Loss of parla Function Results in Inactivity, Olfactory Impairment, and Dopamine Neuron Loss in Zebrafish
Abstract
1. Introduction
2. Material and Methods
2.1. Animal Care and Husbandry
2.2. CRISPR Design, sgRNAs Synthesis and Microinjections
2.3. Description of parla Mutation
2.4. Collection of Zebrafish Brain Tissue
2.5. Histology and Immunohistochemistry (IHC)
2.6. RNA Isolation, cDNA Synthesis and qRT-PCR
2.7. Swimming Activity
2.8. Olfactory Function
2.9. Statistical Analysis
3. Results
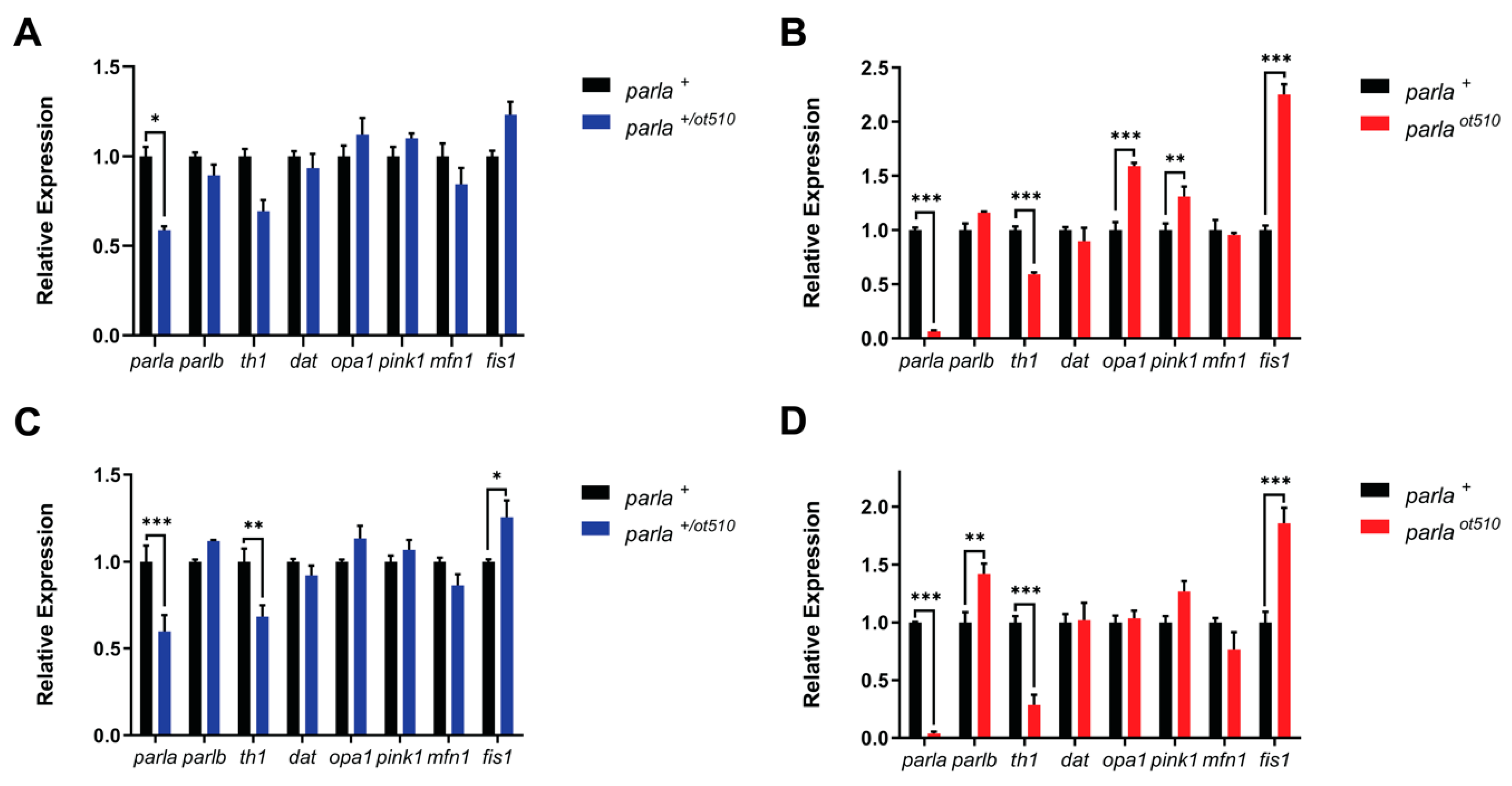
3.1. Gene Expression Analysis
3.1.1. Adult Brain
3.1.2. Larvae
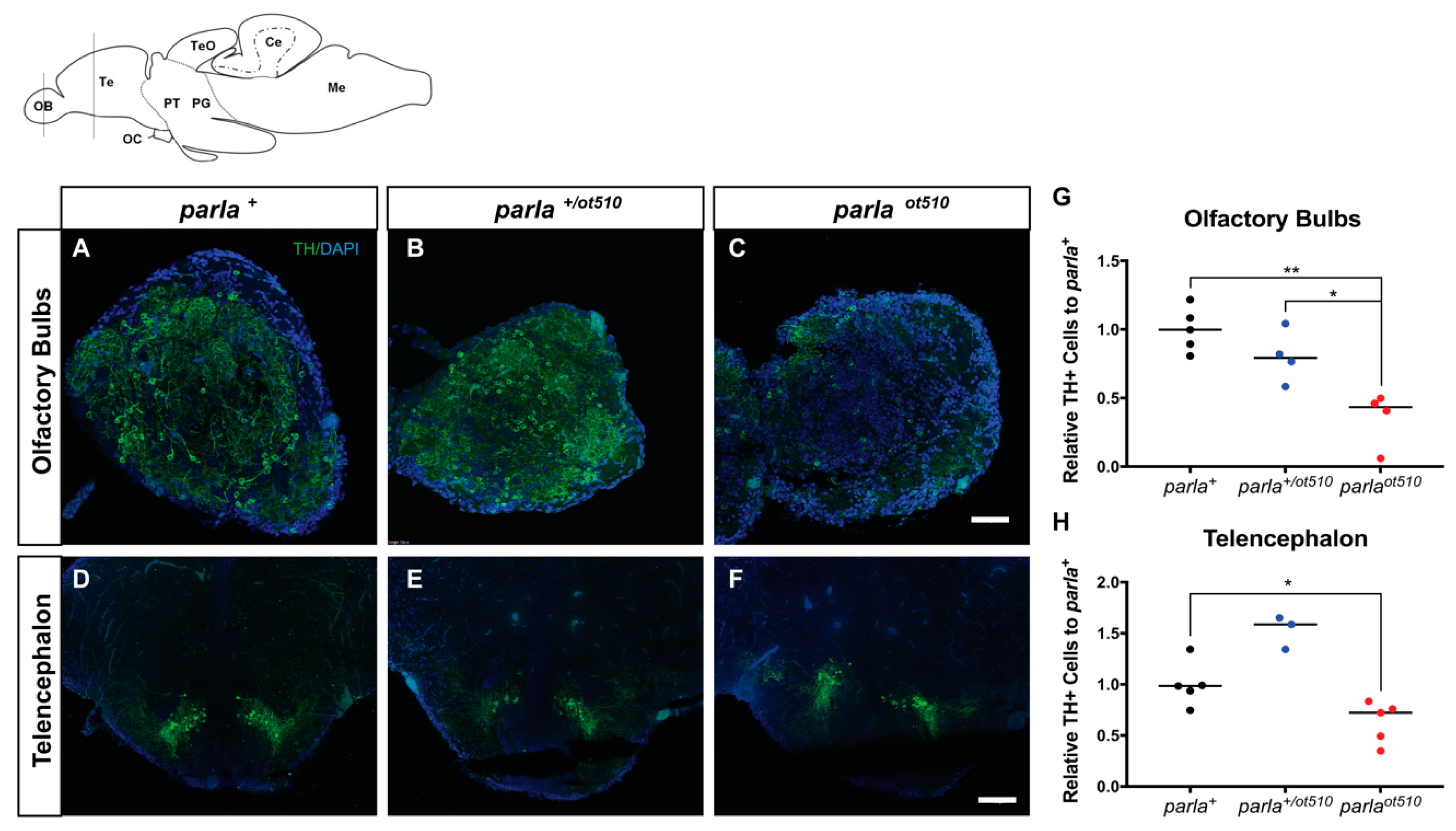
3.2. DA Neuronal Loss in the Telencephalon and Olfactory Bulb
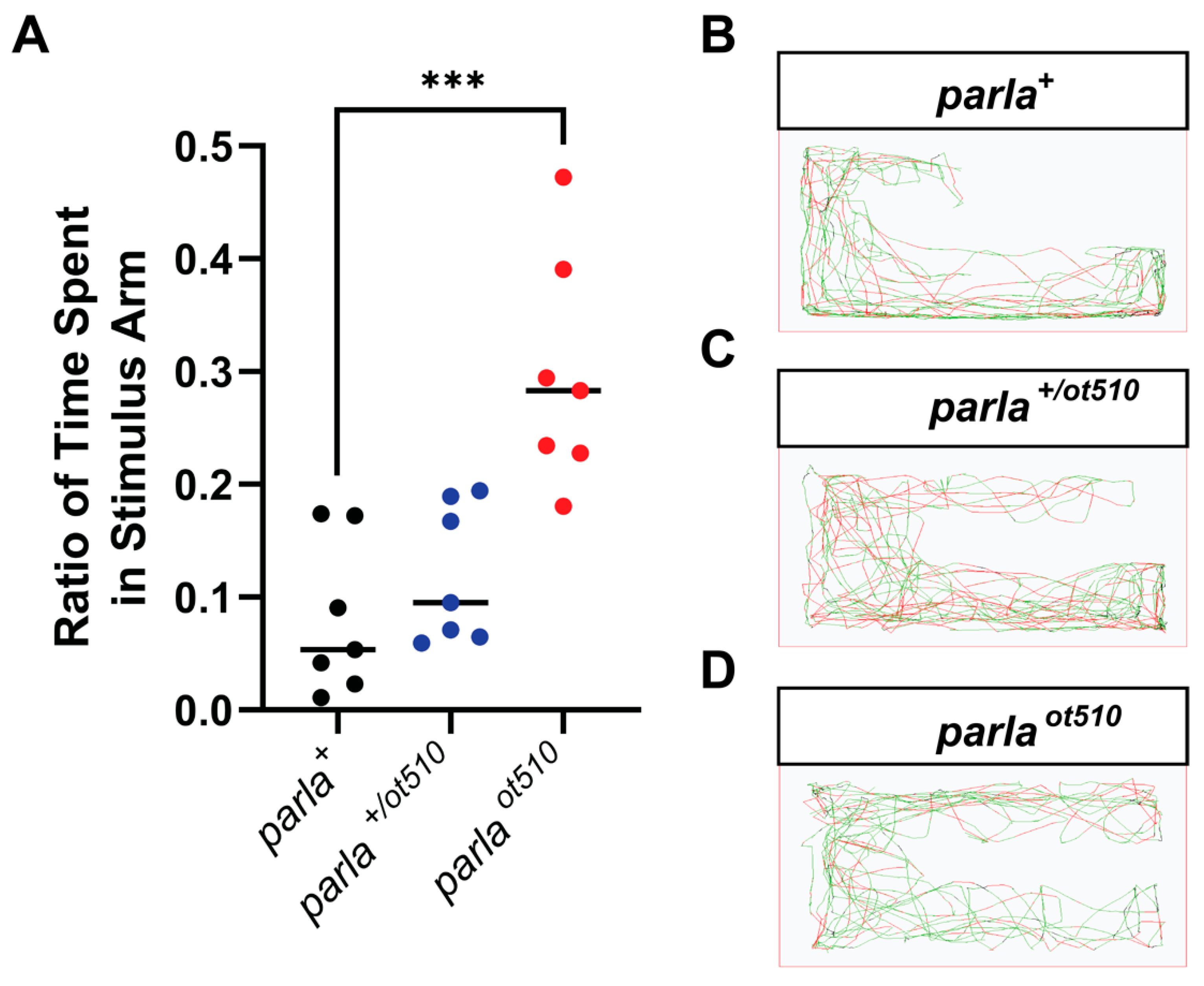
3.3. Effects on Olfactory Function
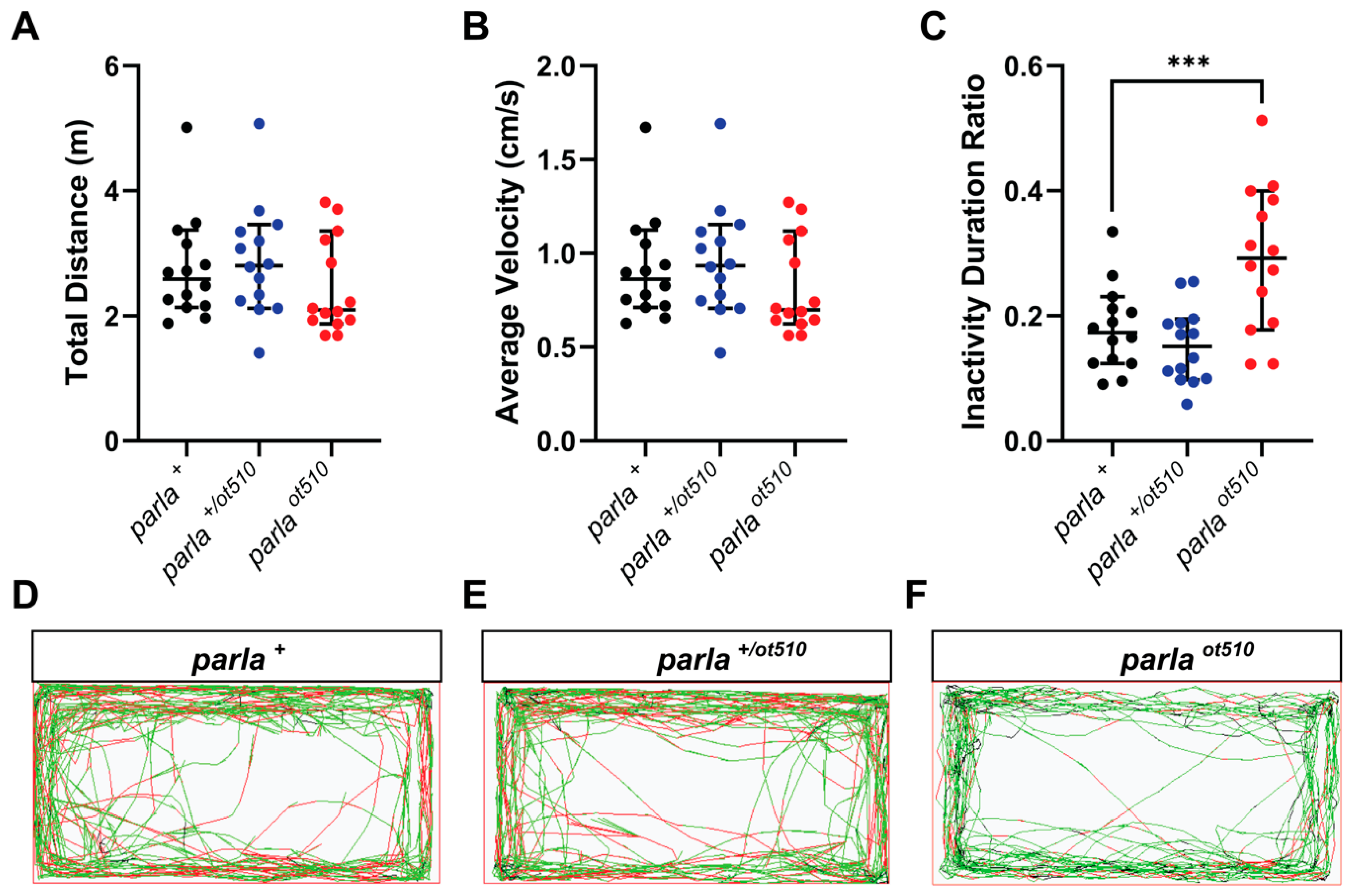
3.4. Effects on Locomotion
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lang, A.E. A critical appraisal of the premotor symptoms of Parkinson’s disease: Potential usefulness in early diagnosis and design of neuroprotective trials. Mov. Disord. 2011, 26, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Haehner, A.; Hummel, T.; Reichmann, H. Olfactory Loss in Parkinson’s Disease. Park. Dis. 2011, 2011, 1–6. [Google Scholar] [CrossRef]
- Heller, J.; Dogan, I.; Schulz, J.B.; Reetz, K. Evidence for Gender Differences in Cognition, Emotion and Quality of Life in Parkinson’s Disease? Aging Dis. 2013, 5, 63–75. [Google Scholar] [CrossRef]
