bioenergetics and progressive mitochondrial dysfunction in the brain were detected. There was a reduction in the mitochondrial membrane potential and adenosine triphosphate (ATP) levels, and respiratory complex activity was decreased [72]. Despite the reduction in fission factor Mtp18, the G309D-Pink1 mice showed normal mitochondrial morphology and mass; only after proteasomal stress did they show reduced fission and increased perinuclear aggregation of the mitochondria, perhaps due to the failed activation of mitophagy [72].
Another Pink1 KO mouse was generated by the deletion of a large portion of the gene, from exons 2 to 5, and primary neuronal cultures were prepared [73]. The major finding of that study conducted on in vitro cortical neurons was an age-dependent reduction in long-term viability and higher cytotoxicity indices compared to the control neurons, with an increase in cell death and apoptosis [73]. Using the same Pink1 KO transgenic mouse, Gandhi and colleagues explored the role of PINK1 in maintaining mitochondrial function in in vitro neurons [65]. They reported that the loss of PINK1 function in primary cortical and midbrain neurons increased Ca2+ levels, causing mitochondrial Ca2+ overload and excess ROS production. PINK1 deficiency was also associated with reduced glucose uptake that resulted in the global impairment of respiration and the loss of mitochondrial membrane potential, which sensitized the mitochondria to the opening of the mitochondrial permeability transition pore (mPTP) [65].
3.2. Rat Model
Pink1 KO rats were generated using ZFN technology [34,74]. In their preliminary characterization, Sun and colleagues performed a quantitative autoradiography analysis of DA pre-synaptic markers and post-synaptic receptors in the striatum of 6-month-old mice, and they found altered DA regulation. The density of DA D2 and D3 receptors was significantly increased in the striatum of transgenic rats, while no remarkable changes in the density of DA D1 receptor, VMAT2 or DAT were observed [74]. Later, Dave and colleagues performed a better phenotypic characterization of the model [34], showing that the loss of the Pink1 gene produced robust behavioral dysfunction and a severe loss of SNc DA neurons. Motor impairment in movement, strength and coordination started at 4 months of age and preceded nigral neuronal loss. The loss of DA neurons was rapid between 4 and 8 months of age, with a decrease of about 25% seen at 6 months and 50% at 8 months of age [34]. The loss of midbrain DA neurons in this model was confirmed by a later study: the number of TH-positive neurons in the SNc was decreased, as well as the size of the SNc at 9 months of age [75]. While nigral neurodegeneration did not correlate with changes in the striatal TH or α-syn immunoreactivity within the SNc, striatum or other brain areas, there was a 2–3-fold increase in DA and 5-HT striatal content at 8 months of age [34]. Pink1 KO rats also displayed metabolic alterations and mitochondrial dysfunction [75]; selected brain metabolomic markers produced in the mitochondria or interacting with the mitochondrial products were altered. In the striatum, aspartate was found to be increased while taurine and creatinine were decreased; myoinositol was found to be decreased in the cortex.
Regarding mitochondria, an analysis of the expression of electron transport chain (ETC) complex I, III, IV and V subunits revealed a decrease in the striatum of Pink1 KO rats, while the complex II subunit levels were unchanged. Proton leak was increased in the brain mitochondria of Pink1 KO rats at 4 and 9 months of age, suggesting increased ROS generation. This is consistent with the increased ROS signaling and altered stress pathway observed in the striatum of 4-month-old Pink1 KO animals [75]. Furthermore, the striatal glycolytic protein levels were higher, suggesting increased glycolysis. Additionally, the mitochondrial trafficking and dynamics were altered in the striatum of Pink1-deficient mice at 4 months of age: dynamin related protein-1 (DRP1) was increased while mitofusin-2 (MFN2) was decreased; these results paralleled increased mitochondrial fission and fragmentation [75].
Grant and coauthors evaluated the effect of the loss of PINK1 function on vocalization, oromotor and limb function, and neurodegenerative features [76]. They showed that Pink1-deficient rats developed the early and progressive impairment of their vocalization: already, at 2 months of age, the rats produced calls which were lower in intensity and bandwidth compared to WT, while the peak frequency was decreased at 6 months of age. The loss of function of PINK1 also caused oromotor deficits, including variable licking behavior and irregular and inconsistent biting patterns. In contrast to the vocalization and licking deficits, which emerged early, sensorimotor deficits occurred at 8 months of age with a reduction in spontaneous activity and an increase in the time needed to travel a tapered beam. Though no differences in TH immunoreactivity were found in the striatum and in the SNc at 8 months of age, Pink1 KO rats showed a reduction in TH immunoreactivity in the LC. Furthermore, insoluble α-syn aggregates were found in the SNc, in the periaqueductal gray (PAG) and in the LC of 8-month-old Pink1 KO rats.
Pultorak and coauthors explored the consequences of the vocalization impairment observed in Pink1 KO rats on social communication [76] as a measure of female approach behavior [77]. By presenting recordings of male Pink1 KO and normal WT ultrasound vocalizations (USVs) to female rat listeners, the authors observed that the female rats were less motivated by the male Pink1 KO USVs than the WT male USVs. Females exposed to Pink1 KO USVs showed decreased neuronal activity in the nucleus accumbens, a region implicated in auditory processing and sexual motivation. The results demonstrated that vocalization deficits in a PINK1-PD related model compromised communication [77]. The studies showed that Pink1 KO rats can be considered a promising model to study PINK1-associated PD.
3.3. Zebrafish (Danio rerio)
The zebrafish pink1 gene (NM_001008628) is located on chromosome 23 and shares 53% identity with humans. It encodes a 574 aa protein which is 54% identical to the human PINK1 protein. An analysis of the zebrafish primary sequence revealed a putative mitochondrial targeting domain at the N-terminus of the protein and a putative serine/threonine protein kinase active site, which is highly similar to human PINK1 [78]. The model of pink1 deficiency in zebrafish was created by using morpholino (MO) antisense; this model exhibits a severe developmental phenotype, including structural alterations in the axonal scaffold and a decrease in the number of central DA neurons. The phenotype was rescued by the overexpression of WT human PINK1 mRNA [78]. MO KD of the zebrafish pink1 ortholog added evidence for the importance of this gene in the control of oxidative stress and mitochondrial function. The downregulation of Pink1 expression resulted in mitochondrial dysfunction, with a reduction in the mitochondrial membrane potential, augmented ROS levels, and the activation of the apoptotic signaling pathway, including increased caspase-3 activity. Furthermore, pink1 KD was associated with the elevated activity of the mitochondrial protein glycogen synthase kinase 3 β (GSK-3β), whereas its inhibition via nonspecific (LiCl) and specific (SB216763) inhibitors partially rescued the phenotypes in pink1 morphant zebrafish [78].
Priyadarshini et al. found that the expression of 177 genes was altered in Pink1 morphants. Among the canonical pathways identified, the most affected was hypoxia-inducible factor (HIF) signaling with the strong downregulation of hif1α mRNA. Transforming Growth Factor-β (TGF-β) signaling, retinoic acid receptor activation, and the biogenesis of the mitochondria were also altered [79]. Catalase enzyme activity was decreased, coupled with catalase and SOD3 transcript downregulation. These enzymes are part of the antioxidant defense system. Both catalase and superoxide dismutase are ROS-scavenging enzymes; consistently, their downregulation was associated with an increase in ROS levels. The Pink1 morphants showed an abnormally lower heart rate but no gross morphological phenotype. Following pink1 KD, ferrochelatase expression was also upregulated, leading to the increased expression of vascular endothelial growth factor (VEGF) and erythropoiesis. The increased erythropoiesis effect was rescued by the administration of the antioxidants N-acetyl cysteine (NAC) and L-glutathione reduced (LGR), while the abnormalities of the gene expression profiles were rescued by pink1 mRNA injection [79]. In another study, the MO-mediated KD of pink1 function did not cause large alterations in the number of dopaminergic neurons in the ventral diencephalon, though the patterning of these neurons and their projections was perturbed [8]. This was accompanied by locomotor dysfunction, a weak or absent response to tactile stimuli, and reduced swimming behavior. These defects were rescued by the expression of exogenous Pink1 or the administration of the DA D1 receptor agonist SFK-38393 [8].
Another study reported no morphological or behavioral deficits in pink1 morphant zebrafish [80]. However, an increased susceptibility to MPTP-induced motor disturbance was observed: the DA neuron clusters of pink1-deficient zebrafish were more sensitive to MPTP toxicity. In addition, the translation inhibition of Pink1 reduced the th1 and th2 mRNA forms without affecting the levels of dat mRNA, and this effect was rescued by the injection of pink1 mRNA. Although there was a decrease in the number of TH-positive neurons in the DA diencephalic cluster, the normal DAT levels suggested that the downregulation of Pink1 may cause a decline in essential mRNAs and proteins (e.g., TH) in the absence of effects on neuronal survival [80]. The increased susceptibility to MPTP underlined the importance of Pink1 in oxidative stress: exposure to hydrogen peroxide (H2O2) dramatically upregulated the expression of pink1 mRNA and decreased th2 mRNA in zebrafish. Both effects could be reverted by treatment with the antioxidant LGR [93]. LGR could also rescue, along with pink1 mRNA, th1 and th2 expression, which were lost in Pink1 morphants [80,93].
A pink1 mutant zebrafish line (pink1−/−) with a premature stop mutation in exon 7 (Y431*) in the pink1 homolog was found in ENU mutagenesis libraries [55]. This mutation resulted in a truncated Pink1 protein, with the loss of its C-terminus and part of its kinase domain, leading to the inactivation of Pink1 catalytic activity and decreased mRNA stability [81]. While these adult pink1−/− fishes did not display obvious behavioral abnormalities, pink1−/− embryos at 5 days postfertilization (dpf) showed a marked decrease in their number of DA neurons and a reduction in their mitochondrial complex I activity [55]. Another study confirmed that Pink1 deficiency resulted in a progressive loss of DA neurons from early development (5 dpf) to adulthood (18 months) [81]. Pink1−/− embryos did not display any overt morphological abnormalities and the expression of several neurodevelopmental markers was unchanged compared to WT embryos. These results contrasted with findings by Anichtchik et al., who observed a severe developmental phenotype with major generalized neurodevelopmental abnormalities in the MO-mediated knock down of pink1 in zebrafish embryos [78]. Despite the absence of overt morphological abnormalities, Pink1 deficiency resulted in severe alterations in mitochondrial function and morphology: a reduction in mitochondrial complex I and III activity and enlarged mitochondria were observed during early development and adulthood. The expression of TigarB, the zebrafish ortholog of the human glycolysis and apoptosis regulator TIGAR, was markedly increased in the pink1 null mutant. The antisense-mediated inactivation of TigarB normalized the mitochondrial function. Pink1 mutant larvae also displayed a marked increase in microglial activation, but the inactivation of the microglia failed to rescue the DA neuron loss, suggesting that microglial activation may be a downstream mechanism involved in DA neuron loss in Pink1 deficiency [81].
3.4. Medaka Fish (Oryzias latipes)
A single ortholog of the human PINK1 gene was identified in the medaka genome: the medaka pink1 gene has 8 exons and encodes a protein consisting of 577 amino acids which has 54.1% homology to human PINK1. The kinase domain is highly conserved between the two species.
Several genetic models were created using the targeting-induced local lesions in genomes (TILLING) method [82]. Matsui and coauthors generated a pink1 mutant medaka fish by screening the TILLING library. Among the 14 mutations found in exons 2 and 3 of the pink1 gene, the nonsense mutation Q178X, which resulted in the disruption of the kinase domain and the degradation of pink1 mRNA, was selected to generate homozygous mutants (pink1Q178X/Q178X) [82]. The pink1 mutant grew normally at first, without any obvious morphological abnormalities or developmental disorders; they showed a normal phenotype for germ-cell lineage, skeletal muscle and mitochondrial morphology. Then, in the late-adult stage, the frequency of their spontaneous swimming movements dramatically decreased and death occurred at 12 months, in a lifespan shorter than that of the control fishes. Although the loss of Pink1 caused the dysregulation of the DA metabolism with a decrease in the amount of 3,4-dihydroxyphenylacetic acid (DOPAC), the mutants showed no evident defect in the number or morphology of their DA neurons [82].
3.5. Drosophila melanogaster
The Drosophila pink1 gene (CG4523) encodes a polypeptide of 721 amino acids (about 80 kDa in molecular mass). Similar to human Pink1, a structural analysis of the Drosophila Pink1 protein revealed an MTS and a serine/threonine kinase domain that shares 60% similarity and 42% amino acid identity with human PINK1. Consistent with the localization of human PINK1, Drosophila Pink1 was also found to localize in mitochondria [83].
Loss-of-function Pink1 mutants exhibited male sterility and mitochondrial defects (e.g., enlarged swollen mitochondria with the loss of the outer membrane and reduced ATP levels). The phenotype was restored by Pink1 expression. Furthermore, mitochondrial dysfunction led to the degeneration of flight muscles and the mild loss of DA neurons [83]. Pink1 mutant flies shared marked phenotypic similarities with Parkin mutants: abnormally positioned wings, a crushed thorax, disorganized muscle fibers with enlarged mitochondria, muscle cell apoptosis, impaired flight ability, a severely reduced climbing rate and complete male sterility [39,40,41,43]. Clark and coauthors observed a similar phenotype after the removal of pink1 in Drosophila: male sterility, apoptotic muscle degeneration, defects in the mitochondrial morphology (fragmented mitochondrial cristae) and a reduction in ATP, but no change in the number of DA neurons. Pink1 mutants showed increased sensitivity to multiple stresses, including oxidative stress. These phenotypes were fully suppressed by pink1 expression [84]. Morais and coauthors, using previously characterized pink1 mutant flies [83,84], observed that Drosophila pink1 mutant flies fail to maintain normal synaptic transmission and show the impaired mobilization of synaptic vesicles from the reserve pool during rapid stimulation [85]. This was caused by synaptic ATP depletion. The expression of human PINK1, but not its clinical mutant G309D, rescued the mobilization of reserve pool vesicles. The absence of Pink1 caused deficits in the mitochondrial membrane potential and defects in the catalytic activity of ETC complex I, but no alteration in mitochondrial morphology [85].
Another study reported that the Drosophila pink1 KO model showed a severe reduction in mitochondrial respiration driven by the ETC in the mitochondria, resulting from a decrease in ETC complex I and IV enzymatic activity [86]; in addition, the assembly of ETC complex I was compromised, as previously reported [85]. As a consequence, the pink1-KO flies also displayed reduced mitochondrial ATP synthesis [86]. It was previously observed that DRP1, a key molecule in mitochondrial fission, can rescue mitochondrial fission and morphological abnormalities in pink1-KO flies [94,95]. Liu et al. observed that DRP1 expression restored the ATP synthesis rate, rescued complex I and IV activity deficits, and increased the amount of assembled ETC complex I in pink1-KO flies [86]. Yang and coauthors used the transgenic RNAi approach to knock down the expression of pink1 in Drosophila [87]. The inhibition of Pink1 function in Drosophila severely reduced their lifespan. Pink1 RNAi flies also showed defects in their wing posture, reduced climbing ability, and abolished flight capacity by 10 days of age. Their muscle pathology was characterized by the severe disruption of muscle integrity, followed by the degeneration of selected indirect flight muscles with extensive DNA fragmentation, preceded by mitochondrial dysfunction. The ATP level was reduced, the mitochondria were grossly swollen, lacked electron-dense material, and showed the disintegration of the cristae. The inactivation of Pink1 led to a degeneration of TH-positive neurons and a reduction in brain DA content [87].
3.6. Caenorhabditis elegans
The C. elegans ortholog of human PINK1 was identified: it was named pink-1 and, like human PINK1, it consists of two characteristic motifs: an N-terminal MTS and a serine/threonine kinase domain that shares 36% identity and 54% similarity to human PINK1 [88]. Sämann et al. characterized a deletion mutant of pink-1. The pink-1(tm1779) mutants harbor a 350-bp deletion that eliminates part of the promoter region and the first two exons, including the proposed transcriptional and translational start sites, resulting in complete functional loss [88]. The authors demonstrated that the loss of C. elegans pink-1 resulted in increased paraquat sensitivity and decreased oxidative stress response. This dysfunction was accompanied by mitochondrial phenotypes characterized by a reduction in the mitochondrial cristae length (12% in muscle cells and more than 30% in neurons). Furthermore, the pink-1(tm1779) mutants displayed defects in the axonal outgrowth of the canal-associated (CAN) neurons, a pair of migratory neurons in the midbody region of the worm that are frequently used as an indicator of axonal guidance and cell migration defects [88].
