BET Proteins Regulate Expression of Osr1 in Early Kidney Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Handling
2.2. Metanephric Kidney Culture
2.3. Genotyping
- Tomato/EGFP forward 5′ CTC TGC TGC CTC CTG GCT TCT 3′
- Tomato/EGFP reverse wildtype 5′ CGA GGC GGA TCA CAA GCA ATA 3′
- Tomato/EGFP reverse mutant 5′ TCA ATG GGC CGG GGT CGT T3′
- Cre forward 5′ GCA TTA CCG GTC GAT GCA ACG AGT GAT GAG 3′
- Cre reverse 5′ GAG TGA ACG AAC CTG GTC GAA ATC AGT GCG 3′
2.4. Whole Mount Immunofluorescence Staining
2.5. Imaging
2.6. ISH
| Brd2 | Forward: CGCGGGACGCGTAGCCTTCTCTGCTGTATGAGGG Reverse: CGCGGGGCGGCCGCGAGGACTAGCTGGGGAACCAGG |
| Brd3 | Forward: CGCGGGACGCGTCAGCCAGCAGACAGCTCA Reverse: CGCGGGGCGGCCGCTCTGTCTTCAGCCCCTGC |
| Brd4 (P0) | Forward: CGCGGGACGCGTCGAGGGAGGAAAGAAACAGGGG Reverse: CGCGGGGCGGCCGCTGGCTACCACTTCATGGTCAGG |
| Brd4 (whole mount) | Forward: CGCGGGACGCGTAAAGAAGCGCTTGGAAAACA Reverse: CGCGGGGCGGCCGCGATGCCACTGCAGCACTTTA |
| Cited1 | Forward: CGCGGGACGCGTATGCCAACCAGGAGATGAAC Reverse: CGCGGGGCGGCCGCCAACAGAATCGGTGGCTTTT |
| Eya1 | Forward: CGCGGGACGCGTTGCATATGGGCAAACACAGT Reverse: CGCGGGGCGGCCGCCCAGGTCCCAGATGAACACT |
| Lhx1 | Forward: CGCGGGACGCGTCAGTGTCGCCAAAGAGAACA Reverse: CGCGGGGCGGCCGCACAAATGGTTCCCGTAGCTG |
| Osr1 | Forward: CGCGGGACGCGTACAGAAATGGGCAGCAAAAC Reverse: CGCGGGGCGGCCGCGCGAGGCTTGGTCTTAAGTG |
| Pax8 | Forward: CGCGGGACGCGTATACACCTCTGGGACGCAAC Reverse: CGCGGGGCGGCCGCGCTTGGCCTTGATGTAGAGC |
| Ret | Forward: CGCGGGACGCGTTTGGTCCAGGTCAACAACAA Reverse: CGCGGGGCGGCCGCAGATGCCGTAGCCTGCTTTA |
| Six2 | Forward: CGCGGGACGCGTGGACCCACTGCAGCATCACC Reverse: CGCGGGGCGGCCGCTTCAGGTGCTTCTGGGGTGCAG |
| Wt1 | Forward: CGCGGGACGCGTGCCTTCACCTTGCACTTCTC Reverse: CGCGGGGCGGCCGCGCTGAAGGGCTTTTCACTTG |
2.7. qPCR
2.8. RNA-Sequencing
2.9. ChIP-Seq
2.10. Statistics
3. Results
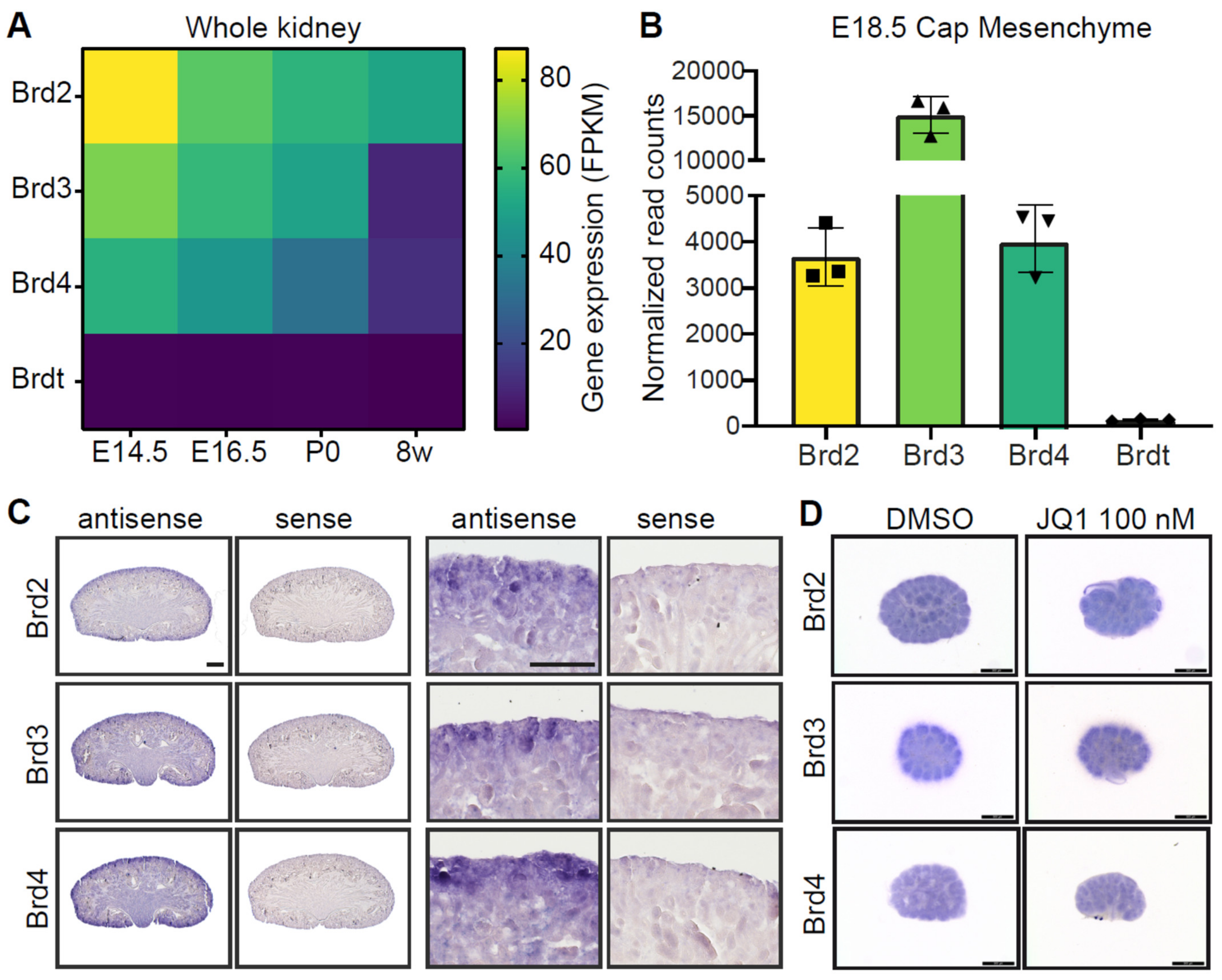
3.1. BET Proteins Are Expressed in the Developing Kidney
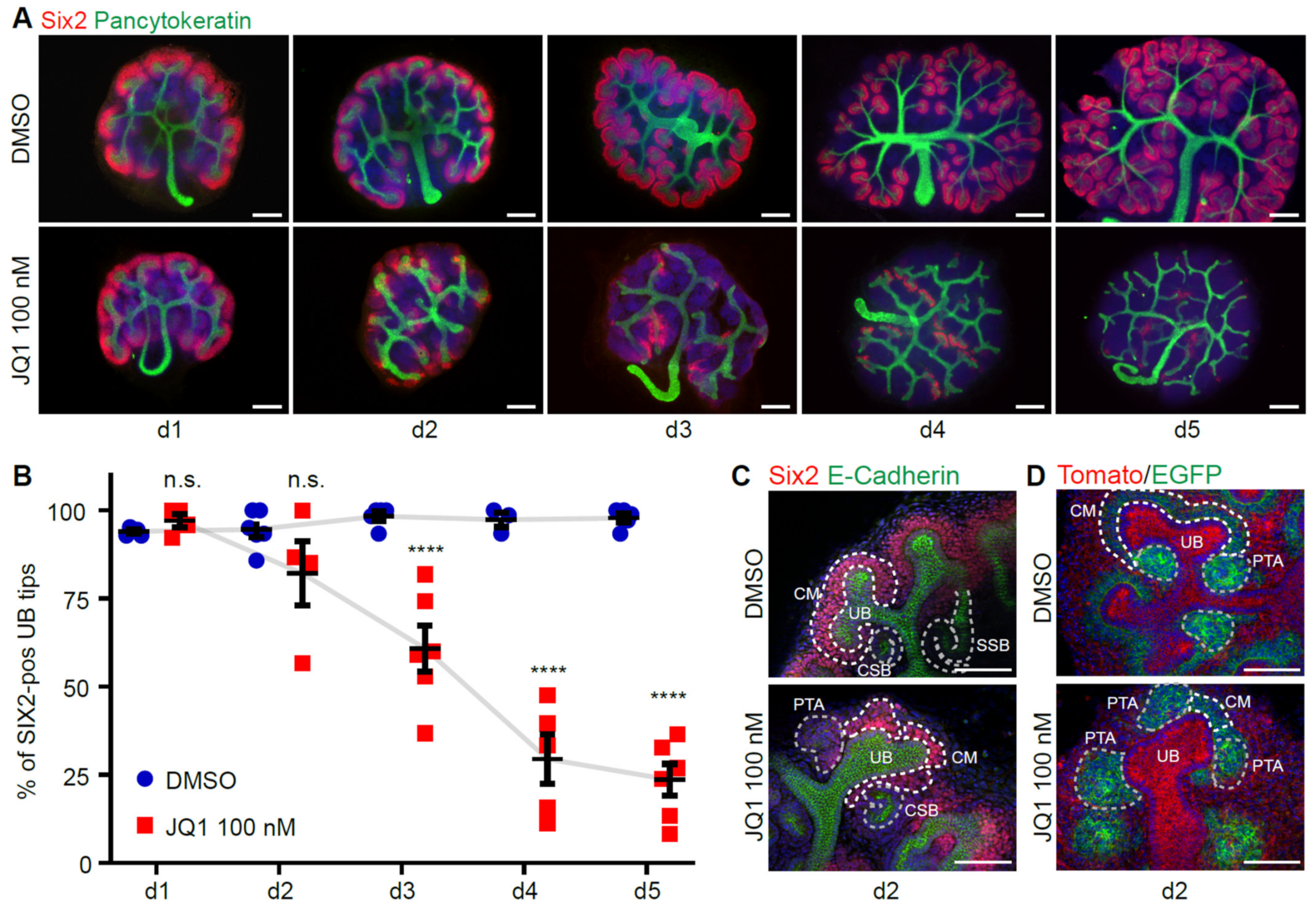
3.2. Inhibition of BET Proteins Impairs Metanephric Kidney Development