- Klein, C.; Westenberger, A. Genetics of Parkinson’s Disease. Cold Spring Harb. Perspect. Med. 2012, 2, a008888. [Google Scholar] [CrossRef] [PubMed]
- Cerri, S.; Mus, L.; Blandini, F. Parkinson’s Disease in Women and Men: What’s the Difference? J. Park. Dis. 2019, 9, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Cookson, M.R. The biochemistry of Parkinson’s disease. Annu. Rev. Biochem. 2005, 74, 29–52. [Google Scholar] [CrossRef]
- Farrer, M.J. Genetics of Parkinson disease: Paradigm shifts and future prospects. Nat. Rev. Genet. 2006, 7, 306–318. [Google Scholar] [CrossRef]
- Whitworth, A.J.; Lee, J.R.; Ho, V.M.-W.; Flick, R.; Chowdhury, R.; McQuibban, G.A. Rhomboid-7 and HtrA2/Omi act in a common pathway with the Parkinson’s disease factors Pink1 and Parkin. Dis. Models Mech. 2008, 1, 168–174. [Google Scholar] [CrossRef]
- Westermann, B. Bioenergetic role of mitochondrial fusion and fission. Biochim. Biophys. Acta (BBA) Bioenerg. 2012, 1817, 1833–1838. [Google Scholar] [CrossRef]
- Mishra, P.; Chan, D.C. Metabolic regulation of mitochondrial dynamics. J. Cell Biol. 2016, 212, 379–387. [Google Scholar] [CrossRef] [PubMed]
- Twig, G.; Elorza, A.; Molina, A.A.J.; Mohamed, H.; Wikstrom, J.D.; Walzer, G.; Stiles, L.; Haigh, S.E.; Katz, S.; Las, G.; et al. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 2008, 27, 433–446. [Google Scholar] [CrossRef] [PubMed]