In 2015, Luz et al. analyzed the mitochondrial morphology and the fundamental parameters of the mitochondrial respiratory chain in the C. elegans pink-1(tm1779) model [89]. No differences were observed in the basal oxygen consumption rate (OCR), but treatment with N,N′-dicyclohexylcarbodiimide (DCCD) caused a major reduction in OCR coupled to ATP consumption. Furthermore, exposure to the mitochondrial uncoupler carbonyl cyanide-p-trifluoromethoxyphenylhydrazone (FCCP) caused a marked increase in OCR above basal levels, while no differences were observed in the maximal or spare respiratory capacity. Pink-1-deficient nematodes showed severe proton leakage and a highly fused mitochondrial network that exhibited larger aspect ratios compared to the WT nematodes, which is consistent with the loss of mitophagy and a reduction in the mitochondrial turnover [89]. In line with these findings, elevated mitochondrial ROS generation, decreased ATP levels and decreased mitochondrial membrane potential, suggestive of proton leak, were recently reported in the same model [90]. The authors also found increased basal OCR and a fragmented and disorganized mitochondrial network morphology. They observed that in pink-1-deficient nematodes, mitophagy is not activated under stress, which leads to the accumulation of dysfunctional and damaged mitochondria and a shortened lifespan [90]. In agreement with these findings, pink-1 silencing abolished the detectable removal of ultraviolet C (UVC)-induced damaged mitochondrial DNA (mtDNA) due to a lack of mitophagy [91,92].
4. PARK7: The Parkinsonism-Associated Deglycase Gene (DJ-1)
The PARK7 or DJ-1 gene (OMIM 606324), localized on chromosome 1p36.23, was discovered to cause AR EOPD in 2001 [96]. DJ-1 mutations are quite rare: the frequency is 0% to 1% in early-onset PD cohorts [97]. Molecular analyses for DJ-1 mutations have identified homozygous and compound heterozygous mutations, including missense, truncating and splice-site mutations, and large deletions [4,98,99]. The DJ-1 protein is a redox-sensitive molecular chaperone protein, ubiquitously and highly expressed in brain areas and extra-cerebral tissues, localized in the cell cytosol, nucleus and mitochondria [100,101,102]. Some DJ-1 mutations cause a loss-of-function of the DJ-1 protein, thus inducing the instability of the DJ-1 dimer, while other mutations induce a lack of DJ-1 expression [103,104]. Studies have shown that DJ-1 mutations can generate ROS, leading to oxidative stress, mitochondrial dysfunction and cell death [4,105], but the precise mechanisms by which DJ-1 deficiency leads to PD remain elusive [4,105] (Table 3).

Table 3.
Observed phenotypes in animal models of PARK7-linked Parkinson’s disease.
4.1. Mouse Model
The first Dj-1 KO mouse model was generated by replacing exons 3–5 with a neomycin (Neo) selectable cassette. These Dj-1 null mice were viable and fertile, and did not display any gross neuronal abnormalities or motor deficits observed on the pole, the open field or the adhesive removal test [106]. A stereological analysis of their SNc DA neurons showed no alterations in the number of TH-positive neurons; striatal immunostaining showed no change in the TH fiber density or DAT level. The quantification of the DA by High Performance Liquid Chromatography (HPLC) revealed no change in the striatal region [106]. However, the mice were more susceptible to MPTP-induced neuron loss, and the restoration of DJ-1 expression via adenoviral vector delivery mitigated this phenotype. The authors also showed that the overexpression of DJ-1 via an adenoviral vector in WT mice prevented MPTP-induced neuronal loss and protected the animals against neurodegeneration in the SNc [106]. Using the same mouse model, Zhou and coauthors reported that Dj-1 null mice cannot perform the running wheel or the rotarod test at the same intensity as WT animals [107]. A second mouse model was generated by Chen and coauthors in 2005 by deleting 9.3-kb genomic DNA, including the first five exons and part of the promoter region of the Dj-1 gene. The mice were healthy and fertile, and appeared indistinguishable from the WT at birth. No motor deficits were detected at any age on the rotarod test, whereas the open field test revealed impairment at 11 months of age, and the tape removal task disclosed nigrostriatal dysfunction at 5 and 11 months [103]. Voltammetry studies of these Dj-1 null mice showed an increase in the evoked DA release in the dorsal striatum but not in the nucleus accumbens core and shell, and the HPLC dosage in the striatum displayed an age-dependent increase in the DA content [103]. Immunohistochemical analyses showed no differences in the TH, DAT or VMAT2 protein levels, no death of DA neurons, and no α-syn- or ubiquitin-positive inclusions were found in the SNc [103].
Another Dj-1 KO mouse was generated by Goldberg et al. in 2005 by targeting Dj-1 exon 2 [99]. Similarly to the other Dj-1 mouse models, this one did not display DA neuron loss or a deficit in the DA levels in the basal ganglia. The mice had normal TH activity, as measured by L-DOPA quantification determined by HPLC in the presence of an inhibitor of L-DOPA decarboxylase, and they did not have α-syn- or ubiquitin-positive inclusions [99]. Unlike the other models, this model showed a reduction in the stimulated DA release upon amperometry analysis, suggesting altered dopaminergic neurotransmission. The electrophysiological analysis of the glutamatergic transmission showed that LTP induction was normal but LTD was absent. Furthermore, the mice exhibited defects in their locomotor activity in the open field, rotarod and startle tests at 3 months of age [99]. Later, in 2007, Manning-Boğ and colleagues generated another Dj-1 KO mouse model that targeted the intron between exons 6 and 7. They reported that these Dj-1 KO mice showed no change in the number of TH-positive and Nissl-stained nigral cells compared to the WT mice, and no change in the levels of DA and its metabolites, DOPAC or homovanillic acid (HVA). Moreover, no behavioral impairment was observed. However, the mice showed alterations in their striatal DA transmission: a marked increase in striatal DAT was detected in synaptosomal fractions from null Dj-1 mice, and this increase in cell surface DAT was confirmed by autoradiographic studies [108].
Altogether, studies on Dj-1 KO mice converge in concluding that Dj-1 KO mice do not show the hallmark of PD found in DJ-1 mutated patients, i.e., the loss of SNc DA neurons and the loss of nigrostriatal fibers. The underlying reason could be genetic compensation, a frequent phenomenon in the KO mouse model [48]. However, because subtle dysfunction features were observed, these models might be useful to study early dysfunction induced by the loss of DJ-1 function.
4.2. Rat Model
Under the sponsorship of The Michael J. Fox Foundation for Parkinson’s Research, which supports the generation, characterization and distribution of rat genetic models to accelerate PD research [122], Dj-1 KO rats were generated using ZFN Technology [34,74]. The Dj-1 KO rats showed a 50% loss of SNc DA neurons at 8 months of age [34,109]. The Dj-1 KO rats also showed a marked reduction in TH-positive neurons in the LC at 8 months of age [110]. The TH immunoreactivity in the striatum was unchanged [34]. In the striatum, ultra-high performance spectroscopy showed an increase in the DA and 5-HT content at 8 months [34]. The quantitative radiography analysis of the striatum showed no change in the DAT density, but an increase in VMAT2, D1, D2, and D3 DA receptors between 4 and 8 months of age [74,109]. Dj-1 KO rats also showed an increase in bodyweight [110,111]. Behavioral tests showed motor impairment in their movement, strength and coordination between 6 and 8 months of age [34]. The cylinder test revealed that Dj-1 KO rats took more forelimb and hindlimb steps than their WT counterparts starting from 4 to 13 months of age, whereas the adhesive removal test disclosed no differences in their sensorimotor function [111]. Dj-1 KO rats showed no anxiety or depression at 4, 8 or 17 months of age when tested with an elevated-plus maze or with the sucrose preference test [111]. A decrease in sucrose preference is an indication of anhedonia, which is a core feature of depression. The Dj-1 KO rats showed no signs of anhedonia, and indeed they drank more sucrose with respect to the WT, with and without stress playing a factor, which probably indicates some abnormality in the neuroendocrine system [111]. In conclusion, while Dj-1 KO mice do not show nigral degeneration [99,103,106], Dj-1 KO rats exhibit progressive nigral neurodegeneration and motor impairments. This difference is in line with findings from rat models based on the genetic modification of α-syn. The overexpression of mutated Snca induces severe DA loss in the rat, whereas a similar effect has not been reported after the overexpression of the same gene in mice [123]. The underlying reasons are unclear; a plausible explanation is that these differences stem from the physiological, anatomical, biochemical and pharmacological differences between rats and mice [124].
4.3. Zebrafish (Danio rerio)
Zebrafish dj-1 is evolutionarily conserved: it is 83% identical and 89% similar to human DJ-1, and is widely expressed in neurons [125]. In adult zebrafish brains, Dj-1 is present in the DA neurons of the olfactory bulbs, the telencephalon and each of the DA nuclei of the diencephalic groups [125]. In addition, Dj-1 is expressed in the LC and other hindbrain catecholaminergic neuronal groups [125]. Bretaud and colleagues, in 2007, generated KO zebrafish dj-1 using the microinjection of antisense MO oligonucleotides into zebrafish embryos [112]. The dj-1 KO zebrafish embryos showed no loss of DA neurons in the basal condition, but a pronounced reduction in the number of DA neurons was evident after exposure to H2O2 or proteasome inhibitor MG132. Dj-1 KO had increased levels of p53 and the proapoptotic protein Bax, and MG132 exposure further enhanced the p53 and Bax expression in dj-1 KO zebrafish embryos [112]. Later, in 2009, Baulac and colleagues generated another dj-1 KO zebrafish model with MO oligonucleotides directed against the 5′ untranslated region of dj-1. This model was similar to Bretaud’s model and it showed no change in the DA neurons of embryos, but it did show greater sensitivity to exposure to H2O2, with an increase in the number of apoptotic cells, as observed by TUNEL (terminal deoxynucleotidyl transferase dUTP nick end labeling) staining [113].
In 2019, Edson and colleagues used the CRISPR-Cas9 method to generate a dj-1 KO zebrafish model. This dj-1 KO zebrafish model underwent normal development until the adult stages, was viable and fertile, but beyond age 3 months the fishes tended toward a smaller body size and lower mass [114]. With age (16 months), there was a severe reduction in the TH levels and DA content in the brain, and in the complex I activity in the skeletal muscle. A proteomic analysis of the early adult brains revealed that about 5% of the 4091 identified proteins were influenced by the lack of Dj-1. The dysregulated proteins were mainly those involved in mitochondrial metabolism, mitophagy, stress response, redox regulation and inflammation [114].
Hughes and colleagues, in 2020, used an unbiased computational method to record the movement of dj-1 KO fishes and found that the dj-1 KO zebrafish displays an overall reduction in movement resembling bradykinesia: a reduction in the distance travelled, velocity, time spent moving and duration of a swimming episode. In order to investigate the changes in gene expression associated with Dj-1 loss, an RNA-seq on adult zebrafish brains disclosed many metabolic changes [115]. Overall, these results suggest that the zebrafish is a useful model for studying the functions of Dj-1 in vivo and the consequences of dj-1 mutations.
4.4. Drosophila melanogaster
Drosophila melanogaster has two genes—dj-1α (CG6646) and dj-1β (CG1349)—that share substantial homology with human DJ-1 (dj-1α shares 56% identity and 70% similarity with the human protein; dj-1β shares 52% identity and 69% similarity) [116,119]. In adult flies, dj-1β is expressed at similar levels in male and female heads, brains and bodies, while dj-1α is expressed at detectable levels only in males, in the testes in particular, and at low levels in the adult head [116,119]. In order to address the biological function of dj-1 in PD with the Drosophila model, studies were conducted using dj-1β KO or double-KO (DKO) [116,117,119]. Drosophila dj-1β KO demonstrated the extended survival of DA neurons and resistance to oxidative stress induced by paraquat [116,117]; however, it showed increased sensitivity to H2O2-induced toxicity [116,117]. In addition, dj-1β KO displayed reduced taste sensitivity under normal food conditions with behavioral feeding assays, defects in the ability to form associative memories with taste memory assays [118], and decreased climbing ability [117]. Although dj-1β KO showed resistance to exposure to paraquat stress, a further loss of climbing activity occurred after repeated paraquat exposure [117]. The DKO lines were viable and fertile, and they displayed a normal lifespan. Though the model did not show differences in the number of DA neurons, it displayed increased sensitivity to chemical agents like H2O2 and paraquat, and much higher sensitivity to rotenone [119]. Hence, dj-1 mutant flies can be considered an interesting model to study motor impairment in DJ1-associated PD.
4.5. Caenorhabditis elegans
C. elegans possesses two distinct dj-1 paralogs: djr-1.1 and djr-1.2 [120]. Ved and colleagues, in 2005, generated a KD of the djr-1.1 gene [45] and found that C. elegans KDs for the djr-1.1 gene were far more sensitive to rotenone exposure than the control nematodes. Therapies that stimulated complex II function and inhibited apoptosis (D-β-hydroxybutyrate and tauroursodeoxycholic acid) provided protection against complex I inhibition [45]. Another group generated double mutants for the djr-1.1 and djr-1.2 genes. They exposed C. elegans to Pseudomonas aeruginosa and investigated their innate immunity. They found that the loss of djr-1.1 and the djr-1.2 function stimulated the phosphorylation of p38MAPK/PMK-1, a key step in inflammatory signaling. The findings underscored the importance of DJ-1 for the regulation of innate immune responses, and they suggested that DJ-1 loss-of-function may promote neuroinflammation in PD [120].
Another research group generated worms with djr deletions: djr-1.1 or djr-1.2 alone, and djr double deletion [121]. The researchers showed that the deletion of djr-1.2 alone or djr-1.1 and djr-1.2 together led to a marked increase in sensitivity to Mn and decreased survival and lifespan after acute Mn exposure. Djr-1.2 overexpression was sufficient to restore protection against Mn-induced lethality in djr double deletion mutants [121]. In addition, the overexpression of DAF-16, the C. elegans ortholog of human FOXO, which functions as a transcription factor for antioxidant response, fully reversed the lifespan shortening caused by djr-1.2 deletion and Mn exposure [121]. In order to investigate the role of DJR-1.2 in DA signaling, the researchers employed an assay to detect spontaneous movement in the dauer state, and they observed that djr-1.2 deletion alone greatly increased dauer movement; this phenomenon is compatible with reduced dopaminergic signaling. Furthermore, djr-1.2 deletion combined with Mn exposure increased the dauer movement, suggesting that DA signaling is further impaired by Mn exposure [121].
5. PARK9: The ATPase 13A2 Gene (ATP13A2)
A mutation of the ATP13A2 gene (OMIM 606693), located on the 1p36.13 locus and first described in 1994, was associated with a rare hereditary form of autosomal recessive juvenile onset Parkinsonism in the Kufor-Rakeb community in Jordan [126]. The ATP13A2 gene encodes a transmembrane endo/lysosomal-associated protein belonging to the P5 type transport ATPase subfamily [127,128,129]. ATP13A2 is ubiquitously expressed, with the strongest expression in the brain and in the SNc [129]. Studies have shown that ATP13A2 is involved in autophagy and the lysosomal pathways [130,131]. It plays a role in the metabolism of Mn2+ and Zn2+ [127,132,133], and in mitochondrial pathways [134]. ATP13A2 can also regulate the metabolism of α-syn, one of the major components of Lewy bodies [135] (Table 4).

Table 4.
Observed phenotype in animal models of PARK9-linked Parkinson’s disease (ATP13A2).
5.1. Mouse Model
Atp13a2 KO mice were created to study the role of ATP13A2 in PD. Atp13a2 KO mice grew normally and were fertile. The sensorimotor function in this model was measured using tests sensitive to various types of motor dysfunction, including challenging beam traversal, spontaneous activity, gait, and nest building. The sensorimotor function did not differ between the genotypes at 5–12 months of age; however, at 20–29 months the Atp13a2 KO mice displayed severe impairment on all of the motor tests [136]. Histopathology showed no change in the number of DA neurons in the SNc or in the striatal DA levels in aged Atp13a2 KO mice [136,137]. Nonetheless, the model disclosed many progressive neuropathological changes: the early onset of gliosis at 1 month of age, lipofuscinosis at 3 months, the accumulation of lysosome-associated membrane glycoprotein (LAMP) 1 and 2, and lysosomal lipid bis(monoacylglycero)phosphate (BMP) at 6 months [131,135,136,137]. At 12 months of age, the Atp13a2 KO mice showed the aggregation of ubiquitinated proteins and p62 and aberrant processing of the lysosomal protease cathepsin D (CATD) in many brain regions [137]. The Atp13a2 KO mice also displayed the accumulation of insoluble α-syn in the hippocampus at 18-20 months, but no change in the α-syn protein solubility in either the cortex or the cerebellum [136]. The Atp13a2 KO mice did not present a substantial impairment of their spontaneous activity between 5 and 15 months of age, while differences emerged on the challenging beam traversal and the gait test between 20 and 29 months of age [136].