3.3. BET Inhibition Leads to Depletion of Nephron Progenitor Cells
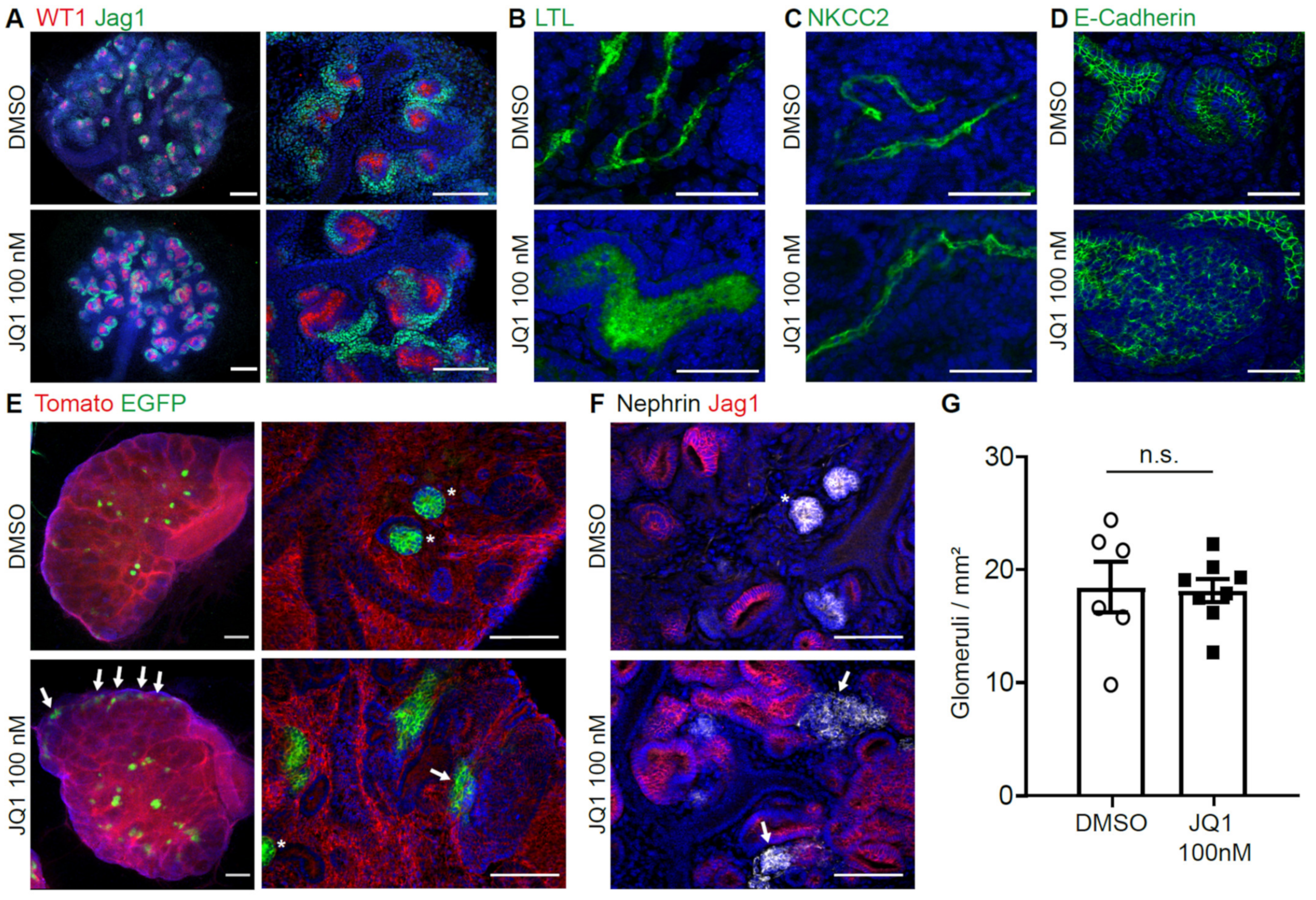
3.4. BET Inhibition Leads to Abnormal Nephron Differentiation
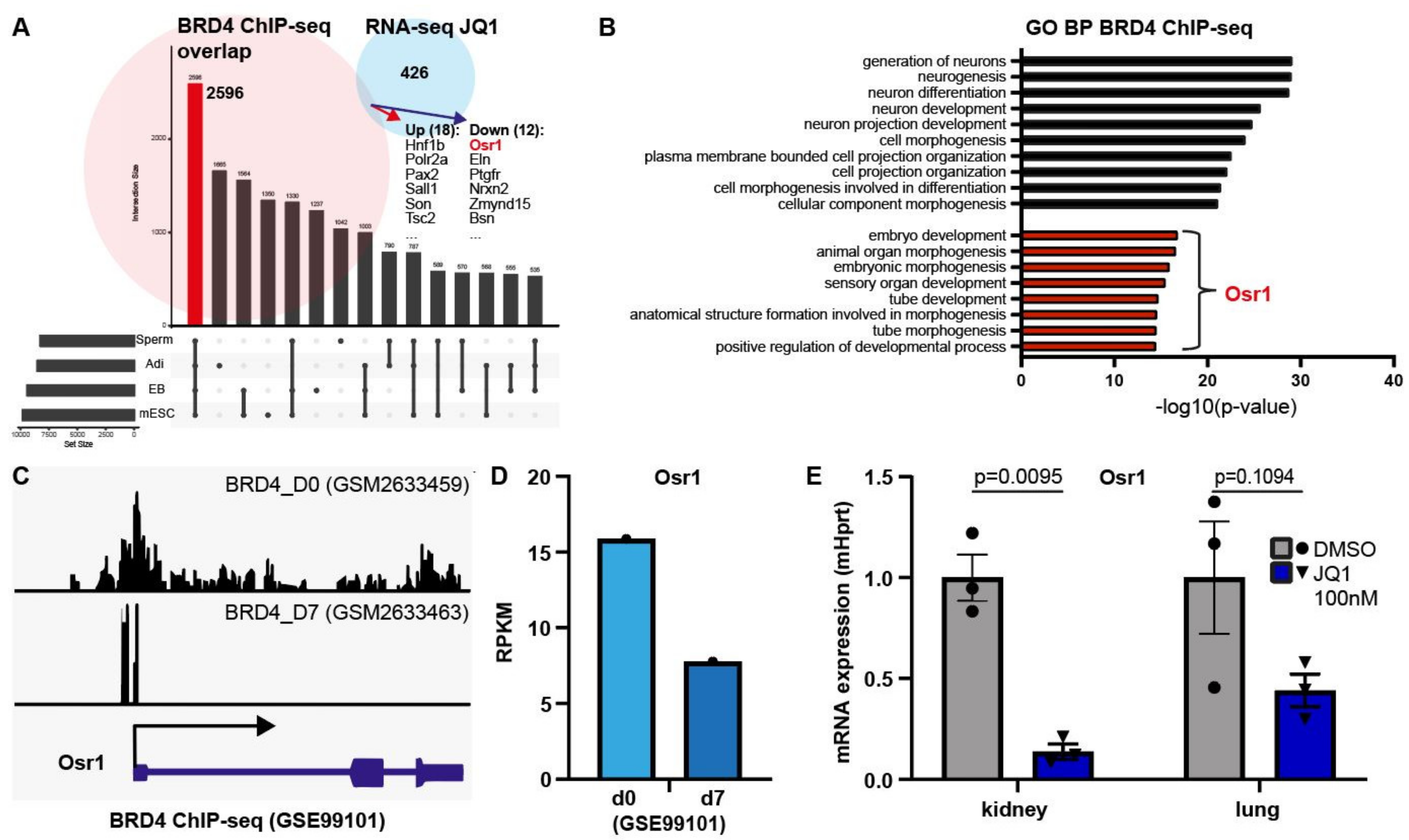
3.5. Loss of BET Protein Activity Leads to Downregulation of Osr1
3.6. BRD4 Regulates Osr1 Expression
4. Discussion
5. Summary
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Luyckx, V.A.; Brenner, B.M. The clinical importance of nephron mass. J. Am. Soc. Nephrol. 2010, 21, 898–910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brenner, B.M.; Garcia, D.L.; Anderson, S. Glomeruli and blood pressure. Less of one, more the other? Am. J. Hypertens. 1988, 1, 335–347. [Google Scholar] [CrossRef]
- Hinchliffe, S.A.; Sargent, P.H.; Howard, C.V.; Chan, Y.F.; van Velzen, D. Human intrauterine renal growth expressed in absolute number of glomeruli assessed by the disector method and Cavalieri principle. Lab. Investig. 1991, 64, 777–784. [Google Scholar] [PubMed]
- Keijzer-Veen, M.G.; Schrevel, M.; Finken, M.J.; Dekker, F.W.; Nauta, J.; Hille, E.T.; Frolich, M.; van der Heijden, B.J.; Dutch, P.-C.S.G. Microalbuminuria and lower glomerular filtration rate at young adult age in subjects born very premature and after intrauterine growth retardation. J. Am. Soc. Nephrol. 2005, 16, 2762–2768. [Google Scholar] [CrossRef] [Green Version]
- Luyckx, V.A.; Perico, N.; Somaschini, M.; Manfellotto, D.; Valensise, H.; Cetin, I.; Simeoni, U.; Allegaert, K.; Vikse, B.E.; Steegers, E.A.; et al. A developmental approach to the prevention of hypertension and kidney disease: A report from the Low Birth Weight and Nephron Number Working Group. Lancet 2017, 390, 424–428. [Google Scholar] [CrossRef] [Green Version]
- White, S.L.; Perkovic, V.; Cass, A.; Chang, C.L.; Poulter, N.R.; Spector, T.; Haysom, L.; Craig, J.C.; Salmi, I.A.; Chadban, S.J.; et al. Is low birth weight an antecedent of CKD in later life? A systematic review of observational studies. Am. J. Kidney Dis. 2009, 54, 248–261. [Google Scholar] [CrossRef]