- Narendra, D.P.; Jin, S.M.; Tanaka, A.; Suen, D.-F.; Gautier, C.A.; Shen, J.; Cookson, M.R.; Youle, R.J. PINK1 Is Selectively Stabilized on Impaired Mitochondria to Activate Parkin. PLoS Biol. 2010, 8, e1000298. [Google Scholar] [CrossRef]
- Pickrell, A.M.; Youle, R.J. The Roles of PINK1, Parkin, and Mitochondrial Fidelity in Parkinson’s Disease. Neuron 2015, 85, 257–273. [Google Scholar] [CrossRef]
- Shi, G.; Lee, J.R.; Grimes, D.A.; Racacho, L.; Ye, D.; Yang, H.; Ross, O.A.; Farrer, M.; McQuibban, G.A.; Bulman, D.E. Functional alteration of PARL contributes to mitochondrial dysregulation in Parkinson’s disease. Hum. Mol. Genet. 2011, 20, 1966–1974. [Google Scholar] [CrossRef] [PubMed]
- Meissner, C.; Lorenz, H.; Hehn, B.; Lemberg, M.K. Intramembrane protease PARL defines a negative regulator of PINK1- and PARK2/Parkin-dependent mitophagy. Autophagy 2015, 11, 1484–1498. [Google Scholar] [CrossRef]
- Cipolat, S.; Rudka, T.; Hartmann, D.; Costa, V.; Serneels, L.; Craessaerts, K.; Metzger, K.; Frezza, C.; Annaert, W.; D’Adamio, L.; et al. Mitochondrial Rhomboid PARL Regulates Cytochrome c Release during Apoptosis via OPA1-Dependent Cristae Remodeling. Cell 2006, 126, 163–175. [Google Scholar] [CrossRef]
- Robea, M.A.; Balmus, I.-M.; Ciobica, A.; Strungaru, S.; Plavan, G.; Gorgan, L.D.; Savuca, A.; Nicoara, M. Parkinson’s Disease-Induced Zebrafish Models: Focussing on Oxidative Stress Implications and Sleep Processes. Oxidative Med. Cell. Longev. 2020, 2020, 1–15. [Google Scholar] [CrossRef]