5.2. Zebrafish (Danio rerio)
The zebrafish has two proteins with high homology with human ATP13A: zgc:136762 and zgc:63781, with 1170 and 1177 amino acids, respectively; zgc:136762 is closely related to human ATP13A2, with 50% homology and 69% similarity, while zgc:63781 is phylogenetically closer to human ATP13A1, sharing 73% identity and 83% similarity. The human ATP13A2 has two functional domains, an E1–E2 ATPase domain and a haloacid dehalogenase (HAD-like) domain, which are important for the biological role of the protein. Both domains are present in the zebrafish zgc:136762 sequence. The Atp13a2 protein is widely expressed throughout the body of the embryo; during embryonic development, its expression becomes more restricted to the brain area [138]. In order to obtain zebrafish atp13a2 KO, MOs were used to inhibit the correct splicing of atp13a2 mRNA: the complete abrogation of Atp13a2 led to embryonic lethality, while the partial abrogation of atp13a2 allowed the offspring to survive. Partial gene KD embryos showed a postural defect, i.e., a curved phenotype at 48 h post-fertilization (hpf). In addition, the KD of atp13a2 in embryos at 7 dpf caused a distinct movement pattern characterized by general movement latency and an abnormal response to environmental stimuli, as observed by the swimming pattern/locomotor activity in the light on–light off paradigm [138].
Heins-Marroquin and coauthors used zebrafish lines carrying atp13a2 mutant alleles (atp13a2sa14250 or atp13a2sa18624) generated by ENU mutagenesis. Each allele carries a single non-sense mutation (2153 T > A or 2457 T > G) resulting in a premature stop codon in exon 20 or exon 22, respectively. Both mutations were predicted to cause loss-of-function. Homozygous atp13a2sa18624 mutants were able to reach adulthood without any obvious morphological or behavioral abnormalities [139]. As Atp13a2 seems to play an important role in heavy metal homeostasis, the authors tested the effect of Mn on atp13a2sa18624+/+, atp13a2sa18624−/+ and atp13a2sa18624−/− larvae. Exposure to MnCl2 was found to cause neurodegeneration in zebrafish [144]. At 5 dpf, in the presence of Mn2+, WT and heterozygous larvae showed a moderate phenotype characterized mainly by an underdeveloped swimming bladder. In contrast, the homozygous atp13a2 mutants were highly sensitive to Mn2+ and displayed multiple abnormalities in addition to swimming bladder underdevelopment, including pericardial edema, movement loss and spinal curvature. Similar results were obtained in the second mutant line (atp13a2sa14250) upon exposure to Mn2+. Furthermore, compared to WT, 64% of the atp13a2sa18624−/− larvae showed large apoptotic areas throughout the CNS (brain and dorsal spine) after exposure to Mn2+ [139].
Recently, another atp13a2 deficient zebrafish was generated by introducing a 10 bp deletion in exon 2 of atp13a2 using CRISPR/Cas9 gene editing [140]. This atp13a2 KO zebrafish model showed, at 4 and 12 months of age, a severe loss of DA neurons in the posterior tuberculum (64% at 4 months and 37% at 12 months) and norepinephrine neurons in the LC (52% at 4 months and 40% at 12 months), as observed by the immunostaining of TH-positive neurons [140]. Furthermore, the Western blotting, electron microscopy and liquid-chromatography tandem mass spectrometry (LC-MS/MS) of the brain tissues revealed that the atp13a2 KO zebrafish had CATD deficiency, lysosomal dysfunction and intracellular vesicle trafficking dysfunction, which are all relevant to PD [140].
5.3. Medaka Fish (Oryzias latipes)
The medaka fish has also been used to study Atp13a2. By screening the TILLING library, Matsui and colleagues found a mutation, “IVS13, T-C, +2”, [141]. The mutation resulted in an abnormal splicing pattern, which was almost identical to that observed in human PARK9 patients in which 111-bp exon 13 has been skipped, causing the delocalization of the Atp13a2 protein in the endoplasmic reticulum rather than in the lysosome [10,141]. Atp13a2 “IVS13, T-C, +2” mutant medaka had a shorter lifespan and exhibited more spontaneous swimming movement at 4 months but no differences in swimming at 12 months. They showed a reduction in the number of TH-positive neurons in the middle diencephalon and in the TH-positive fiber density in the telencephalon at 8 and 12 months. Furthermore, a reduction in their DA content was observed at 12 months. At 8 and 12 months, there were fewer noradrenergic neurons in the medulla oblongata [141]. Mutant medaka disclosed decreased CATD protein levels and activity, and fingerprint-like subcellular structures in the brain, indicating an autophagy/lysosome disorder [141].
5.4. Caenorhabditis elegans
The ortholog of the human ATP13A2 gene in C. elegans is the W08D2.5/catp-6 gene. The catp-6(ok3473) mutant bears a deletion of 900 bp that results in a substantial loss of catp-6 mRNA [142]. C. elegans catp-6 mutants showed decreased locomotion, a severe delay in the rate of development [142] and higher mortality in midlife [143]. Furthermore, catp-6 mutants displayed an alteration in their iron homeostasis caused by the major down-regulation of the core genes required for metabolizing iron [142]. Young (L4 larvae to day 2 adults) and middle-aged (day 4 to day 5 adults) catp-6 mutants also displayed altered Zn homeostasis [143].
In order to assess the autophagy and lysosomal function in these mutants, Anand and coauthors quantified the protein levels of autophagosome-(cleaved LGG-1/LC3-II) and lysosome-specific aspartyl protease ASP-3/CATD, respectively. The protein level of cleaved LGG-1/LC3-II was severely reduced in the catp-6 mutants, suggesting a defect in their early autophagosome formation. The lysosomal function was assessed by monitoring the levels of the premature (P-CATD) and the mature (M-CATD) form of the pH-dependent lysosomal enzyme aspartyl protease ASP-3/CATD: the P-CATD levels were far higher in the catp-6 mutants than in the WT worms, suggesting the reduced conversion of P-CATD to M-CATD. The Catp-6 mutant worms also showed the low mRNA expression of several key genes required for autophagy and lysosomal function [142]. Altogether, these results show that the loss of catp-6 affected autophagy and lysosomal function. Furthermore, the catp-6 mutants presented multiple defects in their mitochondrial function, including a reduced mitochondrial membrane potential and maximal respiration rate, as well as increased sensitivity to rotenone, which could be rescued by treatment with iron chelators [142].
6. PARK14: The Phospholipase A2 Group VI Gene (PLA2G6)
The phospholipase A2 group VI (PLA2G6) gene (OMIM 612953) is located on chromosome 22 (22q13.1); it encodes a calcium-independent phospholipase A2 enzyme (PLA2G6) which is widely expressed in the brain, particularly in dendrites and axon terminals [145]. Mainly involved in the homeostatic processes of cell membranes, the protein selectively hydrolyzes glycerophospholipids to release free fatty acids [146]. Recessive mutations in PLA2G6 were found to induce alterations in phospholipid metabolism and abnormal iron accumulation, which ultimately led to infantile neuroaxonal dystrophy (INAD), atypical neuroaxonal dystrophy (ANAD) and neurodegeneration with brain iron accumulation (NBIA). These diseases cause neurodegeneration in early life [147]. PLA2G6 mutations (e.g., D331Y, R365Q, R714Q, R747W) were also found to be associated with familial forms of PD (PARK14) characterized by AR inheritance and early-onset dystonia-Parkinsonism [148]. Among these mutations, the compound heterozygous mutations p.F72L/p.R635Q and p.Q452X/p.R635, and the homozygous mutation D331Y are associated with AR EOPD [149,150]. Other less common mutations associated with PARK14 phenotypes are R37X, F72L, A80T, M358I, Q452X, A499T, T572I, R632W, N659S, D739H and P806R [151]. Generally, PARK14 patients show psychomotor deterioration, cerebellar ataxia and dystonia, as well as tremors, rigidity, bradykinesia and autonomic dysfunction [151]. Immunohistochemical studies have shown the presence of Lewy bodies, neurofibrillary tangles and neuropil threads, and the loss of DA neurons in the SNc, along with the accumulation of phosphorylated α-syn in the nerve cell bodies and processes of the gastrointestinal tract and cardiac sympathetic nerves [151] (Table 5).

Table 5.
Observed phenotypes in animal models of PARK14-linked Parkinson’s disease (PLA2G6).
6.1. Mouse Model
In 2008, Shinzawa and colleagues generated and characterized Pla2g6 KO mice [152]. In their first year of life, the animals showed no developmental deficits, but their fertility was reduced (especially among males). By the age of 2 years, motor dysfunctions (especially the hindlimb clasping reflex during tail suspension), abnormal gait and poor performance in the hanging wire grip test were observed [152]. The Pla2g6 KO animals showed the degeneration of nigrostriatal DA neurons and the focal loss of TH and DAT nerve terminals in the striatum (from 56 weeks of age) [152]. The Pla2g6 KO animals also showed prominent axonal degeneration and the widespread presence of spheroids and vacuoles throughout the CNS and the peripheral nervous system (PNS) [153]. In Pla2g6 KO mice, Sumi-Akamaru and colleagues observed atrophic axons accompanied by mild neuronal loss in the late stages of the disease [154]. They reported the strong expression of α-syn in the mitochondrial outer membrane protein (TOM20)-positive neuronal granules before the onset of motor dysfunction [154].
In 2010, Basselin and coworkers investigated whether the brain metabolism of docosahexaenoic acid (DHA), the substrate of the PLA2G6 enzyme, was reduced in the early stages of the disease. To this aim, 4-month-old Pla2g6−/−, Pla2g6+/− and Pla2g6+/+ mice were administered radiolabeled unesterified [1-14C] DHA, and its metabolism (incorporation coefficients and rates) was determined by quantitative autoradiography. Pla2g6−/− and Pla2g6+/− mice showed a severe baseline reduction in DHA metabolism compared to the Pla2g6+/+ mice, including 4-month-old mice which lacked histopathological or neurological alterations [155].
Zhou and colleagues developed a novel transgenic mouse model with the constitutive deletion of exon 2 of the Pla2g6 gene (Pla2g6 ex2KO) resulting in the expression of a truncated ex2KO Pla2g6 protein which lacks the first 178 amino acids in the N-terminus [156]. The genetic truncation of the N-terminus did not affect the protein catalytic activity [156]. Mouse embryonic fibroblasts (MEFs) from Pla2g6 ex2KO mice exhibited functional impairment in stored–operated Pla2g6-dependent Ca2+ signaling, with the substantial depletion of intracellular Ca2+ stores. Pla2g6 ex2KO mice developed progressive age-dependent motor dysfunction starting from 16 months of age. The mice displayed impairment in motor coordination, e.g., a progressive age-dependent increase in the number of missteps on the balance beam test; the results from the pole and the rotarod test further confirmed severe PD-like motor dysfunction in these animals. Interestingly, the Pla2g6 ex2KO mice were responsive to L-DOPA: the administration of L-DOPA markedly improved their motor coordination in an age- and dose-dependent manner. The analysis of the SNc area of the brain from aged Pla2g6 ex2KO animals revealed a progressive age-dependent loss of SNc DA neurons: no changes were observed at 8 months of age, whereas over 30% of the DA neurons in the SNc of ex2KO mice was lost by 16 months, and over 50% by 24 months of age. In addition, the DA neurons in the SNc of PLA2g6 ex2KO showed marked autophagy dysfunction, i.e., the accumulation of LC3 and an increased autophagosome number [156].
In 2019, Chiu and colleagues generated a KI mouse model harboring the homozygous human PLA2G6 D331Y (GAC→TAC) mutation, which associates with Parkinsonism in humans (Pla2g6D331Y/D331Y). The aim was to study the neurological and neuropathological changes associated with this mutation, as well as the molecular mechanisms leading to DA neuron degeneration [157]. The model showed early-onset neuronal death in the SNc, as observed by a reduction in the TH-positive neurons in the SNc at 6 and 9 months of age and a decreased density of striatal TH staining at 9 months, indicating the degeneration of nigrostriatal DA terminals. Lewy bodies were found in the SNc of 9-month-old Pla2g6D331Y/D331Y mice, along with the upregulated protein expression of α-syn and phosphorilated α-syn, while 6- to 12-month-old Pla2g6D331Y/D331Y mice exhibited a progressive PD phenotype characterized by slowness of movement, hypoactivity, and impaired motor coordination and performance. L-DOPA treatment reversed the hypoactivity. A homozygous D331Y Pla2g6 mutation led to abnormalities in the mitochondrial ultrastructure in the SNc, i.e., the disrupted structure of the mitochondrial cristae and smaller mitochondria in the Pla2g6D331Y/D331Y mice compared to the WT mice. The model revealed multiple mitochondrial dysfunctions in the SNc, including decreased mitochondrial complex I and III activity, a lower ATP level, increased ROS production and lipid peroxidation, and the upregulation of the cytosolic level of cytochrome c. In addition, homozygous D331Y Pla2g6 mutation activated the mitochondrial apoptotic pathway and induced endoplasmic reticular stress and mitophagy dysfunction. The study demonstrated that homozygous D331Y Pla2g6 mutation was able to cause DA neuron degeneration in the SNc and induce an early-onset PD phenotype in the mice [157].
6.2. Drosophila melanogaster
Drosophila melanogaster was used to study the pathological effects of pla2g6 mutations. iPLA2-VIA, the Drosophila homolog of PLA2G6, shows 51% identity and 67% positivity with the human gene [159]. Lin and colleagues generated a KO fly model of pla2g6 (iPLA2-VIA KO). While the lack of iPLA2-VIA did not modify the phospholipid composition of the brain, it increased the production of ceramides (thus affecting membrane fluidity), which led to impaired synaptic transmission and neurodegeneration, resulting in a shortened lifespan [146]. In 2019, Mori and colleagues reported impaired neurotransmission in the early developmental stages of iPLA2-VIA KO flies, which was associated with the progressive degeneration of DA neurons and the formation of α-syn aggregates [158].
7. PARK15: The F-Box Protein 7 Gene (FBXO7)
Homozygous or compound heterozygous mutations in the F-box only protein 7 (FBXO7) gene located on chromosome 22q12.3 are associated with Parkinsonian-pyramidal syndrome (PPS) and AR EOPD (PARK15, OMIM 260300) [160,161]. FBXO7-mutated patients show early-onset bradykinesia, tremors, rigidity, pyramidal tract signs and L-DOPA response [160,161,162,163]. FBXO7 encodes F-box protein 7 (FBXO7), which is a member of the F-box-containing protein (FBP) family. These proteins function as adaptors for the SKP1/cullin-1/F-box protein E3 ubiquitin ligase complex to recognize substrates and facilitate their ubiquitination [160]. FBXO7 is broadly expressed in human tissues, but is particularly enriched in the bone marrow, liver, kidney, testis and thyroid gland. FBXO7 is also expressed in several regions of the human brain: its expression is high in the cerebral cortex, globus pallidum, thalamus, striatum and substantia nigra, but low in the hippocampus and cerebellar cortex [164]. PD-associated mutations in FBXO7 can induce impairment in mitochondrial homeostasis by disrupting mitochondrial mitophagy. FBXO7 deficiency is also associated with an increased mitochondrial NADH redox index, the impaired activity of complex I in the ETC, reduced mitochondrial membrane potential and ATP contents, and increased cytosolic ROS production [165,166] (Table 6).

Table 6.
Observed phenotypes in animal models of PARK15-linked Parkinson’s disease (FBXO7).
7.1. Mouse Model
Vingill et al. generated a conventional FbxO7 KO mouse in which the fourth exon of the FbxO7 gene was deleted, resulting in a truncated protein [167]. At postnatal day (P) 5, there was no difference in the body or brain weight between the FbxO7 KO and the control littermates, while at P18 the FbxO7 KO mice displayed a substantially lower body and brain weight, and obvious motor deficits. The FbxO7 KO mice died a premature death starting from postnatal week 4 [167]. At P18, the FbxO7 KO mice displayed a moderate increase in cell death in the cortex, but no difference in TH-positive neurons in the SNc or change in the DA level and its metabolites in the striatum. Further neuropathological investigation of the FbxO7 KO brain revealed no a-syn protein deposits, though astrogliosis was observed in the cortex, because the area occupied by the glia fibrillary acidic protein (GFAP)-positive astrocytes was increased [167].