- Luyckx, V.A.; Brenner, B.M. Clinical consequences of developmental programming of low nephron number. Anat. Rec. 2020, 303, 2613–2631. [Google Scholar] [CrossRef]
- Diehm, C.J.; Lumbers, E.R.; Weatherall, L.; Keogh, L.; Eades, S.; Brown, A.; Smith, R.; Johnson, V.; Pringle, K.G.; Rae, K.M. Assessment of Fetal Kidney Growth and Birth Weight in an Indigenous Australian Cohort. Front. Physiol. 2017, 8, 1129. [Google Scholar] [CrossRef] [Green Version]
- Schell, C.; Wanner, N.; Huber, T.B. Glomerular development—shaping the multi-cellular filtration unit. Semin. Cell. Dev. Biol. 2014, 36, 39–49. [Google Scholar] [CrossRef] [Green Version]
- Dressler, G.R. Advances in early kidney specification, development and patterning. Development 2009, 136, 3863–3874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Short, K.M.; Combes, A.N.; Lefevre, J.; Ju, A.L.; Georgas, K.M.; Lamberton, T.; Cairncross, O.; Rumballe, B.A.; McMahon, A.P.; Hamilton, N.A.; et al. Global quantification of tissue dynamics in the developing mouse kidney. Dev. Cell 2014, 29, 188–202. [Google Scholar] [CrossRef] [Green Version]
- Kopan, R.; Chen, S.; Little, M. Nephron progenitor cells: Shifting the balance of self-renewal and differentiation. Curr. Top. Dev. Biol. 2014, 107, 293–331. [Google Scholar]
- Costantini, F.; Kopan, R. Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev. Cell 2010, 18, 698–712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindstrom, N.O.; Tran, T.; Guo, J.; Rutledge, E.; Parvez, R.K.; Thornton, M.E.; Grubbs, B.; McMahon, J.A.; McMahon, A.P. Conserved and Divergent Molecular and Anatomic Features of Human and Mouse Nephron Patterning. J. Am. Soc. Nephrol 2018, 29, 825–840. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wanner, N.; Vornweg, J.; Combes, A.; Wilson, S.; Plappert, J.; Rafflenbeul, G.; Puelles, V.G.; Rahman, R.U.; Liwinski, T.; Lindner, S.; et al. DNA Methyltransferase 1 Controls Nephron Progenitor Cell Renewal and Differentiation. J. Am. Soc. Nephrol. 