- Metzger, J.M.; Emborg, M.E. Autonomic Dysfunction in Parkinson Disease and Animal Models. Clin. Auton. Res. 2019, 29, 397–414. [Google Scholar] [CrossRef]
- Godoy, R.; Hua, K.; Kalyn, M.; Cusson, V.-M.; Anisman, H.; Ekker, M. Dopaminergic neurons regenerate following chemogenetic ablation in the olfactory bulb of adult Zebrafish (Danio rerio). Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef] [PubMed]
- Rink, E.; Wullimann, M.F. Development of the catecholaminergic system in the early zebrafish brain: An immunohistochemical study. Dev. Brain Res. 2002, 137, 89–100. [Google Scholar] [CrossRef]
- Tay, T.L.; Ronneberger, O.; Ryu, S.; Nitschke, R.; Driever, W. Comprehensive catecholaminergic projectome analysis reveals single-neuron integration of zebrafish ascending and descending dopaminergic systems. Nat. Commun. 2011, 2, 171. [Google Scholar] [CrossRef]
- Noble, S.; Ismail, A.; Godoy, R.; Xi, Y.; Ekker, M. Zebrafish Parla- and Parlb-deficiency affects dopaminergic neuron patterning and embryonic survival. J. Neurochem. 2012, 122, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Montague, T.G.; Cruz, J.M.; Gagnon, J.A.; Church, G.M.; Valen, E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014, 42, W401–W407. [Google Scholar] [CrossRef]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Ahkmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; Schier, A.F. Efficient Mutagenesis by Cas9 Protein-Mediated Oligonucleotide Insertion and Large-Scale Assessment of Single-Guide RNAs. PLoS ONE 2014, 9, e98186. [Google Scholar] [CrossRef] [PubMed]