Because the systemic loss of FbxO7 caused premature death, perhaps due to a negative impact on the peripheral organs, the authors generated and characterized conditional FbxO7 KO mice. In detail, they first generated FbxO7-floxed animals (FbxO7fl/fl) carrying loxP-flanked exon 4, then homozygous conditional FbxO7fl/fl animals were bred to a NEX (neuronal helix-loop-helix protein-1)-Cre line [169] to delete Fbx07 from the pyramidal neurons of the cortex and the hippocampus (Nex-Cre;fl/fl mice) or B6.Cg-Tg(TH-cre) 1Tmd/J line [170] to delete the Fbx07 gene from the TH-expressing neurons (TH-Cre;fl/fl mice) [167]. The conditional Nex-Cre;fl/fl mice were viable and lived for at least 4 months. They showed a normal bodyweight at 2 months of age, after which their growth stagnated. At 2 and 4 months, they displayed spasticity and progressive motor coordination deficits, which resembled the pyramidal tract signs seen in PARK15-mutated patients. The mice also showed an increase in GFAP-positive cells and astrogliosis in the cortex [167]. TH-Cre;fl/fl mice lived longer than the previous mice, because most of them lived for at least 12 months. At age 2 months, the mice showed no motor deficit, but later (at 6 months) motor deficits began to appear on the rotarod test; by 12 months their overall health and motor performance had further worsened. The TH-Cre;fl/fl mice displayed slow movements, reduced mobility and alterations in many fine gait parameters; however, unlike the Nex-Cre;fl/fl mice, they retained their coordination ability. The deletion of Fbxo7 in the catecholaminergic neurons caused symptoms reminiscent of the bradykinesia and rigidity seen in PARK15 patients. The histological analysis of the brain revealed no changes in the number of DA neurons in either the 2-month-old or the 12-month-old TH-Cre;fl/fl mice, though there was a 50% reduction in the DA content in the striatum at 2 and 12 months of age. Neuropathological examination revealed an increase in astrogliosis in the SNc [167].
In conclusion, although the systemic and specific neuronal loss of the Fbxo7 gene in mice does not cause DA neuron degeneration, severe motor deficits reminiscent of the symptoms seen in PARK15 patients were observed in the conventional and the conditional FbxO7 KO mice. These models are therefore useful to study early- and late-onset motor impairments characterizing the PPS associated with mutations in the FBXO7 gene.
7.2. Zebrafish (Danio rerio)
In the zebrafish genome, a single homolog to human FBXO7, termed zFbxo7, is annotated. The zFbxo7 protein is expressed throughout embryonic development until adulthood; it is abundantly present in the brain and the liver, but scarce in the heart and the kidney. It is ubiquitously expressed in the brain: high levels are found in the neurons of the olfactory bulb and the diencephalon, intermediate levels in the cerebellum and the medulla oblongata, and low levels in the optic tectum and the habenula [168]. The first vertebrate animal model of PARK15 was generated by Zhao and colleagues in 2012 by the MO-mediated KD of zFbxo7; it is the only zebrafish model created to date [168]. These researchers generated two non-overlapping zFbxo7 MOs, one targeting the ATG translation initiation site (ATG-MO) and the other targeting the exon2/intron2 splice site (SP-MO) of zFbxo7, respectively. Both MOs were able to knock down zFbxo7 protein expression. A range of morphological phenotypes, spanning from mild (ATG-MO-Mild and SP-MO-Mild morphants) to severe (ATG-MO-Severe and SP-MO-Severe morphants), was observed at 72 hpf, including curly tails, heart edema and heart malformations; the severity of the phenotype was correlated with the silencing of the zFbxo7 protein. A significant reduction (40%) in the number of TH-positive neurons was seen in the SP-MO-severe morphants compared to the WT zebrafish, while the number of DAT-positive neurons was considerably reduced in the ATG-MO and the SP-MO morphants, and more dramatically in the morphants with a more severe phenotype (ATG-MO-Severe and SP-MO-Severe) [168]. Because the ATG-MO-Severe and SP-MO-Severe morphants showed scarcely any motor activity, the researchers focused the locomotor analysis on the ATG-MO-Mild and SP-MO-Mild phenotypes: the morphants displayed a markedly decreased swimming velocity, which was improved by exposure to the DA agonist apomorphine [168]. Because it reproduces the pathological (DA neuronal loss) and behavioral (locomotor disturbances) hallmarks of human Parkinsonism, the model provides a valid tool for investigating the mechanisms underlying selective DA neuronal death in AR EOPD.
7.3. Drosophila melanogaster
Zhou and colleagues generated a transgenic Drosophila model overexpressing human WT FBXO7 in the DA neurons [166]. The model had a normal lifespan, but by 30–40 days began to show locomotor deficits in its flight and climbing ability. At 40 days there was a loss of DA neurons, especially in the paired posterior medials 1 and 2 (PPM1/2) and paired posterior medial 3 (PPM3) DA neuron clusters [166]. The overexpression of the human WT FBXO7 protein also led to fbxo7 protein aggregation and a mitochondrial deficit, with the swelling of the muscle mitochondria, broken mitochondrial cristae, and the accumulation of high-density materials in the swollen mitochondria [166]. Thus, the results suggest that the overexpression of FBXO7 can be detrimental to DA neurons in Drosophila.
8. PARK19: The DnaJ Heat Shock Protein Family (Hsp40) Member C6 Gene (DNAJC6)
The DNAJC6 (DnaJ (Hsp40) homolog, subfamily C, member 6) gene (OMIM 615528) is located on chromosome 1p31.3 and encodes the brain-specific isoform of AUXILIN. AUXILINS have a well-established role in clathrin-mediated endocytosis (CME), which is responsible for the uptake of material into the cells through clathrin-coated vesicles [4,171]. Mutations of DNAJC6 are associated with AR EOPD [171,172,173]. Homozygous DNAJC6 c.801-2A > G mutation was discovered in 2012 in two patients of a consanguineous family with ARJP [172]. Other disease-associated DNAJC6 variants were discovered, including p.Q734X, p.R927G, c.2223A > T, p.L209P, p.M133L, p.R619C, p.F839Lfs*22 and c.2038 + 3A > G [171,173,174]. The AUXILIN-1 protein has a high homology with the cyclin G-associated kinase (GAK) protein. Though they share a multidomain structure, GAK has an additional N-terminal kinase domain, and it is ubiquitously expressed, whereas AUXILIN-1 is neuron-specific and enriched in the nerve terminals [175,176,177]. Both AUXILIN-1 and GAK are members of the J domain family of proteins, which are characterized by the canonical histidine-proline-aspartic acid motif that binds to HSC70 [175,176,177] (Table 7).

Table 7.
Observed phenotypes in animal models of PARK19-linked Parkinson’s disease (DNAJC6).
8.1. Mouse Model
Conventional Dnajc6 KO mice were generated in 2010 [177]. The rate of early postnatal mortality among the mutant mice was high, but the surviving pups had an apparently normal lifespan. The birth weight of the KO and heterozygous mice was lower than that of WT pups. Female Dnajc6 KO mice also showed delayed sexual maturity [177]. Because AUXILIN-1 and GAK are highly homologous proteins, Yim and colleagues studied the Dnajc6 KO mice to determine whether GAK was up-regulated and could compensate for the absence of AUXILIN-1. A Western blot analysis showed that Dnajc6 KO embryos and 3–5-week-old mice had higher GAK levels in the brain than WT mice. In contrast, the GAK levels in nonneuronal tissues (e.g., spleen, liver, kidney, testes) were not very different between the Dnajc6 KO and WT mice [177]. The study also showed a relationship between the brain GAK levels and bodyweight: the bodyweight was lower in the Dnajc6 KO pups with no up-regulation of GAK, whereas the Dnajc6 KO pups with the up-regulation of GAK showed no change in their bodyweight compared to the WT [177]. However, the analysis of the brain tissue of 1-week-old Dnajc6 KO mice showed no change in the endocytic synaptic proteins dynamin 1, synaptojanin 1, epsin, eps15, amphiphysin 1 or Hsc70 [177]. Because the study was performed before the identification of DNAJC6 as the gene associated with AR EOPD, it did not investigate whether the Dnajc6 KO mice displayed a loss of DA neurons or motor impairment.
8.2. Drosophila melanogaster
Two different RNAi lines in Drosophila were generated by silencing the GAK homolog auxilin (aux): auxR16182 and auxR103426 [178]. The downregulation of auxilin in Drosophila decreased the lifespan and caused an age-dependent reduction in the DA neuron number [178]. Drosophila auxR16182 and auxR103426 showed no obvious alteration in the mitochondrial morphology in the DA neurons, but did show increased sensitivity to oxidative stress upon paraquat exposure [178]. Furthermore, auxR16182 and auxR103426 displayed locomotor deficits in climbing ability [178]. The model therefore provides an interesting tool to study the consequences of auxilin/GAK silencing.
9. PARK20: The Synaptojanin 1 Gene (SYNJ1)
Mutations in the gene SYNJ1 (OMIM 615530) are associated with AR EOPD [4,179,180,181,182,183]. Patients with a mutation in SYNJ1 develop progressive tremors, dystonia, rigidity and cognitive decline [180]. The SYNJ1 gene is located on 21q22.2 and encodes the protein SYNAPTOJANIN-1, an important phosphoinositide phosphatase enriched in nerve terminals that regulates synaptic vesicle recycling [180,184] (Table 8).

Table 8.
Observed phenotype in animal models of PARK20-linked Parkinson’s disease (SYNJ1).
Mouse Model
Synj1 KO mice were generated in 1999. The KO mice were indistinguishable from WT mice at birth, but within a few hours they could be easily identified by the severely low milk content in their stomach: 85% died within 24 h and the remaining 15% died within 15 days. At 10 days of age, the Synj1 KO mice weighed less than half of the weight the WT mice, showed severe weakness and ataxia, and displayed generalized convulsions upon stimulation by nociceptive stimuli [185]. The Synj1 KO mice did not show abnormal levels of SYNAPTOJANIN-1 interactors (amphiphysin 1 and 2, Grb2, SH3P4, SH3P8, SH3P13), proteins involved in synaptic vesicle endocytosis (dynamin 1, clathrin, AP-2, AP180, Eps15, epsin1, auxilin, Hsc70), proteins implicated in the synaptic vesicle cycle (synapsin 1, rab3a, rab5, actin), and enzymes involved in phosphatidylinositol (PI) metabolism, i.e., PI(4)P 5-kinase type II and PI 3-kinase. Electron microscopy revealed structural changes in the synapses and many clathrin-coated vesicles in the cytomatrix-rich area surrounding the synaptic vesicle cluster. Electrophysiological experiments on the CA1 hippocampal area showed impaired synaptic vesicle recycling, with the depression of synaptic transmission after prolonged high-frequency stimulation and a corresponding delayed recovery after the interruption of the stimulus [185].
More recently, heterozygous Synj1+/− mice were investigated as a potential model of early-onset PD. The Synj1+/− mice had a normal bodyweight and lifespan [186]. At 7 and 12 months, they showed a reduction in DA, DOPAC, and HVA content in the striatum, as determined by HPLC analysis. Immunohistochemical analysis revealed a progressive reduction in DA terminals from 3 to 18 months of age, which reached 75% at 18 months [186]. Furthermore, immunofluorescence for phospho-Ser129 (pS129) α-syn showed an accumulation of pS129 α-syn in the cortex, the striatum and the midbrain in Synj1+/− mice at 18 months [186]. Synj1+/− mice also showed age-dependent motor impairment [186]. These findings suggest that Synj1 heterozygous deletion leads to PD-like pathologies in mice.
10. PARK23: The Vacuolar Protein-Sorting 13 Homolog C Gene (VPS13C)
Vacuolar protein sorting (VPS) is evolutionarily conserved, and was identified in Saccharomyces cerevisiae. Human VPS13 (hVPS13) consists of a group of four ubiquitously expressed proteins (hVPS13A, hVPS13B, hVPS13C, hVPS13D). Although the molecular function of hVPS13 proteins is not yet well understood, studies on fibroblasts, neurons differentiated from patients’ fibroblasts, and organisms such as yeast and fruit flies all suggest that they might play a role in a variety of processes, including the regulation of vesicle trafficking, cell-to-cell interactions, mitochondrial functions, actin cytoskeleton organization and autophagy [187,188,189].
Several mutations identified in hVPS13 genes have been associated with the onset of neurological and developmental disorders. Mutations in the hVPS13A (9q21.2) and hVPS13B (8q22.2) genes lead to recessive autosomal genetic disorders, including chorea-acanthocytosis (ChAc) and Cohen’s syndrome, respectively [190,191,192]. Mutations in hVPS13D (15q22.2) are associated with ataxia/spastic paraplegia. Polymorphisms or mutations of the hVPS13C (1p36) gene have been found to be associated with an increased risk of type 2 diabetes or young adult-onset PD (PARK23, OMIM 616840), respectively [193].
Notably, the link between hVPS13C mutations and PARK23 is a recent discovery. A genome-wide association study (GWAS), involving a cohort of 13,708 PD patients and 95,282 controls, identified the hVPS13C gene as a new risk variant for PD in Caucasian and East Asian populations [194]. Lesage and colleagues performed homozygosity mapping and exome sequencing studies of 66 individuals with PD (23 family members and 43 isolated subjects) and 39 unaffected relatives, and discovered a homozygous T-to-G transversion in the hVPS13C gene (intron 61) of a Turkish woman with AR EOPD, and heterozygous truncating mutations (4-bp insertion in exon 11 and G-to-T transversion in exon 69 or 1-bp deletion in exon 43 and G-to-C transversion in exon 37) in the hVPS13C gene of two French individuals with EOPD. The individuals harboring the hVPS13C mutations showed a peculiar form of EOPD characterized by rapid, severe disease progression and early cognitive decline. The post-mortem examination of the brain showed numerous Lewy bodies in the brainstem, limbic system and many cortical areas, typically observed in the brain of patients with α-synucleinopathy [193]. One year later, the whole-exome sequencing (WES) of 80 individuals with sporadic EOPD showed a heterozygous hVPS13C mutation in one of the patients [195]. Additional studies led to the discovery of a large homozygous genomic deletion (50 coding exons) in the hVPS13C gene in a patient with clinical symptoms of Parkinsonism [196]. Finally, a recent paper described seven new hVPS13C mutations in a large cohort of Chinese patients with EOPD [197]. Unfortunately, no animal model has been developed yet for VPS13C.
11. Discussion
From the experimental data presented in this review, it is clear that, to date, none of the developed experimental models can be considered a perfect model of PD. However, many of these models reproduce some of the key features of the disease and, for this reason, they can be considered as useful tools for the study of pathogenetic mechanisms and for the screening of neuroprotective therapies (Figure 2). In general, the murine KO models, which in theory—from the genetic point of view—are those that best reproduce AR transmission pathologies, are those that, unexpectedly, have often shown milder or even absent phenotypes. One plausible explanation is that, often, genetic compensation occurs in response to gene knocking-out [48]. Mouse models expressing genes bearing mutations associated with the disease have shown more evident PD-associated phenotypes, and for this reason they can be considered valid models to study the toxicity induced by mutant proteins.
Figure 2.
Animal models for the study of autosomal recessive Parkinson’s disease.
Studies in Drosophila mutant flies have often revealed that the deletion/silencing of genes associated with AR PD leads to a phenotype that mirrors many of the neuropathological features found in human tissues; in particular, in flies, the mitochondrial dysfunction phenotype is often evident. In this context, the data on PD models are in agreement with studies carried out on other fly models of neurodegenerative diseases that identify Drosophila as a useful model genetic system for the understanding of neurodegeneration [198].
Studies on the teleost fishes zebrafish and medaka have highlighted that, in these models, the deletion/silencing of genes associated with AR PD often leads to the spontaneous death of DA neurons or to a higher susceptibility to toxic substances. This key neuropathological hallmark of PD is often lacking in other organisms, raising the possibility that teleost fishes have a vulnerability of DA neurons more similar to humans.
Finally, C. elegans, the less-used model to date, has produced interesting results for PD modeling, although more detailed studies will be necessary to confirm the validity of these models.
In conclusion, although none of the models described mirror all the characteristics of PD, the efforts made in the last 20 years by many laboratories have allowed us to create and characterize a multitude of models, and each of these can be useful in selected contexts. Taking into account the strengths and limitations of each model, the use of multiple models remains the best strategy to validate a pathogenetic mechanism or to test a therapeutic approach.
Author Contributions
First draft: G.B., M.R., F.C., C.M.G.D.L., E.B., L.Z. Revision for important intellectual content: G.B., M.R., F.V., F.M., J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the ITALIAN MINISTRY OF HEALTH grant number GR–2016–02361366.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Figures created with BioRender.com (accessed on 28 April 2021).