2019, 30, 63–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fuhrmann, L.; Lindner, S.; Hauser, A.T.; Hose, C.; Kretz, O.; Cohen, C.D.; Lindenmeyer, M.T.; Sippl, W.; Jung, M.; Huber, T.B.; et al. Effects of Environmental Conditions on Nephron Number: Modeling Maternal Disease and Epigenetic Regulation in Renal Development. Int. J. Mol. Sci. 2021, 22, 4157. [Google Scholar] [CrossRef]
- Wang, F.; Ngo, J.; Li, Y.; Liu, H.; Chen, C.H.; Saifudeen, Z.; Sequeira-Lopez, M.L.S.; El-Dahr, S.S. Targeted disruption of the histone lysine 79 methyltransferase Dot1L in nephron progenitors causes congenital renal dysplasia. Epigenetics 2020, 16, 1235–1250. [Google Scholar] [CrossRef]
- Liu, H.; Chen, S.; Yao, X.; Li, Y.; Chen, C.H.; Liu, J.; Saifudeen, Z.; El-Dahr, S.S. Histone deacetylases 1 and 2 regulate the transcriptional programs of nephron progenitors and renal vesicles. Development 2018, 145, dev153619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borck, P.C.; Guo, L.W.; Plutzky, J. BET Epigenetic Reader Proteins in Cardiovascular Transcriptional Programs. Circ. Res. 2020, 126, 1190–1208. [Google Scholar] [CrossRef]
- Belkina, A.C.; Denis, G.V. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer 2012, 12, 465–477. [Google Scholar] [CrossRef] [Green Version]
- Houzelstein, D.; Bullock, S.L.; Lynch, D.E.; Grigorieva, E.F.; Wilson, V.A.; Beddington, R.S. Growth and early postimplantation defects in mice deficient for the bromodomain-containing protein Brd4. Mol. Cell Biol. 2002, 22, 3794–3802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, E.; Wang, X.; Wen, D.; Greenberg, D.A.; Wolgemuth, D.J. Double bromodomain-containing gene Brd2 is essential for embryonic development in mouse. Dev. Dyn. 2009, 238, 908–917. [Google Scholar] [CrossRef] [Green Version]