- Den Dunnen, J.T.; E Antonarakis, S. Nomenclature for the description of human sequence variations. Qual. Life Res. 2001, 109, 121–124. [Google Scholar] [CrossRef]
- Tang, R.; Dodd, A.; Lai, D.; McNabb, W.C.; Love, D.R. Validation of Zebrafish (Danio rerio) Reference Genes for Quantitative Real-time RT-PCR Normalization. Acta Biochim. Biophys. Sin. 2007, 39, 384–390. [Google Scholar] [CrossRef]
- Goedhart, J. Leaving the Bar in Five Steps. The Node. 24 March 2017. Available online: https://thenode.biologists.com/leaving-bar-five-steps/research/ (accessed on 28 December 2020).
- McQuibban, G.A.; Saurya, S.; Freeman, M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nat. Cell Biol. 2003, 423, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Vaz, R.L.; Outeiro, T.F.; Ferreira, J.J. Zebrafish as an Animal Model for Drug Discovery in Parkinson’s Disease and Other Movement Disorders: A Systematic Review. Front. Neurol. 2018, 9, 347. [Google Scholar] [CrossRef] [PubMed]
- McQuibban, G.A.; Lee, J.R.; Zheng, L.; Juusola, M.; Freeman, M. Normal Mitochondrial Dynamics Requires Rhomboid-7 and Affects Drosophila Lifespan and Neuronal Function. Curr. Biol. 2006, 16, 982–989. [Google Scholar] [CrossRef]