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| (pS129) α-syn | phospho-Ser129 α-synuclein |
| 5-HT | serotonin |
| 6-OHDA | 6-hydroxydopamine |
| ADE | anterior deirids neurons |
| AM | anteromedial |
| ANAD | atypical neuroaxonal dystrophy |
| AR EOPD | autosomal recessive early-onset Parkinson’s Disease |
| AR | autosomal-recessive |
| ARJP | autosomal recessive Juvenile Parkinsonism |
| ARPD | autosomal recessive Parkinson’s disease |
| ATP | adenosine triphosphate |
| BAC | bacterial artificial chromosome |
| BMP | lysosomal lipid Bis(monoacylglycero)phosphate |
| CAN | canal-associated neurons |
| CATD | cathepsin D |
| CEP | chepalic sensilla neurons |
| ChAc | chorea-acanthocytosis syndrome |
| CME | clathrin-mediated endocytosis |
| CNS | central nervous system |
| DA | dopamine |
| DAT | dopamine transporter |
| DCCD | dicyclohexylcarbodiimide |
| DHA | docosahexaenoic acid |
| DKO | double-KO |
| DL | dorsolateral |
| DM | dorsomedial |
| DOPAC | 3,4-dihydroxyphenylacetic acid |
| Dpf | days postfertilization |
| DRP1 | dynamin related protein-1 |
| ENU | N-ethyl-N-nitrosourea |
| ETC | electron transport chain |
| FBP | F-box-containing protein |
| FBXO7 | F-box protein 7 |
| FCCP | carbonyl cyanide-p-trifluoromethoxyphenylhydrazone |
| GAK | cyclin G-associated kinase protein |
| GFAP | glia fibrillary acidic protein |
| GFP | green fluorescent protein |
| GSK-3β | glycogen synthase kinase 3 β |
| GWAS | genome-wide association study |
| H2O2 | hydrogen peroxide |
| HAD | haloacid dehalogenase domain |
| HIF | hypoxia-inducible factor |
| Hpf | hours post-fertilization |
| HPLC | high performance liquid chromatography |
| HVA | homovanillic acid |
| IMS | mitochondrial intermembrane space |
| INAD | infantile neuroaxonal dystrophy |
| KD | knock-down |
| KI | knock-in |
| KO | knock-out |
| LAMP | lysosome-associated membrane glycoprotein |
| LC | locus coeruleus |
| LC-MS/MS | liquid-chromatography tandem mass spectrometry |
| LGR | L-glutathione reduced |
| LTD | long-term depression |
| LTP | long-term potentiation |
| MAO | monoamine oxidase |
| M-CATD | mature form of pH dependent lysosomal enzyme aspartyl protease ASP-3/CATD |
| MEFs | mouse embryonic fibroblasts |
| MFN2 | mitofusin-2 |
| Mn | manganese |
| MO | morpholino |
| MPP | mitochondrial processing peptidase |
| MPP+ | 1-methyl-4-phenylpyridinium |
| MPTP | 1-methyl-4-phenyl-1,2,3,6-tetrapyridine |
| mPTP | mitochondrial permeability transition pore |
| mtDNA | mitochondrial DNA |
| MTS | mitochondrial targeting sequence |
| NAC | N-acetyl cysteine |
| NBIA | neurodegeneration with brain iron accumulation |
| NEO | neomycin |
| NEX | neuronal helix-loop-helix protein-1 |
| OCR | oxygen consumption rate |
| P | postnatal day |
| PAG | periaqueductal gray |
| PARL | presenilin-associated rhomboid-like protease |
| P-CATD | premature form of pH dependent lysosomal enzyme aspartyl protease ASP-3/CATD |
| PD | Parkinson’s disease |
| PDE | posterior deirids neurons |
| PI | phosphatidylinositol |
| PINK1 | (PTEN)-induced kinase 1 |
| PNS | peripheral nervous system |
| PPM1/2 | paired posterior medial 1 and 2 DA neuron clusters |
| PPM3 | paired posterior medial 3 DA neuron cluster |
| PPS | Parkinsonian-pyramidal syndrome |
| rAAV2/8 | recombinant type 2 adeno-associated viral vector pseudotyped with type 8 capsid |
| RNAi | RNA interference |
| RONS | reactive oxygen and nitrogen species |
| ROS | reactive oxygen species |
| SNc | substantia nigra pars compacta |
| β-PEA | β-phenylenthylamine |
| TGF-β | transforming growth factor-β |
| TH | tyrosine hydroxylase |
| TILLING | targeting induced local lesions in genomes |
| TIM23 | translocase of the inner mitochondrial membrane |
| TMD | transmembrane domain |
| TOM | translocase of the outer mitochondrial membrane |
| TOM20 | mitochondrial outer membrane protein |
| TUNEL | terminal deoxynucleotidyl transferase dUTP nick end labeling |
| USVs | ultrasound vocalizations |
| UVC | ultraviolet C |
| VEGF | vascular endothelial growth factor |
| VMAT2 | vesicular monoamine transporter type-2 |
| VPS | vacuolar protein sorting |
| WES | whole-exome sequencing |
| WT | wild type |
| ZFN | zinc finger nuclease |
| α-syn | α-synuclein |
References
- GBD 2016 Parkinson’s Disease Collaborators. Global, regional, and national burden of Parkinson’s disease, 1990–2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet Neurol. 2018, 17, 939–953. [Google Scholar] [CrossRef]
- Shulman, J.M.; De Jager, P.L.; Feany, M.B. Parkinson’s disease: Genetics and pathogenesis. Annu. Rev. Pathol. 2011, 6, 193–222. [Google Scholar] [CrossRef]
- Obeso, J.A.; Stamelou, M.; Goetz, C.G.; Poewe, W.; Lang, A.E.; Weintraub, D.; Burn, D.; Halliday, G.M.; Bezard, E.; Przedborski, S.; et al. Past, present, and future of Parkinson’s disease: A special essay on the 200th Anniversary of the Shaking Palsy. Mov. Disord. 2017, 32, 1264–1310. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Wang, P.; Jankovic, J. The genetics of Parkinson disease. Ageing Res. Rev. 2018, 42, 72–85. [Google Scholar] [CrossRef]
- Vingill, S.; Connor-Robson, N.; Wade-Martins, R. Are rodent models of Parkinson’s disease behaving as they should? Behav. Brain Res. 2018, 352, 133–141. [Google Scholar] [CrossRef]
- Chia, S.J.; Tan, E.K.; Chao, Y.X. Historical Perspective: Models of Parkinson’s Disease. Int. J. Mol. Sci. 2020, 21, 2464. [Google Scholar] [CrossRef]
- Vaz, R.L.; Outeiro, T.F.; Ferreira, J.J. Zebrafish as an animal model for drug discovery in Parkinson’s disease and other movement disorders: A systematic review. Front. Neurol. 2018, 9, 347. [Google Scholar] [CrossRef]
- Xi, Y.; Ryan, J.; Noble, S.; Yu, M.; Yilbas, A.E.; Ekker, M. Impaired dopaminergic neuron development and locomotor function in zebrafish with loss of pink1 function. Eur. J. Neurosci. 2010, 31, 623–633. [Google Scholar] [CrossRef]
- Blandini, F.; Armentero, M.T. Animal models of Parkinson’s disease. FEBS J. 2012, 279, 1156–1166. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Uemura, N.; Yamakado, H.; Takeda, S.; Takahashi, R. Exploring the pathogenetic mechanisms underlying Parkinson’s disease in medaka fish. J. Parkinsons Dis. 2014, 4, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Yu, J. Modeling Parkinson’s disease in drosophila: What have we learned for dominant traits? Front. Neurol. 2018, 9, 228. [Google Scholar] [CrossRef]
- Cooper, J.F.; Van Raamsdonk, J.M. Modeling Parkinson’s disease in C. elegans. J. Parkinsons Dis. 2018, 8, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Asakawa, S.; Hattori, N.; Matsumine, H.; Yamamura, Y.; Minoshima, S.; Yokochi, M.; Mizuno, Y.; Shimizu, N. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature 1998, 392, 605–608. [Google Scholar] [CrossRef]
- Hattori, N.; Matsumine, H.; Asakawa, S.; Kitada, T.; Yoshino, H.; Elibol, B.; Brookes, A.J.; Yamamura, Y.; Kobayashi, T.; Wang, M.; et al. Point mutations (Thr240Arg and Gln311Stop) [correction of Thr240Arg and Ala311Stop] in the Parkin gene. Biochem. Biophys. Res. Commun. 1998, 249, 754–758. [Google Scholar] [CrossRef]
- Klein, C.; Schlossmacher, M.G. The genetics of Parkinson disease: Implications for neurological care. Nat. Clin. Pract. Neurol. 2006, 2, 136–146. [Google Scholar] [CrossRef]
- Grunewald, A.; Kasten, M.; Ziegler, A.; Klein, C. Next-generation phenotyping using the parkin example: Time to catch up with genetics. JAMA Neurol. 2013, 70, 1186–1191. [Google Scholar] [CrossRef]
- Mitsuyama, S.; Ohtsubo, M.; Minoshima, S.; Shimizu, N. The KM-parkin-DB: A sub-set mutationview database specialized for PARK2 (PARKIN) variants. Hum. Mutat. 2015, 36, E2430–E2440. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.W.; Hang, L.; Yao, T.P.; Lim, K.L. Parkin regulation and neurodegenerative disorders. Front. Aging Neurosci. 2015, 7, 248. [Google Scholar] [CrossRef] [PubMed]
- Cookson, M.R. Parkin’s substrates and the pathways leading to neuronal damage. Neuromol. Med. 2003, 3, 1–13. [Google Scholar] [CrossRef]
- Hattori, N.; Mizuno, Y. Pathogenetic mechanisms of parkin in Parkinson’s disease. Lancet 2004, 364, 722–724. [Google Scholar] [CrossRef]
- Mata, I.F.; Lockhart, P.J.; Farrer, M.J. Parkin genetics: One model for Parkinson’s disease. Hum. Mol. Genet. 2004, 13, R127–R133. [Google Scholar] [CrossRef]
- Itier, J.M.; Ibanez, P.; Mena, M.A.; Abbas, N.; Cohen-Salmon, C.; Bohme, G.A.; Laville, M.; Pratt, J.; Corti, O.; Pradier, L.; et al. Parkin gene inactivation alters behaviour and dopamine neurotransmission in the mouse. Hum. Mol. Genet. 2003, 12, 2277–2291. [Google Scholar] [CrossRef]
- Goldberg, M.S.; Fleming, S.M.; Palacino, J.J.; Cepeda, C.; Lam, H.A.; Bhatnagar, A.; Meloni, E.G.; Wu, N.; Ackerson, L.C.; Klapstein, G.J.; et al. Parkin-deficient mice exhibit nigrostriatal deficits but not loss of dopaminergic neurons. J. Biol. Chem. 2003, 278, 43628–43635. [Google Scholar] [CrossRef] [PubMed]
- Palacino, J.J.; Sagi, D.; Goldberg, M.S.; Krauss, S.; Motz, C.; Wacker, M.; Klose, J.; Shen, J. Mitochondrial dysfunction and oxidative damage in parkin-deficient mice. J. Biol. Chem. 2004, 279, 18614–18622. [Google Scholar] [CrossRef] [PubMed]
- Stichel, C.C.; Zhu, X.R.; Bader, V.; Linnartz, B.; Schmidt, S.; Lubbert, H. Mono- and double-mutant mouse models of Parkinson’s disease display severe mitochondrial damage. Hum. Mol. Genet. 2007, 16, 2377–2393. [Google Scholar] [CrossRef]
- Perez, F.A.; Palmiter, R.D. Parkin-deficient mice are not a robust model of parkinsonism. Proc. Natl Acad. Sci. USA 2005, 102, 2174–2179. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Chiba, T.; Nishiyama, S.; Kakiuchi, T.; Tsukada, H.; Hatano, T.; Fukuda, T.; Yasoshima, Y.; Kai, N.; Kobayashi, K.; et al. Decline of striatal dopamine release in parkin-deficient mice shown by ex vivo autoradiography. J. Neurosci. Res. 2006, 84, 1350–1357. [Google Scholar] [CrossRef] [PubMed]
- von Coelln, R.; Thomas, B.; Savitt, J.M.; Lim, K.-L.; Sasaki, M.; Hess, E.J.; Dawson, V.; Dawson, T.M. Loss of locus coeruleus neurons and reduced startle in parkin null mice. Proc. Natl. Acad. Sci. USA 2004, 101, 10744–10749. [Google Scholar] [CrossRef]
- Shin, J.-H.; Ko, H.S.; Kang, H.; Lee, Y.; Lee, Y.-I.; Pletinkova, O.; Troconso, J.C.; Dawson, V.; Dawson, T.M. PARIS (ZNF746) repression of PGC-1alpha contributes to neurodegeneration in Parkinson’s disease. Cell 2011, 144, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Stevens, D.A.; Lee, Y.; Kang, H.C.; Lee, B.D.; Lee, Y.I.; Bower, A.; Jiang, H.; Kang, S.; Andrabi, S.A.; Dawson, V.L.; et al. Parkin loss leads to PARIS-dependent declines in mitochondrial mass and respiration. Proc. Natl. Acad. Sci. USA 2015, 112, 11696–11701. [Google Scholar] [CrossRef]
- Lorenzetti, D.; Antalffy, B.; Vogel, H.; Noveroske, J.; Armstrong, D.; Justice, M. The neurological mutant quaking(viable) is Parkin deficient. Mamm. Genome 2004, 15, 210–217. [Google Scholar] [CrossRef] [PubMed]
- Ebersole, T.A.; Chen, Q.; Justice, M.J.; Artzt, K. The quaking gene product necessary in embryogenesis and myelination combines features of RNA binding and signal transduction proteins. Nat. Genet. 1996, 12, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.-H.; Fleming, S.M.; Meurers, B.; Ackerson, L.C.; Mortazavi, F.; Lo, V.; Hernandez, D.; Sulzer, D.; Jackson, G.R.; Maidment, N.T.; et al. Bacterial artificial chromosome transgenic mice expressing a truncated mutant parkin exhibit age-dependent hypokinetic motor deficits, dopaminergic neuron degeneration, and accumulation of proteinase K-resistant alpha-synuclein. J. Neurosci. 2009, 29, 1962–1976. [Google Scholar] [CrossRef] [PubMed]
- Dave, K.D.; De Silva, S.; Sheth, N.P.; Ramboz, S.; Beck, M.J.; Quang, C.; Switzer, R.C.; Ahmad, S.O.; Sunkin, S.M.; Walker, D.; et al. Phenotypic characterization of recessive gene knockout rat models of Parkinson’s disease. Neurobiol. Dis. 2014, 70, 190–203. [Google Scholar] [CrossRef]
- Gemechu, J.M.; Sharma, A.; Yu, D.; Xie, Y.; Merkel, O.M.; Moszczynska, A. Characterization of dopaminergic system in the striatum of young adult Park2-/- knockout rats. Sci. Rep. 2018, 8, 1517. [Google Scholar] [CrossRef]
- Stauch, K.L.; Villeneuve, L.M.; Purnell, P.R.; Pandey, S.; Guda, C.; Fox, H.S. SWATH-MS proteome profiling data comparison of DJ-1, Parkin, and PINK1 knockout rat striatal mitochondria. Data Brief. 2016, 9, 589–593. [Google Scholar] [CrossRef][Green Version]
- Van Rompuy, A.S.; Lobbestael, E.; Van der Perren, A.; Van den Haute, C.; Baekelandt, V. Long-term overexpression of human wild-type and T240R mutant Parkin in rat substantia nigra induces progressive dopaminergic neurodegeneration. J. Neuropathol. Exp. Neurol. 2014, 73, 159–174. [Google Scholar] [CrossRef]
- Flinn, L.; Mortiboys, H.; Volkmann, K.; Koster, R.W.; Ingham, P.W.; Bandmann, O. Complex I deficiency and dopaminergic neuronal cell loss in parkin-deficient zebrafish (Danio rerio). Brain 2009, 132, 1613–1623. [Google Scholar] [CrossRef] [PubMed]
- Greene, J.C.; Whitworth, A.J.; Kuo, I.; Andrews, L.A.; Feany, M.B.; Pallanck, L.J. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc. Natl. Acad. Sci. USA 2003, 100, 4078–4083. [Google Scholar] [CrossRef]