- Gyuris, A.; Donovan, D.J.; Seymour, K.A.; Lovasco, L.A.; Smilowitz, N.R.; Halperin, A.L.; Klysik, J.E.; Freiman, R.N. The chromatin-targeting protein Brd2 is required for neural tube closure and embryogenesis. Biochim. Biophys. Acta 2009, 1789, 413–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Prakash, C.; Sum, C.; Gong, Y.; Li, Y.; Kwok, J.J.; Thiessen, N.; Pettersson, S.; Jones, S.J.; Knapp, S.; et al. Bromodomain-containing protein 4 (BRD4) regulates RNA polymerase II serine 2 phosphorylation in human CD4+ T cells. J. Biol. Chem. 2012, 287, 43137–43155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; He, N.; Zhou, Q. Brd4 recruits P-TEFb to chromosomes at late mitosis to promote G1 gene expression and cell cycle progression. Mol. Cell Biol. 2008, 28, 967–976. [Google Scholar] [CrossRef] [Green Version]
- Filippakopoulos, P.; Qi, J.; Picaud, S.; Shen, Y.; Smith, W.B.; Fedorov, O.; Morse, E.M.; Keates, T.; Hickman, T.T.; Felletar, I.; et al. Selective inhibition of BET bromodomains. Nature 2010, 468, 1067–1073. [Google Scholar] [CrossRef] [Green Version]
- Matzuk, M.M.; McKeown, M.R.; Filippakopoulos, P.; Li, Q.; Ma, L.; Agno, J.E.; Lemieux, M.E.; Picaud, S.; Yu, R.N.; Qi, J.; et al. Small-molecule inhibition of BRDT for male contraception. Cell 2012, 150, 673–684. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Rellinger, E.J.; Kim, K.W.; Craig, B.T.; Romain, C.V.; Qiao, J.; Chung, D.H. Bromodomain and extraterminal inhibition blocks tumor progression and promotes differentiation in neuroblastoma. Surgery 2015, 158, 819–826. [Google Scholar] [CrossRef] [Green Version]
- Moeller, M.J.; Sanden, S.K.; Soofi, A.; Wiggins, R.C.; Holzman, L.B. Podocyte-specific expression of cre recombinase in transgenic mice. Genesis 2003, 35, 39–42. [Google Scholar] [CrossRef] [Green Version]
- Muzumdar, M.D.; Tasic, B.; Miyamichi, K.; Li, L.; Luo, L. A global double-fluorescent Cre reporter mouse. Genesis 2007, 45, 593–605. [Google Scholar] [CrossRef]
- Kobayashi, A.; Valerius, M.T.; Mugford, J.W.; Carroll, T.J.; Self, M.; Oliver, G.; McMahon, A.P. Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 2008, 3, 169–181. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Araoka, T.; Wu, J.; Liao, H.K.; Li, M.; Lazo, M.; Zhou, B.; Sui, Y.; Wu, M.Z.; Tamura, I.; et al. 3D Culture Supports Long-Term Expansion of Mouse and Human Nephrogenic Progenitors. Cell Stem Cell 2016, 19, 516–529. [Google Scholar] [CrossRef] [Green Version]