- Spinazzi, M.; Radaelli, E.; Horré, K.; Arranz, A.M.; Gounko, N.V.; Agostinis, P.; Maia, T.M.; Impens, F.; Morais, V.A.; Lopez-Lluch, G.; et al. PARL deficiency in mouse causes Complex III defects, coenzyme Q depletion, and Leigh-like syndrome. Proc. Natl. Acad. Sci. USA 2019, 116, 277–286. [Google Scholar] [CrossRef]
- Candy, J.; Collet, C. Two tyrosine hydroxylase genes in teleosts. Biochim. Biophys. Acta Gene Struct. Expr. 2005, 1727, 35–44. [Google Scholar] [CrossRef]
- Barrios, J.P.; Wang, W.-C.; England, R.; Reifenberg, E.; Douglass, A.D. Hypothalamic Dopamine Neurons Control Sensorimotor Behavior by Modulating Brainstem Premotor Nuclei in Zebrafish. Curr. Biol. 2020, 30, 4606–4618.e4. [Google Scholar] [CrossRef] [PubMed]
- Kalyn, M.; Hua, K.; Noor, S.M.; Wong, C.E.D.; Ekker, M. Comprehensive Analysis of Neurotoxin-Induced Ablation of Dopaminergic Neurons in Zebrafish Larvae. Biomedicines 2019, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Ross, G.W.; Petrovitch, H.; Abbott, R.D.; Tanner, C.M.; Popper, J.; Masaki, K.; Launer, L.; White, L.R. Association of olfactory dysfunction with risk for future Parkinson’s disease. Ann. Neurol. 2008, 63, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.M.; Lazarou, M.; Wang, C.; Kane, L.A.; Narendra, D.P.; Youle, R.J. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J. Cell Biol. 2010, 191, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Meissner, C.; Lorenz, H.; Weihofen, A.; Selkoe, D.J.; Lemberg, M.K. The mitochondrial intramembrane protease PARL cleaves human Pink1 to regulate Pink1 trafficking. J. Neurochem. 2011, 117, 856–867. [Google Scholar] [CrossRef]