- Whitworth, A.J.; Theodore, D.A.; Greene, J.C.; Benes, H.; Wes, P.D.; Pallanck, L.J. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Proc. Natl. Acad. Sci. USA 2005, 102, 8024–8029. [Google Scholar] [CrossRef]
- Pesah, Y.; Pham, T.; Burgess, H.; Middlebrooks, B.; Verstreken, P.; Zhou, Y.; Mark Harding, H.B. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development 2004, 131, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Lu, R.; Ouyang, X.; Ho, M.W.L.; Chia, W.; Yu, F.; Lim, K.-L. Drosophila overexpressing parkin R275W mutant exhibits dopaminergic neuron degeneration and mitochondrial abnormalities. J. Neurosci. 2007, 27, 8563–8570. [Google Scholar] [CrossRef]
- Cha, G.-H.; Kim, S.; Park, J.; Lee, E.; Kim, M.; Lee, S.B.; Kim, J.M.; Chung, J.; Cho, K.S. Parkin negatively regulates JNK pathway in the dopaminergic neurons of Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 10345–10350. [Google Scholar] [CrossRef]
- Sang, T.K.; Chang, H.Y.; Lawless, G.M.; Ratnaparkhi, A.; Mee, L.; Ackerson, L.C.; Maidment, N.T.; Krantz, D.E.; Jackson, G.R. A Drosophila model of mutant human parkin-induced toxicity demonstrates selective loss of dopaminergic neurons and dependence on cellular dopamine. J. Neurosci. 2007, 27, 981–992. [Google Scholar] [CrossRef] [PubMed]
- Ved, R.; Saha, S.; Westlund, B.; Perier, C.; Burnam, L.; Sluder, A.; Hoener, M.; Rodrigues, C.M.P.; Alfonso, A.; Steer, C.; et al. Similar patterns of mitochondrial vulnerability and rescue induced by genetic modification of alpha-synuclein, parkin, and DJ-1 in Caenorhabditis elegans. J. Biol. Chem. 2005, 280, 42655–42668. [Google Scholar] [CrossRef]
- Bornhorst, J.; Chakraborty, S.; Meyer, S.; Lohren, H.; Brinkhaus, S.G.; Knight, A.L.; Caldwell, K.; Caldwell, G.; Karst, U.; Schwerdtle, T.; et al. The effects of pdr1, djr1.1 and pink1 loss in manganese-induced toxicity and the role of alpha-synuclein in C. elegans. Metallomics 2014, 6, 476–490. [Google Scholar] [CrossRef]
- Poulopoulos, M.; Levy, O.A.; Alcalay, R.N. The neuropathology of genetic Parkinson’s disease. Mov. Disord. 2012, 27, 831–842. [Google Scholar] [CrossRef] [PubMed]
- El-Brolosy, M.A.; Stainier, D.Y.R. Genetic compensation: A phenomenon in search of mechanisms. PLoS Genet. 2017, 13, e1006780. [Google Scholar] [CrossRef] [PubMed]
- Pascual, A.; Hidalgo-Figueroa, M.; Piruat, J.I.; Pintado, C.O.; Gomez-Diaz, R.; Lopez-Barneo, J. Absolute requirement of GDNF for adult catecholaminergic neuron survival. Nat. Neurosci. 2008, 11, 755–761. [Google Scholar] [CrossRef] [PubMed]
- Ramsey, C.P.; Giasson, B.I. Identification and characterization of a novel endogenous murine parkin mutation. J. Neurochem. 2010, 113, 402–417. [Google Scholar] [CrossRef] [PubMed]
- Kitada, T.; Asakawa, S.; Minoshima, S.; Mizuno, Y.; Shimizu, N. Molecular cloning, gene expression, and identification of a splicing variant of the mouse parkin gene. Mamm. Genome 2000, 11, 417–421. [Google Scholar] [CrossRef]
- Regoni, M.; Cattaneo, S.; Mercatelli, D.; Novello, S.; Passoni, A.; Bagnati, R.; Davoli, E.; Croci, L.; Consalez, G.G.; Albanese, F.; et al. Pharmacological antagonism of kainate receptor rescues dysfunction and loss of dopamine neurons in a mouse model of human parkin-induced toxicity. Cell Death Dis. 2020, 11, 963. [Google Scholar] [CrossRef]
- Creed, R.B.; Goldberg, M.S. New developments in genetic rat models of Parkinson’s disease. Mov. Disord. 2018, 33, 717–729. [Google Scholar] [CrossRef] [PubMed]
- Cookson, M.R.; Lockhart, P.J.; McLendon, C.; O’Farrell, C.; Schlossmacher, M.; Farrer, M.J. RING finger 1 mutations in Parkin produce altered localization of the protein. Hum. Mol. Genet. 2003, 12, 2957–2965. [Google Scholar] [CrossRef]
- Bandmann, O.; Burton, E.A. Genetic zebrafish models of neurodegenerative diseases. Neurobiol. Dis. 2010, 40, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, Y.; Takeda, S.; Furutani-Seiki, M.; Kamei, Y.; Todo, T.; Sasado, T.; Deguchi, T.; Kondoh, H.; Mudde, J.; Yamazoe, M.; et al. Generation of medaka gene knockout models by target-selected mutagenesis. Genome Biol. 2006, 7, R116. [Google Scholar] [CrossRef] [PubMed]
- Valente, E.M.; Abou-Sleiman, P.M.; Caputo, V.; Muqit, M.M.; Harvey, K.; Gispert, S.; Ali, Z.; del Turco, D.; Bentivoglio, A.R.; Healy, D.G.; et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science 2004, 304, 1158–1160. [Google Scholar] [CrossRef]
- Voigt, A.; Berlemann, L.A.; Winklhofer, K.F. The mitochondrial kinase PINK1: Functions beyond mitophagy. J. Neurochem. 2016, 139, 232–239. [Google Scholar] [CrossRef] [PubMed]
- Silvestri, L.; Caputo, V.; Bellacchio, E.; Atorino, L.; Dallapiccola, B.; Valente, E.M.; Casari, G. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum. Mol. Genet. 2005, 14, 3477–3492. [Google Scholar] [CrossRef]
- Meissner, C.; Lorenz, H.; Weihofen, A.; Selkoe, D.J.; Lemberg, M.K. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 2011, 117, 856–867. [Google Scholar] [CrossRef]
- Pickrell, A.M.; Youle, R.J. The roles of PINK1, parkin, and mitochondrial fidelity in Parkinson’s disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef]
- Sim, C.H.; Gabriel, K.; Mills, R.D.; Culvenor, J.G.; Cheng, H.C. Analysis of the regulatory and catalytic domains of PTEN-induced kinase-1 (PINK1). Hum. Mutat. 2012, 33, 1408–1422. [Google Scholar] [CrossRef]
- Zhou, C.; Huang, Y.; Shao, Y.; May, J.; Prou, D.; Perier, C.; Dauer, W.; Schon, E.A.; Przedborski, S. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc. Natl. Acad. Sci. USA 2008, 105, 12022–12027. [Google Scholar] [CrossRef]
- Springer, W.; Kahle, P.J. Regulation of PINK1-Parkin-mediated mitophagy. Autophagy 2011, 7, 266–278. [Google Scholar] [CrossRef]
- Gandhi, S.; Wood-Kaczmar, A.; Yao, Z.; Plun-Favreau, H.; Deas, E.; Klupsch, K.; Downward, J.; Latchman, D.S.; Tabrizi, S.; Wood, N.; et al. PINK1-associated Parkinson’s disease is caused by neuronal vulnerability to calcium-induced cell death. Mol. Cell. 2009, 33, 627–638. [Google Scholar] [CrossRef] [PubMed]
- Trancikova, A.; Tsika, E.; Moore, D.J. Mitochondrial dysfunction in genetic animal models of Parkinson’s disease. Antioxid. Redox Signal. 2012, 16, 896–919. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Falkenburger, B.H.; Schulz, J.B.; Tieu, K.; Xu, Z.; Xia, X.G. Silencing of the Pink1 gene expression by conditional RNAi does not induce dopaminergic neuron death in mice. Int. J. Biol. Sci. 2007, 3, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Gautier, C.A.; Kitada, T.; Shen, J. Loss of PINK1 causes mitochondrial functional defects and increased sensitivity to oxidative stress. Proc. Natl. Acad. Sci. USA 2008, 105, 11364–11369. [Google Scholar] [CrossRef]
- Kitada, T.; Pisani, A.; Porter, D.R.; Yamaguchi, H.; Tscherter, A.; Martella, G.; Bonsi, P.; Zhang, C.; Pothos, E.; Shen, J. Impaired dopamine release and synaptic plasticity in the striatum of PINK1-deficient mice. Proc. Natl. Acad. Sci. USA 2007, 104, 11441–11446. [Google Scholar] [CrossRef]
- Glasl, L.; Kloos, K.; Giesert, F.; Roethig, A.; Di Benedetto, B.; Kuhn, R. Pink1-deficiency in mice impairs gait, olfaction and serotonergic innervation of the olfactory bulb. Exp. Neurol. 2012, 235, 214–227. [Google Scholar] [CrossRef]
- Kelm-Nelson, C.A.; Brauer, A.F.L.; Barth, K.J.; Lake, J.M.; Sinnen, M.L.K.; Stehula, F.J. Characterization of early-onset motor deficits in the Pink1-/- mouse model of Parkinson disease. Brain Res. 2018, 1680, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Gispert, S.; Ricciardi, F.; Kurz, A.; Azizov, M.; Hoepken, H.-H.; Becker, D.; Voos, W.; Leuner, K.; Müller, W.E.; Kudin, A.P.; et al. Parkinson phenotype in aged PINK1-deficient mice is accompanied by progressive mitochondrial dysfunction in absence of neurodegeneration. PLoS ONE 2009, 4, e5777. [Google Scholar] [CrossRef]
- Wood-Kaczmar, A.; Gandhi, S.; Yao, Z.; Abramov, A.Y.; Miljan, E.A.; Keen, G. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS ONE 2008, 3, e2455. [Google Scholar] [CrossRef]
- Sun, J.; Kouranova, E.; Cui, X.; Mach, R.H.; Xu, J. Regulation of dopamine presynaptic markers and receptors in the striatum of DJ-1 and Pink1 knockout rats. Neurosci. Lett. 2013, 557, 123–128. [Google Scholar] [CrossRef]
- Villeneuve, L.M.; Purnell, P.R.; Boska, M.D.; Fox, H.S. Early expression of Parkinson’s disease-related mitochondrial abnormalities in PINK1 knockout rats. Mol. Neurobiol. 2016, 53, 171–186. [Google Scholar] [CrossRef] [PubMed]
- Grant, L.M.; Kelm-Nelson, C.; Hilby, B.L.; Blue, K.V.; Rajamanickam, E.S.P.; Pultorak, J.D.; Fleming, S.M.; Ciucci, M.R. Evidence for early and progressive ultrasonic vocalization and oromotor deficits in a PINK1 gene knockout rat model of Parkinson’s disease. J. Neurosci. Res. 2015, 93, 1713–1727. [Google Scholar] [CrossRef]
- Pultorak, J.D.; Kelm-Nelson, C.A.; Holt, L.R.; Blue, K.V.; Ciucci, M.R.; Johnson, A.M. Decreased approach behavior and nucleus accumbens immediate early gene expression in response to Parkinsonian ultrasonic vocalizations in rats. Soc. Neurosci. 2016, 11, 365–379. [Google Scholar] [CrossRef]
- Anichtchik, O.; Diekmann, H.; Fleming, A.; Roach, A.; Goldsmith, P.; Rubinsztein, D.C. Loss of PINK1 function affects development and results in neurodegeneration in zebrafish. J. Neurosci. 2008, 28, 8199–8207. [Google Scholar] [CrossRef]
- Priyadarshini, M.; Tuimala, J.; Chen, Y.C.; Panula, P. A zebrafish model of PINK1 deficiency reveals key pathway dysfunction including HIF signaling. Neurobiol. Dis. 2013, 54, 127–138. [Google Scholar] [CrossRef]
- Sallinen, V.; Kolehmainen, J.; Priyadarshini, M.; Toleikyte, G.; Chen, Y.C.; Panula, P. Dopaminergic cell damage and vulnerability to MPTP in Pink1 knockdown zebrafish. Neurobiol. Dis. 2010, 40, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Flinn, L.J.; Keatinge, M.; Bretaud, S.; Mortiboys, H.; Matsui, H.; De Felice, E.; Woodroof, H.I.; Brown, L.; McTighe, A.; Soellner, R.; et al. TigarB causes mitochondrial dysfunction and neuronal loss in PINK1 deficiency. Ann. Neurol. 2013, 74, 837–847. [Google Scholar] [CrossRef]
- Matsui, H.; Taniguchi, Y.; Inoue, H.; Kobayashi, Y.; Sakaki, Y.; Toyoda, A. Loss of PINK1 in medaka fish (Oryzias latipes) causes late-onset decrease in spontaneous movement. Neurosci. Res. 2010, 66, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Lee, S.B.; Lee, S.; Kim, Y.; Song, S.; Kim, S. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature 2006, 441, 1157–1161. [Google Scholar] [CrossRef] [PubMed]
- Clark, I.E.; Dodson, M.W.; Jiang, C.; Cao, J.H.; Huh, J.R.; Seol, J.H.; Yoo, S.J.; Hay, B.A.; Guo, M. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature 2006, 441, 1162–1166. [Google Scholar] [CrossRef] [PubMed]
- Morais, V.; Verstreken, P.; Roethig, A.; Smet, J.; Snellinx, A.; Vanbrabant, M.; Haddad, D.; Frezza, C.; Mandemakers, W.; Vogt-Weisenhorn, D.; et al. Parkinson’s disease mutations in PINK1 result in decreased Complex I activity and deficient synaptic function. EMBO Mol. Med. 2009, 1, 99–111. [Google Scholar] [CrossRef]
- Liu, W.; Acin-Perez, R.; Geghman, K.D.; Manfredi, G.; Lu, B.; Li, C. Pink1 regulates the oxidative phosphorylation machinery via mitochondrial fission. Proc. Natl. Acad. Sci. USA 2011, 108, 12920–12924. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Gehrke, S.; Imai, Y.; Huang, Z.; Ouyang, Y.; Wang, J.W. Mitochondrial pathology and muscle and dopaminergic neuron degeneration caused by inactivation of Drosophila Pink1 is rescued by Parkin. Proc. Natl. Acad. Sci. USA 2006, 103, 10793–10798. [Google Scholar] [CrossRef]
- Samann, J.; Hegermann, J.; von Gromoff, E.; Eimer, S.; Baumeister, R.; Schmidt, E. Caenorhabditits elegans LRK-1 and PINK-1 act antagonistically in stress response and neurite outgrowth. J. Biol. Chem. 2009, 284, 16482–16491. [Google Scholar] [CrossRef]
- Luz, A.L.; Rooney, J.P.; Kubik, L.L.; Gonzalez, C.P.; Song, D.H.; Meyer, J.N. Mitochondrial morphology and fundamental parameters of the mitochondrial respiratory chain are altered in Caenorhabditis elegans strains deficient in mitochondrial dynamics and homeostasis processes. PLoS ONE 2015, 10, e0130940. [Google Scholar] [CrossRef]
- Palikaras, K.; Lionaki, E.; Tavernarakis, N. Coordination of mitophagy and mitochondrial biogenesis during ageing in C. elegans. Nature 2015, 521, 525–528. [Google Scholar] [CrossRef]
- Bess, A.S.; Crocker, T.L.; Ryde, I.T.; Meyer, J.N. Mitochondrial dynamics and autophagy aid in removal of persistent mitochondrial DNA damage in Caenorhabditis elegans. Nucleic Acids Res. 2012, 40, 7916–7931. [Google Scholar] [CrossRef]
- Bess, A.S.; Leung, M.C.; Ryde, I.T.; Rooney, J.P.; Hinton, D.E.; Meyer, J.N. Effects of mutations in mitochondrial dynamics-related genes on the mitochondrial response to ultraviolet C radiation in developing Caenorhabditis elegans. Worm 2013, 2, e23763. [Google Scholar] [CrossRef]
- Priyadarshini, M.; Orosco, L.A.; Panula, P.J. Oxidative stress and regulation of Pink1 in zebrafish (Danio rerio). PLoS ONE 2013, 8, e81851. [Google Scholar] [CrossRef]
- Yang, Y.; Ouyang, Y.; Yang, L.; Beal, M.F.; McQuibban, A.; Vogel, O.H.; Lu, B. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. USA 2008, 105, 7070–7075. [Google Scholar] [CrossRef]