- Rahmanian, S.; Murad, R.; Breschi, A.; Zeng, W.; Mackiewicz, M.; Williams, B.; Davis, C.A.; Roberts, B.; Meadows, S.; Moore, D.; et al. Dynamics of microRNA expression during mouse prenatal development. Genome Res. 2019, 29, 1900–1909. [Google Scholar] [CrossRef] [Green Version]
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed]
- Hochane, M.; van den Berg, P.R.; Fan, X.; Berenger-Currias, N.; Adegeest, E.; Bialecka, M.; Nieveen, M.; Menschaart, M.; Chuva de Sousa Lopes, S.M.; Semrau, S. Single-cell transcriptomics reveals gene expression dynamics of human fetal kidney development. PLoS Biol. 2019, 17, e3000152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [Green Version]
- Bryant, J.M.; Donahue, G.; Wang, X.; Meyer-Ficca, M.; Luense, L.J.; Weller, A.H.; Bartolomei, M.S.; Blobel, G.A.; Meyer, R.G.; Garcia, B.A.; et al. Characterization of BRD4 during mammalian postmeiotic sperm development. Mol. Cell Biol. 2015, 35, 1433–1448. [Google Scholar] [CrossRef] [Green Version]
- Gonzales-Cope, M.; Sidoli, S.; Bhanu, N.V.; Won, K.J.; Garcia, B.A. Histone H4 acetylation and the epigenetic reader Brd4 are critical regulators of pluripotency in embryonic stem cells. BMC Genom. 2016, 17, 95. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.E.; Park, Y.K.; Park, S.; Jang, Y.; Waring, N.; Dey, A.; Ozato, K.; Lai, B.; Peng, W.; Ge, K. Brd4 binds to active enhancers to control cell identity gene induction in adipogenesis and myogenesis. Nat. Commun. 2017, 8, 2217. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Wang, L.G.; He, Q.Y. ChIPseeker: An R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015, 31, 2382–2383. [Google Scholar] [CrossRef] [Green Version]
- Conway, J.R.; Lex, A.; Gehlenborg, N. UpSetR: An R package for the visualization of intersecting sets and their properties. Bioinformatics 2017, 33, 2938–2940. [Google Scholar] [CrossRef] [Green Version]