- Morais, V.A.; Haddad, D.; Craessaerts, K.; De Bock, P.-J.; Swerts, J.; Vilain, S.; Aerts, L.; Overbergh, L.; Grünewald, A.; Seibler, P.; et al. PINK1 Loss-of-Function Mutations Affect Mitochondrial Complex I Activity via NdufA10 Ubiquinone Uncoupling. Science 2014, 344, 203–207. [Google Scholar] [CrossRef]
- Greene, A.W.; Grenier, K.; A Aguileta, M.; Muise, S.; Farazifard, R.; Haque, M.E.; McBride, H.M.; Park, D.S.; A Fon, E. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012, 13, 378–385. [Google Scholar] [CrossRef]
- Postlethwait, J.; Ruotti, V.; Carvan, M.J.; Tonellato, P.J. Automated Analysis of Conserved Syntenies for the Zebrafish Genome. Zebrafish Dis. Models Chem. Screens 2004, 77, 255–271. [Google Scholar] [CrossRef]
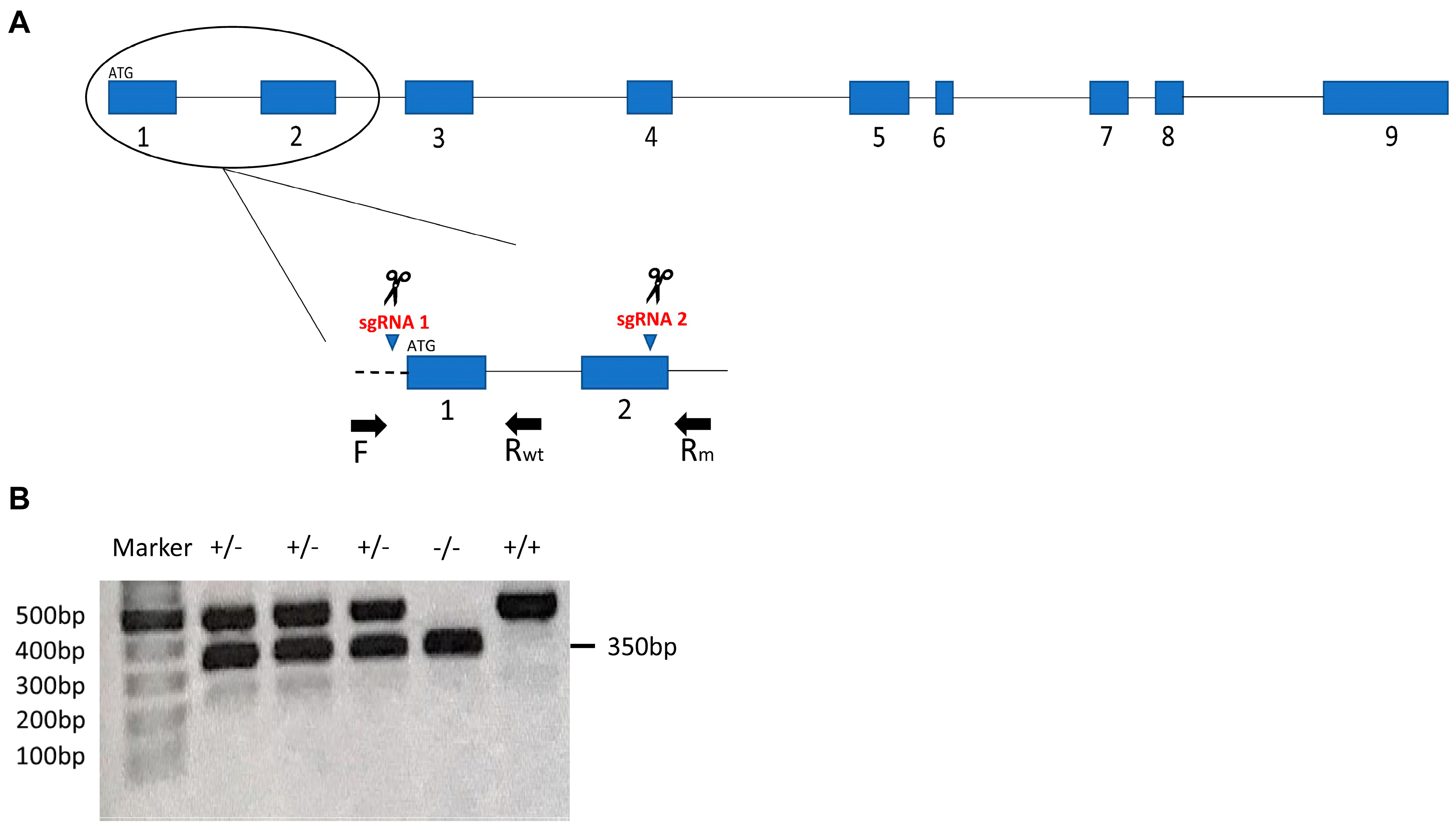
| Region Targeted | Target Sequence | Orientation |
|---|---|---|
| 5′ UTR | GGCGCGCTGTGAGCCGGAAG | 5′-3′ |
| 5′ UTR | GGAAAGCGCAGTTTTATGCA | 5′-3′ |
| Exon 2 | GCCAAATGACTTGGTGGGCC | 3′-5′ |
| Primer | Sequence (5′-3′) |
|---|---|
| F | CTCCAACAGGAGCCGCTATG |
| Rm | CTAGGGATTTTGTGCATGCTTAC |
| Rwt | ATTCATTATCCTGACAGGCC |
| Primer | Forward Sequence (5′-3′) | Reverse Sequence (5′-3′) |
|---|---|---|
| parla | GTGTTTTGTTGTTGGCGGGT | ATGTAGACAGCAGCATCGG |
| parlb | ATCACGCAGCACATCTTCCT | TATTAGTGGCTCGCGCTTCC |
| th1 | GACGGAAGATGATCGGAGACA | AGAAATCGGAACATGGCGG |
| dat | AGACATCTGGGAAGGTGGTG | ACCTGAGCATCATAGAGGCG |