- Deng, H.; Dodson, M.W.; Huang, H.; Guo, M. The Parkinson’s disease genes pink1 and parkin promote mitochondrial fission and/or inhibit fusion in Drosophila. Proc. Natl. Acad. Sci. USA 2008, 105, 14503–14508. [Google Scholar] [CrossRef]
- Van Duijn, C.M.; Dekker, M.C.; Bonifati, V.; Galjaard, R.J.; Houwing-Duistermaat, J.J.; Snijders, P.J. Park7, a novel locus for autosomal recessive early-onset parkinsonism, on chromosome 1p36. Am. J. Hum. Genet. 2001, 69, 629–634. [Google Scholar] [CrossRef] [PubMed]
- Abou-Sleiman, P.M.; Healy, D.G.; Quinn, N.; Lees, A.J.; Wood, N.W. The role of pathogenic DJ-1 mutations in Parkinson’s disease. Ann. Neurol. 2003, 54, 283–286. [Google Scholar] [CrossRef] [PubMed]
- Bonifati, V.; Rizzu, P.; Van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.J.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 2003, 299, 256–259. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, M.S.; Pisani, A.; Haburcak, M.; Vortherms, T.A.; Kitada, T.; Costa, C.; Tong, Y.; Martella, G.; Tscherter, A.; Martins, A.; et al. Nigrostriatal dopaminergic deficits and hypokinesia caused by inactivation of the familial Parkinsonism-linked gene DJ-1. Neuron 2005, 45, 489–496. [Google Scholar] [CrossRef]
- Kahle, P.J.; Waak, J.; Gasser, T. DJ-1 and prevention of oxidative stress in Parkinson’s disease and other age-related disorders. Free Radic. Biol. Med. 2009, 47, 1354–1361. [Google Scholar] [CrossRef]
- Van der Merwe, C.; Jalali Sefid Dashti, Z.; Christoffels, A.; Loos, B.; Bardien, S. Evidence for a common biological pathway linking three Parkinson’s disease-causing genes: Parkin, PINK1 and DJ-1. Eur. J. Neurosci. 2015, 41, 1113–1125. [Google Scholar] [CrossRef]
- Wilhelmus, M.M.; Nijland, P.G.; Drukarch, B.; de Vries, H.E.; van Horssen, J. Involvement and interplay of Parkin, PINK1, and DJ1 in neurodegenerative and neuroinflammatory disorders. Free Radic. Biol. Med. 2012, 53, 983–992. [Google Scholar] [CrossRef]
- Chen, L.; Cagniard, B.; Mathews, T.; Jones, S.; Koh, H.C.; Ding, Y.; Carvey, P.M.; Ling, Z.; Kang, U.; Zhuang, X. Age-dependent motor deficits and dopaminergic dysfunction in DJ-1 null mice. J. Biol. Chem. 2005, 280, 21418–21426. [Google Scholar] [CrossRef]
- Malgieri, G.; Eliezer, D. Structural effects of Parkinson’s disease linked DJ-1 mutations. Protein Sci. 2008, 17, 855–868. [Google Scholar] [CrossRef] [PubMed]
- Cookson, M.R. DJ-1, PINK1, and their effects on mitochondrial pathways. Mov. Disord. 2010, 25, S44–S48. [Google Scholar] [CrossRef]
- Kim, R.H.; Smith, P.D.; Aleyasin, H.; Hayley, S.; Mount, M.P.; Pownall, S. Hypersensitivity of DJ-1-deficient mice to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyrindine (MPTP) and oxidative stress. Proc. Natl. Acad. Sci. USA 2005, 102, 5215–5220. [Google Scholar] [CrossRef]
- Zhou, W.; Barkow, J.C.; Freed, C.R. Running wheel exercise reduces alpha-synuclein aggregation and improves motor and cognitive function in a transgenic mouse model of Parkinson’s disease. PLoS ONE 2017, 12, e0190160. [Google Scholar] [CrossRef] [PubMed]
- Manning-Bog, A.B.; Caudle, W.M.; Perez, X.A.; Reaney, S.H.; Paletzki, R.; Isla, M.Z. Increased vulnerability of nigrostriatal terminals in DJ-1-deficient mice is mediated by the dopamine transporter. Neurobiol. Dis. 2007, 27, 141–150. [Google Scholar] [CrossRef] [PubMed]
- Giangrasso, D.M.; Furlong, T.M.; Keefe, K.A. Characterization of striatum-mediated behavior and neurochemistry in the DJ-1 knock-out rat model of Parkinson’s disease. Neurobiol. Dis. 2020, 134, 104673. [Google Scholar] [CrossRef]
- Yang, K.M.; Blue, K.V.; Mulholland, H.M.; Kurup, M.P.; Kelm-Nelson, C.A.; Ciucci, M.R. Characterization of oromotor and limb motor dysfunction in the DJ1 -/- model of Parkinson disease. Behav. Brain Res. 2018, 339, 47–56. [Google Scholar] [CrossRef] [PubMed]
- Kyser, T.L.; Dourson, A.J.; McGuire, J.L.; Hemmerle, A.M.; Williams, M.T.; Seroogy, K.B. Characterization of motor and non-motor behavioral alterations in the Dj-1 (PARK7) knockout rat. J. Mol. Neurosci. 2019, 69, 298–311. [Google Scholar] [CrossRef]
- Bretaud, S.; Allen, C.; Ingham, P.W.; Bandmann, O. p53-dependent neuronal cell death in a DJ-1-deficient zebrafish model of Parkinson’s disease. J. Neurochem. 2007, 100, 1626–1635. [Google Scholar] [CrossRef]
- Baulac, S.; Lu, H.; Strahle, J.; Yang, T.; Goldberg, M.S.; Shen, J.; Schlossmacher, M.G.; Lemere, C.A.; Lu, Q.; Xia, W. Increased DJ-1 expression under oxidative stress and in Alzheimer’s disease brains. Mol. Neurodegener. 2009, 4, 12. [Google Scholar] [CrossRef]
- Edson, A.J.; Hushagen, H.A.; Froyset, A.K.; Elda, I.; Khan, E.A.; Di Stefano, A. Dysregulation in the Brain protein profile of zebrafish lacking the Parkinson’s disease-related protein DJ-1. Mol. Neurobiol. 2019, 56, 8306–8322. [Google Scholar] [CrossRef] [PubMed]
- Hughes, G.L.; Lones, M.A.; Bedder, M.; Currie, P.D.; Smith, S.L.; Pownall, M.E. Machine learning discriminates a movement disorder in a zebrafish model of Parkinson’s disease. Dis. Model. Mech. 2020, 13. [Google Scholar] [CrossRef]
- Menzies, F.M.; Yenisetti, S.C.; Min, K.T. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr. Biol. 2005, 15, 1578–1582. [Google Scholar] [CrossRef]
- Park, J.; Kim, S.Y.; Cha, G.H.; Lee, S.B.; Kim, S.; Chung, J. Drosophila DJ-1 mutants show oxidative stress-sensitive locomotive dysfunction. Gene 2005, 361, 133–139. [Google Scholar] [CrossRef]
- Poudel, S.; Lee, Y. Impaired taste associative memory and memory enhancement by feeding Omija in Parkinson’s disease fly model. Mol. Cells 2018, 41, 646–652. [Google Scholar] [PubMed]
- Meulener, M.; Whitworth, A.; Armstrong-Gold, C.E.; Rizzu, P.; Heutink, P.; Wes, P.; Pallanck, L.J.; Bonini, N. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson’s disease. Curr. Biol. 2005, 15, 1572–1577. [Google Scholar] [CrossRef] [PubMed]
- Castro, E.M.C.; Waak, J.; Weber, S.S.; Fiesel, F.C.; Oberhettinger, P.; Schütz, M.; Autenrieth, I.B.; Springer, W.; Kahle, P.J. Parkinson’s disease-associated DJ-1 modulates innate immunity signaling in Caenorhabditis elegans. J. Neural Transm. 2010, 117, 599–604. [Google Scholar] [CrossRef]
- Chen, P.; DeWitt, M.R.; Bornhorst, J.; Soares, F.; Mukhopadhyay, S.; Bowman, A.B.; Aschner, M. Age- and manganese-dependent modulation of dopaminergic phenotypes in a C. elegans DJ-1 genetic model of Parkinson’s disease. Metallomics 2015, 7, 289–298. [Google Scholar] [CrossRef]
- Baptista, M.A.; Dave, K.D.; Sheth, N.P.; De Silva, S.N.; Carlson, K.M.; Aziz, Y.N.; Fiske, B.K.; Sherer, T.B.; Frasier, M.A. A strategy for the generation, characterization and distribution of animal models by The Michael, J. Fox Foundation for Parkinson’s Research. Dis. Model. Mech. 2013, 6, 1316–1324. [Google Scholar] [CrossRef] [PubMed]
- Number, S.; Harmuth, F.; Kohl, Z.; Adame, A.; Trejo, M.; Schonig, K. A progressive dopaminergic phenotype associated with neurotoxic conversion of alpha-synuclein in BAC-transgenic rats. Brain 2013, 136, 412–432. [Google Scholar] [CrossRef]
- Ellenbroek, B.; Youn, J. Rodent models in neuroscience research: Is it a rat race? Dis. Model. Mech. 2016, 9, 1079–1087. [Google Scholar] [CrossRef]
- Bai, Q.; Mullett, S.J.; Garver, J.A.; Hinkle, D.A.; Burton, E.A. Zebrafish DJ-1 is evolutionarily conserved and expressed in dopaminergic neurons. Brain Res. 2006, 1113, 33–44. [Google Scholar] [CrossRef]
- Najim al-Din, A.S.; Wriekat, A.; Mubaidin, A.; Dasouki, M.; Hiari, M. Pallido-pyramidal degeneration, supranuclear upgaze paresis and dementia: Kufor-Rakeb syndrome. Acta Neurol. Scand. 1994, 89, 347–352. [Google Scholar] [CrossRef]
- Park, J.S.; Blair, N.F.; Sue, C.M. The role of ATP13A2 in Parkinson’s disease: Clinical phenotypes and molecular mechanisms. Mov. Disord. 2015, 30, 770–779. [Google Scholar] [CrossRef] [PubMed]
- Demirsoy, S.; Martin, S.; Motamedi, S.; van Veen, S.; Holemans, T.; Van den Haute, C. ATP13A2/PARK9 regulates endo-/lysosomal cargo sorting and proteostasis through a novel PI(3, 5)P2-mediated scaffolding function. Hum. Mol. Genet. 2017, 26, 1656–1669. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, A.; Heimbach, A.; Grundemann, J.; Stiller, B.; Hampshire, D.; Cid, L.P. Hereditary parkinsonism with dementia is caused by mutations in ATP13A2, encoding a lysosomal type 5 P-type ATPase. Nat. Genet. 2006, 38, 1184–1191. [Google Scholar] [CrossRef]
- Van Veen, S.; Martin, S.; Van den Haute, C.; Benoy, V.; Lyons, J.; Vanhoutte, R. ATP13A2 deficiency disrupts lysosomal polyamine export. Nature 2020, 578, 419–424. [Google Scholar] [CrossRef]
- Dehay, B.; Ramirez, A.; Martinez-Vicente, M.; Perier, C.; Canron, M.-H.; Doudnikoff, E.; Vital, A.; Vila, M.; Klein, C.; Bezard, E. Loss of P-type ATPase ATP13A2/PARK9 function induces general lysosomal deficiency and leads to Parkinson disease neurodegeneration. Proc. Natl. Acad. Sci. USA 2012, 109, 9611–9616. [Google Scholar] [CrossRef]
- Gitler, A.D.; Chesi, A.; Geddie, M.L.; Strathearn, K.E.; Hamamichi, S.; Hill, K.J. Alpha-synuclein is part of a diverse and highly conserved interaction network that includes PARK9 and manganese toxicity. Nat. Genet. 2009, 41, 308–315. [Google Scholar] [CrossRef]
- Kong, S.M.; Chan, B.K.; Park, J.-S.; Hill, K.J.; Aitken, J.B.; Cottle, L.; Farghaian, H.; Cole, A.R.; Lay, P.; Sue, C.M.; et al. Parkinson’s disease-linked human PARK9/ATP13A2 maintains zinc homeostasis and promotes alpha-Synuclein externalization via exosomes. Hum. Mol. Genet. 2014, 23, 2816–2833. [Google Scholar] [CrossRef]
- Gusdon, A.M.; Zhu, J.; Van Houten, B.; Chu, C.T. ATP13A2 regulates mitochondrial bioenergetics through macroautophagy. Neurobiol Dis. 2012, 45, 962–972. [Google Scholar] [CrossRef] [PubMed]
- Usenovic, M.; Tresse, E.; Mazzulli, J.R.; Taylor, J.P.; Krainc, D. Deficiency of ATP13A2 leads to lysosomal dysfunction, alpha-synuclein accumulation, and neurotoxicity. J. Neurosci. 2012, 32, 4240–4246. [Google Scholar] [CrossRef] [PubMed]
- Schultheis, P.J.; Fleming, S.M.; Clippinger, A.K.; Lewis, J.; Tsunemi, T.; Giasson, B. Atp13a2-deficient mice exhibit neuronal ceroid lipofuscinosis, limited alpha-synuclein accumulation and age-dependent sensorimotor deficits. Hum. Mol. Genet. 2013, 22, 2067–2082. [Google Scholar] [CrossRef] [PubMed]
- Kett, L.R.; Stiller, B.; Bernath, M.M.; Tasset, I.; Blesa, J.; Jackson-Lewis, V. Alpha-Synuclein-independent histopathological and motor deficits in mice lacking the endolysosomal Parkinsonism protein Atp13a2. J. Neurosci. 2015, 35, 5724–5742. [Google Scholar] [CrossRef]
- Lopes da Fonseca, T.; Correia, A.; Hasselaar, W.; van der Linde, H.C.; Willemsen, R.; Outeiro, T.F. The zebrafish homologue of Parkinson’s disease ATP13A2 is essential for embryonic survival. Brain Res. Bull. 2013, 90, 118–126. [Google Scholar] [CrossRef]
- Heins-Marroquin, U.; Jung, P.P.; Cordero-Maldonado, M.L.; Crawford, A.D.; Linster, C.L. Phenotypic assays in yeast and zebrafish reveal drugs that rescue ATP13A2 deficiency. Brain Commun. 2019, 1, fcz019. [Google Scholar] [CrossRef]
- Nyuzuki, H.; Ito, S.; Nagasaki, K.; Nitta, Y.; Matsui, N.; Saitoh, A. Degeneration of dopaminergic neurons and impaired intracellular trafficking in Atp13a2 deficient zebrafish. IBRO Rep. 2020, 9, 1–8. [Google Scholar] [CrossRef]
- Matsui, H.; Sato, F.; Sato, S.; Koike, M.; Taruno, Y.; Saiki, S. ATP13A2 deficiency induces a decrease in cathepsin D activity, fingerprint-like inclusion body formation, and selective degeneration of dopaminergic neurons. FEBS Lett. 2013, 587, 1316–1325. [Google Scholar] [CrossRef] [PubMed]
- Anand, N.; Holcom, A.; Broussalian, M.; Schmidt, M.; Chinta, S.J.; Lithgow, G.J. Dysregulated iron metabolism in C. elegans catp-6/ATP13A2 mutant impairs mitochondrial function. Neurobiol Dis. 2020, 139, 104786. [Google Scholar] [CrossRef]
- Baesler, J.; Kopp, J.F.; Pohl, G.; Aschner, M.; Haase, H.; Schwerdtle, T.; Bornhorst, J. Zn homeostasis in genetic models of Parkinson’s disease in Caenorhabditis elegans. J. Trace Elem. Med. Biol. 2019, 55, 44–49. [Google Scholar] [CrossRef]
- Bakthavatsalam, S.; Das Sharma, S.; Sonawane, M.; Thirumalai, V.; Datta, A. A zebrafish model of manganism reveals reversible and treatable symptoms that are independent of neurotoxicity. Dis Model. Mech. 2014, 7, 1239–1251. [Google Scholar] [CrossRef]
- Polster, B.; Crosier, M.; Lindsay, S.; Hayflick, S. Expression of PLA2G6 in human fetal development: Implications for infantile neuroaxonal dystrophy. Brain Res. Bull. 2010, 83, 374–379. [Google Scholar] [CrossRef]
- Lin, G.; Lee, P.T.; Chen, K.; Mao, D.; Tan, K.L.; Zuo, Z. Phospholipase PLA2G6, a Parkinsonism-associated gene, affects Vps26 and Vps35, retromer function, and ceramide levels, similar to alpha-synuclein gain. Cell Metab. 2018, 28, 605–618.e6. [Google Scholar] [CrossRef]
- Schneider, S.A.; Bhatia, K.P. Rare causes of dystonia parkinsonism. Curr. Neurol. Neurosci. Rep. 2010, 10, 431–439. [Google Scholar] [CrossRef] [PubMed]
- Paisan-Ruiz, C.; Bhatia, K.P.; Li, A.; Hernandez, D.; Davis, M.; Wood, N.; Hardy, J.; Houlden, H.; Singleton, A.B.; Schneider, S.A. Characterization of PLA2G6 as a locus for dystonia-parkinsonism. Ann. Neurol. 2009, 65, 19–23. [Google Scholar] [CrossRef]