- Di Micco, R.; Fontanals-Cirera, B.; Low, V.; Ntziachristos, P.; Yuen, S.K.; Lovell, C.D.; Dolgalev, I.; Yonekubo, Y.; Zhang, G.; Rusinova, E.; et al. Control of embryonic stem cell identity by BRD4-dependent transcriptional elongation of super-enhancer-associated pluripotency genes. Cell Rep. 2014, 9, 234–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bertram, J.F.; Douglas-Denton, R.N.; Diouf, B.; Hughson, M.D.; Hoy, W.E. Human nephron number: Implications for health and disease. Pediatr. Nephrol. 2011, 26, 1529–1533. [Google Scholar] [CrossRef]
- Dawson, M.A.; Prinjha, R.K.; Dittmann, A.; Giotopoulos, G.; Bantscheff, M.; Chan, W.I.; Robson, S.C.; Chung, C.W.; Hopf, C.; Savitski, M.M.; et al. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 2011, 478, 529–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seal, J.; Lamotte, Y.; Donche, F.; Bouillot, A.; Mirguet, O.; Gellibert, F.; Nicodeme, E.; Krysa, G.; Kirilovsky, J.; Beinke, S.; et al. Identification of a novel series of BET family bromodomain inhibitors: Binding mode and profile of I-BET151 (GSK1210151A). Bioorg. Med. Chem. Lett. 2012, 22, 2968–2972. [Google Scholar] [CrossRef]
- Picaud, S.; Leonards, K.; Lambert, J.P.; Dovey, O.; Wells, C.; Fedorov, O.; Monteiro, O.; Fujisawa, T.; Wang, C.Y.; Lingard, H.; et al. Promiscuous targeting of bromodomains by bromosporine identifies BET proteins as master regulators of primary transcription response in leukemia. Sci. Adv. 2016, 2, e1600760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Wu, Y.; Tian, G.; Jiang, Y.; Liu, Z.; Meng, X.; Bao, X.; Feng, L.; Sun, H.; Deng, H.; et al. Chemical Proteomic Profiling of Bromodomains Enables the Wide-Spectrum Evaluation of Bromodomain Inhibitors in Living Cells. J. Am. Chem. Soc. 2019, 141, 11497–11505. [Google Scholar] [CrossRef]