| opa1 | GCTTGAGCCCTTGGAAAAGGAA | TGGCAGGTGATCTTGAGTGTTGT |
| pink1 | GGCAATGAAGATGATGTGGAAC | GGTCGGCAGGACATCAGGA |
| mfn1 | CTGGGTCCCGTCAACGCCAA | ACTGAACCACCGCTGGGGCT |
| fis1 | CCCTGAACCTTCCAGTGTTT | GTCTCTGGAAACGGGTCCTT |
| β-Act1 | CGAGCTGTCTTCCCATCCA | TCACCAACGTAGCTGTCTTTCTG |
| rpl13a1 | TCTGGAGGACTGTAAGAGGTATGC | AGACGCACAATCTTGAGAGCAG |
| ef1a1 | CTGGAGGCCAGCTCAAACAT | ATCAAGAAGAGTAGTACCGCTAGCATTAC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Merhi, R.; Kalyn, M.; Zhu-Pawlowsky, A.; Ekker, M. Loss of parla Function Results in Inactivity, Olfactory Impairment, and Dopamine Neuron Loss in Zebrafish. Biomedicines 2021, 9, 205. https://doi.org/10.3390/biomedicines9020205
Merhi R, Kalyn M, Zhu-Pawlowsky A, Ekker M. Loss of parla Function Results in Inactivity, Olfactory Impairment, and Dopamine Neuron Loss in Zebrafish. Biomedicines. 2021; 9(2):205. https://doi.org/10.3390/biomedicines9020205
Chicago/Turabian StyleMerhi, Rawan, Michael Kalyn, Amanda Zhu-Pawlowsky, and Marc Ekker. 2021. "Loss of parla Function Results in Inactivity, Olfactory Impairment, and Dopamine Neuron Loss in Zebrafish" Biomedicines 9, no. 2: 205. https://doi.org/10.3390/biomedicines9020205
APA StyleMerhi, R., Kalyn, M., Zhu-Pawlowsky, A., & Ekker, M. (2021). Loss of parla Function Results in Inactivity, Olfactory Impairment, and Dopamine Neuron Loss in Zebrafish. Biomedicines, 9(2), 205. https://doi.org/10.3390/biomedicines9020205