- Shi, C.-H.; Tang, B.-S.; Wang, L.; Lv, Z.-Y.; Wang, J.; Luo, L.-Z.; Shen, L.; Jiang, H.; Yan, X.-X.; Pan, Q.; et al. PLA2G6 gene mutation in autosomal recessive early-onset parkinsonism in a Chinese cohort. Neurology 2011, 77, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, H.; Tomiyama, H.; Tachibana, N.; Ogaki, K.; Li, Y.; Funayama, M.; Hashimoto, T.; Takashima, S.; Hattori, N. Phenotypic spectrum of patients with PLA2G6 mutation and PARK14-linked parkinsonism. Neurology 2010, 75, 1356–1361. [Google Scholar] [CrossRef]
- Guo, Y.P.; Tang, B.S.; Guo, J.F. PLA2G6-associated neurodegeneration (PLAN): Review of clinical phenotypes and genotypes. Front. Neurol. 2018, 9, 1100. [Google Scholar] [CrossRef]
- Shinzawa, K.; Sumi, H.; Ikawa, M.; Matsuoka, Y.; Okabe, M.; Sakoda, S.; Tsujimoto, Y. Neuroaxonal dystrophy caused by group VIA phospholipase A2 deficiency in mice: A model of human neurodegenerative disease. J. Neurosci. 2008, 28, 2212–2220. [Google Scholar] [CrossRef]
- Beck, G.; Shinzawa, K.; Hayakawa, H.; Baba, K.; Sumi-Akamaru, H.; Tsujimoto, Y. Progressive axonal degeneration of nigrostriatal dopaminergic neurons in calcium-independent phospholipase A2beta knockout mice. PLoS ONE 2016, 11, e0153789. [Google Scholar] [CrossRef]
- Sumi-Akamaru, H.; Beck, G.; Shinzawa, K.; Kato, S.; Riku, Y.; Yoshida, M.; Fujimura, H.; Tsujimoto, Y.; Sakoda, S.; Mochizuki, H. High expression of alpha-synuclein in damaged mitochondria with PLA2G6 dysfunction. Acta Neuropathol. Commun. 2016, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Basselin, M.; Rosa, A.O.; Ramadan, E.; Cheon, Y.; Chang, L.; Chen, M. Imaging decreased brain docosahexaenoic acid metabolism and signaling in iPLAbeta (VIA)-deficient mice. J. Lipid Res. 2010, 51, 3166–3173. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Yen, A.; Rymarczyk, G.; Asai, H.; Trengrove, C.; Aziz, N. Impairment of PARK14-dependent Ca(2+) signalling is a novel determinant of Parkinson’s disease. Nat. Commun. 2016, 7, 10332. [Google Scholar] [CrossRef] [PubMed]
- Chiu, C.-C.; Lu, C.-S.; Weng, Y.-H.; Chen, Y.-L.; Huang, Y.-Z.; Chen, R.-S.; Cheng, Y.-C.; Huang, Y.-C.; Liu, Y.-C.; Lai, S.-C.; et al. PARK14 (D331Y) PLA2G6 causes early-onset degeneration of substantia nigra dopaminergic neurons by inducing mitochondrial dysfunction, ER stress, mitophagy impairment and transcriptional dysregulation in a knockin mouse model. Mol. Neurobiol. 2019, 56, 3835–3853. [Google Scholar] [CrossRef]
- Mori, A.; Hatano, T.; Inoshita, T.; Shiba-Fukushima, K.; Koinuma, T.; Meng, H.; Kubo, S.-I.; Spratt, S.; Cui, C.; Yamashita, C.; et al. Parkinson’s disease-associated iPLA2-VIA/PLA2G6 regulates neuronal functions and alpha-synuclein stability through membrane remodeling. Proc. Natl. Acad. Sci. USA 2019, 116, 20689–20699. [Google Scholar] [CrossRef] [PubMed]
- Kinghorn, K.J.; Castillo-Quan, J.I.; Bartolome, F.; Angelova, P.R.; Li, L.; Pope, S. Loss of PLA2G6 leads to elevated mitochondrial lipid peroxidation and mitochondrial dysfunction. Brain 2015, 138, 1801–1816. [Google Scholar] [CrossRef]
- Shojaee, S.; Sina, F.; Banihosseini, S.S.; Kazemi, M.H.; Kalhor, R.; Shahidi, G.A. Genome-wide linkage analysis of a Parkinsonian-pyramidal syndrome pedigree by 500 K SNP arrays. Am. J. Hum. Genet. 2008, 82, 1375–1384. [Google Scholar] [CrossRef] [PubMed]
- Di Fonzo, A.; Dekker, M.; Montagna, P.; Baruzzi, A.; Yonova, E.H.; Guedes, L.C.; Szczerbinska, A.; Zhao, T.; Dubbel-Hulsman, L.O.; Wouters, C.; et al. FBXO7 mutations cause autosomal recessive, early-onset parkinsonian-pyramidal syndrome. Neurology 2009, 72, 240–245. [Google Scholar] [CrossRef] [PubMed]
- Lohmann, E.; Coquel, A.-S.; Honoré, A.; Gurvit, H.; Hanagasi, H.; Emre, M.; Leutenegger, A.-L.; Drouet, V.; Sahbatou, M.; Guven, G.; et al. A new F-box protein 7 gene mutation causing typical Parkinson’s disease. Mov. Disord. 2015, 30, 1130–1133. [Google Scholar] [CrossRef]
- Wei, L.; Ding, L.; Li, H.; Lin, Y.; Dai, Y.; Xu, X.; Dong, Q.; Lin, Y.; Long, L. Juvenile-onset parkinsonism with pyramidal signs due to compound heterozygous mutations in the F-Box only protein 7 gene. Parkinsonism Relat. Disord. 2018, 47, 76–79. [Google Scholar] [CrossRef]
- Zhao, T.; De Graaff, E.; Breedveld, G.J.; Loda, A.; Severijnen, L.A.; Wouters, C.H. Loss of nuclear activity of the FBXO7 protein in patients with parkinsonian-pyramidal syndrome (PARK15). PLoS ONE 2011, 6, e16983. [Google Scholar] [CrossRef]
- Delgado-Camprubi, M.; Esteras, N.; Soutar, M.P.; Plun-Favreau, H.; Abramov, A.Y. Deficiency of Parkinson’s disease-related gene Fbxo7 is associated with impaired mitochondrial metabolism by PARP activation. Cell Death Differ. 2017, 24, 2210. [Google Scholar] [CrossRef]
- Zhou, Z.D.; Xie, S.P.; Sathiyamoorthy, S.; Saw, W.T.; Sing, T.Y.; Ng, S.H.; Chua, H.P.H.; Tang, A.M.Y.; Shaffra, F.; Li, Z.; et al. F-box protein 7 mutations promote protein aggregation in mitochondria and inhibit mitophagy. Hum. Mol. Genet. 2015, 24, 6314–6330. [Google Scholar] [CrossRef]
- Vingill, S.; Brockelt, D.; Lancelin, C.; Tatenhorst, L.; Dontcheva, G.; Preisinger, C.; Schwedhelm-Domeyer, N.; Joseph, S.; Mitkovski, M.; Goebbels, S.; et al. Loss of FBXO7 (PARK15) results in reduced proteasome activity and models a parkinsonism-like phenotype in mice. EMBO J. 2016, 35, 2008–2025. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zondervan-van der Linde, H.; Severijnen, L.A.; Oostra, B.A.; Willemsen, R.; Bonifati, V. Dopaminergic neuronal loss and dopamine-dependent locomotor defects in Fbxo7-deficient zebrafish. PLoS ONE 2012, 7, e48911. [Google Scholar] [CrossRef] [PubMed]
- Goebbels, S.; Bormuth, I.; Bode, U.; Hermanson, O.; Schwab, M.H.; Nave, K.A. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis 2006, 44, 611–621. [Google Scholar] [CrossRef]
- Savitt, J.M.; Jang, S.S.; Mu, W.; Dawson, V.L.; Dawson, T.M. Bcl-x is required for proper development of the mouse substantia nigra. J. Neurosci. 2005, 25, 6721–6728. [Google Scholar] [CrossRef]
- Olgiati, S.; Quadri, M.; Fang, M.; Rood, J.P.; Saute, J.A.; Chien, H.F. DNAJC6 mutations associated with early-onset Parkinson’s disease. Ann. Neurol. 2016, 79, 244–256. [Google Scholar] [CrossRef]
- Edvardson, S.; Cinnamon, Y.; Ta-Shma, A.; Shaag, A.; Yim, Y.I.; Zenvirt, S. A deleterious mutation in DNAJC6 encoding the neuronal-specific clathrin-uncoating co-chaperone auxilin, is associated with juvenile parkinsonism. PLoS ONE 2012, 7, e36458. [Google Scholar] [CrossRef] [PubMed]
- Koroglu, C.; Baysal, L.; Cetinkaya, M.; Karasoy, H.; Tolun, A. DNAJC6 is responsible for juvenile parkinsonism with phenotypic variability. Parkinsonism Relat. Disord. 2013, 19, 320–324. [Google Scholar] [CrossRef]
- Shi, C.; Li, F.; Yang, J.; Zhang, S.; Mao, C.; Wang, H. DNAJC6 mutations are not common causes of early onset Parkinson’s disease in Chinese Han population. Neurosci. Lett. 2016, 634, 60–62. [Google Scholar] [CrossRef]
- Kanaoka, Y.; Kimura, S.H.; Okazaki, I.; Ikeda, M.; Nojima, H. GAK: A cyclin G associated kinase contains a tensin/auxilin-like domain. FEBS Lett. 1997, 402, 73–80. [Google Scholar] [CrossRef]
- Ahle, S.; Ungewickell, E. Auxilin, a newly identified clathrin-associated protein in coated vesicles from bovine brain. J. Cell Biol. 1990, 111, 19–29. [Google Scholar] [CrossRef]
- Yim, Y.I.; Sun, T.; Wu, L.G.; Raimondi, A.; De Camilli, P.; Eisenberg, E. Endocytosis and clathrin-uncoating defects at synapses of auxilin knockout mice. Proc. Natl. Acad. Sci. USA 2010, 107, 4412–4417. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; He, Y.; Ou, J.; Zhao, Y.; Li, R.; Cheng, J. Auxilin underlies progressive locomotor deficits and dopaminergic neuron loss in a Drosophila model of Parkinson’s disease. Cell Rep. 2017, 18, 1132–1143. [Google Scholar] [CrossRef]
- Krebs, C.E.; Karkheiran, S.; Powell, J.C.; Cao, M.; Makarov, V.; Darvish, H. The Sac1 domain of SYNJ1 identified mutated in a family with early-onset progressive Parkinsonism with generalized seizures. Hum. Mutat. 2013, 34, 1200–1207. [Google Scholar] [CrossRef]
- Quadri, M.; Fang, M.; Picillo, M.; Olgiati, S.; Breedveld, G.J.; Graafland, J.; Wu, B.; Xu, F.; Erro, R.; Amboni, M.; et al. Mutation in the SYNJ1 gene associated with autosomal recessive, early-onset Parkinsonism. Hum. Mutat. 2013, 34, 1208–1215. [Google Scholar] [CrossRef] [PubMed]
- Kirola, L.; Behari, M.; Shishir, C.; Thelma, B.K. Identification of a novel homozygous mutation Arg459Pro in SYNJ1 gene of an Indian family with autosomal recessive juvenile Parkinsonism. Parkinsonism Relat. Disord. 2016, 31, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Rauschendorf, M.A.; Jost, M.; Stock, F.; Zimmer, A.; Rosler, B.; Rijntjes, M. Novel compound heterozygous synaptojanin-1 mutation causes l-dopa-responsive dystonia-parkinsonism syndrome. Mov. Disord. 2017, 32, 478–480. [Google Scholar] [CrossRef] [PubMed]
- Taghavi, S.; Chaouni, R.; Tafakhori, A.; Azcona, L.J.; Firouzabadi, S.G.; Omrani, M.D.; Jamshidi, J.; Emamalizadeh, B.; Shahidi, G.A.; Ahmadi, M.; et al. A clinical and molecular genetic study of 50 families with autosomal recessive Parkinsonism revealed known and novel gene mutations. Mol. Neurobiol. 2018, 55, 3477–3489. [Google Scholar] [CrossRef] [PubMed]
- McPherson, P.S.; Garcia, E.P.; Slepnev, V.I.; David, C.; Zhang, X.; Grabs, D.; Sossini, W.S.; Bauerfeind, R.; Nemoto, Y.; De Camilli, P. A presynaptic inositol-5-phosphatase. Nature 1996, 379, 353–357. [Google Scholar] [CrossRef]
- Cremona, O.; Di Paolo, G.; Wenk, M.R.; Luthi, A.; Kim, W.T.; Takei, K.; Daniell, L.; Nemoto, Y.; Shears, S.; Flavell, R.A.; et al. Essential role of phosphoinositide metabolism in synaptic vesicle recycling. Cell 1999, 99, 179–188. [Google Scholar] [CrossRef]
- Pan, P.Y.; Sheehan, P.; Wang, Q.; Zhu, X.; Zhang, Y.; Choi, I. Synj1 haploinsufficiency causes dopamine neuron vulnerability and alpha-synuclein accumulation in mice. Hum. Mol. Genet. 2020, 29, 2300–2312. [Google Scholar] [CrossRef] [PubMed]
- Park, J.S.; Thorsness, M.K.; Policastro, R.; McGoldrick, L.L.; Hollingsworth, N.M.; Thorsness, P.E. Yeast Vps13 promotes mitochondrial function and is localized at membrane contact sites. Mol. Biol Cell. 2016, 27, 2435–2449. [Google Scholar] [CrossRef]
- Schmidt, E.M.; Schmid, E.; Munzer, P.; Hermann, A.; Eyrich, A.K.; Russo, A. Chorein sensitivity of cytoskeletal organization and degranulation of platelets. FASEB J. 2013, 27, 2799–2806. [Google Scholar] [CrossRef]
- Kumar, N.; Leonzino, M.; Hancock-Cerutti, W.; Horenkamp, F.A.; Li, P.; Lees, J.A.; Wheeler, H.; Reinisch, K.M.; De Camilli, P. VPS13A and VPS13C are lipid transport proteins differentially localized at ER contact sites. J. Cell Biol. 2018, 217, 3625–3639. [Google Scholar] [CrossRef] [PubMed]
- Peikert, K.; Danek, A.; Hermann, A. Current state of knowledge in Chorea-Acanthocytosis as core Neuroacanthocytosis syndrome. Eur. J. Med. Genet. 2018, 61, 699–705. [Google Scholar] [CrossRef]
- Tomiyasu, A.; Nakamura, M.; Ichiba, M.; Ueno, S.; Saiki, S.; Morimoto, M.; Kobal, J.; Kageyama, Y.; Inui, T.; Wakabayashi, K.; et al. Novel pathogenic mutations and copy number variations in the VPS13A gene in patients with chorea-acanthocytosis. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2011, 156B, 620–631. [Google Scholar] [CrossRef] [PubMed]
- Dulski, J.; Soltan, W.; Schinwelski, M.; Rudzinska, M.; Wojcik-Pedziwiatr, M.; Wictor, L. Clinical variability of neuroacanthocytosis syndromes-a series of six patients with long follow-up. Clin. Neurol. Neurosurg. 2016, 147, 78–83. [Google Scholar] [CrossRef]
- Lesage, S.; Drouet, V.; Majounie, E.; Deramecourt, V.; Jacoupy, M.; Nicolas, A. Loss of VPS13C function in autosomal-recessive Parkinsonism causes mitochondrial dysfunction and increases PINK1/Parkin-dependent mitophagy. Am. J. Hum. Genet. 2016, 98, 500–513. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Pankratz, N.; Lill, C.M.; Do, C.B.; Hernandez, D.G.; Saad, M.; DeStefano, A.L.; Kara, E.; Bras, J.; Sharma, M.; et al. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson’s disease. Nat. Genet. 2014, 46, 989–993. [Google Scholar] [CrossRef]
- Schormair, B.; Kemlink, D.; Mollenhauer, B.; Fiala, O.; Machetanz, G.; Roth, J. Diagnostic exome sequencing in early-onset Parkinson’s disease confirms VPS13C as a rare cause of autosomal-recessive Parkinson’s disease. Clin. Genet. 2018, 93, 603–612. [Google Scholar] [CrossRef]
- Darvish, H.; Bravo, P.; Tafakhori, A.; Azcona, L.J.; Ranji-Burachaloo, S.; Johari, A.H. Identification of a large homozygous VPS13C deletion in a patient with early-onset Parkinsonism. Mov. Disord. 2018, 33, 1968–1970. [Google Scholar] [CrossRef]
- Gu, X.; Li, C.; Chen, Y.; Ou, R.; Cao, B.; Wei, Q.; Hou, Y.; Zhang, L.; Song, W.; Zhao, B.; et al. Mutation screening and burden analysis of VPS13C in Chinese patients with early-onset Parkinson’s disease. Neurobiol. Aging 2020, 94, 311.e1–311.e4. [Google Scholar] [CrossRef] [PubMed]
- Bolus, H.; Crocker, K.; Boekhoff-Falk, G.; Chtarbanova, S. Modeling neurodegenerative disorders in Drosophila melanogaster. Int. J. Mol. Sci. 2020, 21, 3055. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).