- Costantini, F. GDNF/Ret signaling and renal branching morphogenesis: From mesenchymal signals to epithelial cell behaviors. Organogenesis 2010, 6, 252–262. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Liu, H.; Park, J.S.; Lan, Y.; Jiang, R. Osr1 acts downstream of and interacts synergistically with Six2 to maintain nephron progenitor cells during kidney organogenesis. Development 2014, 141, 1442–1452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mugford, J.W.; Sipila, P.; McMahon, J.A.; McMahon, A.P. Osr1 expression demarcates a multi-potent population of intermediate mesoderm that undergoes progressive restriction to an Osr1-dependent nephron progenitor compartment within the mammalian kidney. Dev. Biol. 2008, 324, 88–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liong, S.; Barker, G.; Lappas, M. Bromodomain protein BRD4 is increased in human placentas from women with early-onset preeclampsia. Reproduction 2018, 155, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Lim, R.; Nguyen-Ngo, C.; Lappas, M. Targeting bromodomain-containing proteins to prevent spontaneous preterm birth. Clin. Sci. 2019, 133, 2379–2400. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schreiber, J.; Liaukouskaya, N.; Fuhrmann, L.; Hauser, A.-T.; Jung, M.; Huber, T.B.; Wanner, N. BET Proteins Regulate Expression of Osr1 in Early Kidney Development. Biomedicines 2021, 9, 1878. https://doi.org/10.3390/biomedicines9121878
Schreiber J, Liaukouskaya N, Fuhrmann L, Hauser A-T, Jung M, Huber TB, Wanner N. BET Proteins Regulate Expression of Osr1 in Early Kidney Development. Biomedicines. 2021; 9(12):1878. https://doi.org/10.3390/biomedicines9121878
Chicago/Turabian StyleSchreiber, Janina, Nastassia Liaukouskaya, Lars Fuhrmann, Alexander-Thomas Hauser, Manfred Jung, Tobias B. Huber, and Nicola Wanner. 2021. "BET Proteins Regulate Expression of Osr1 in Early Kidney Development" Biomedicines 9, no. 12: 1878. https://doi.org/10.3390/biomedicines9121878
APA StyleSchreiber, J., Liaukouskaya, N., Fuhrmann, L., Hauser, A.-T., Jung, M., Huber, T. B., & Wanner, N. (2021). BET Proteins Regulate Expression of Osr1 in Early Kidney Development. Biomedicines, 9(12), 1878. https://doi.org/10.3390/biomedicines9121878






