1. Introduction
The tumor microenvironment (TME) is a dynamic and complex niche consisting of both cancer cells (CCs) and supporting stromal cells [
1]. Among them, endothelial cells (ECs) are critical determinants of tumor progression and immune evasion [
2]. ECs’ contribution to tumor development does not only rely on the formation of new blood vessels (angiogenic switch). Indeed, vascular networks are not just passive conduits, and ECs supply a specific pattern of membrane-bound and secreted elements to the surrounding cells (angiocrine switch), thus actively contributing to establishing a unique pro-tumorigenic milieu [
3,
4].
Accumulating evidence indicates that metabolic cooperation exists between CCs and stromal cells, the latter having a crucial role in shaping CC metabolism by shuttling metabolic intermediates to support CC proliferation and survival [
5]. Metabolic reprogramming is indeed a hallmark of CCs [
6]. Nevertheless, ECs metabolism is markedly perturbed in cancer, too [
7,
8]. Tumor-associated ECs (TECs) are indeed activated ECs, which undergo a metabolic rewiring to sustain neo-angiogenesis, the so-called “angiogenic metabolic switch” [
9]. However, changes in EC metabolism also affect the array of soluble factors and metabolites that are released in the TME, thus affecting CC metabolism. In this scenario, CCs and TECs establish a vicious cycle characterized by the bidirectional transfer of a complex network of signals and organic compounds. Targeting the metabolic interplay within the TME has gained attention as a promising strategy in cancer treatment [
10].
Heme, the complex of iron and porphyrin IX, is an essential molecule with an increasingly recognized role in cell energetic metabolism. Heme homeostasis relies on the coordinated activity of enzymes and transporters that modulate heme synthesis, import/export, catabolism, or utilization [
11,
12,
13,
14,
15]. In this context, heme synthesis and heme export have been shown to be co-regulated processes and constitute a unique functional axis interlinked with pathways related to cell energy production [
16]. Heme synthesis starts in mitochondria with the condensation of succinyl-CoA and glycine, a reaction catalyzed by 5-aminolevulinate synthase 1 (ALAS1), the rate-limiting enzyme of the biosynthetic process. Heme export is mainly controlled by the cell membrane transporter Feline Leukemia Virus subgroup C Receptor 1a (FLVCR1a). Interestingly, FLVCR1a inhibition in CCs or TECs leads to ALAS1 inhibition, thus resulting in reduced heme synthesis [
16]. These alterations in heme metabolism, in turn, promote the tricarboxylic acid (TCA) cycle flux and the oxidative phosphorylation (OXPHOS) in proliferating cells as CCs and TECs [
16].
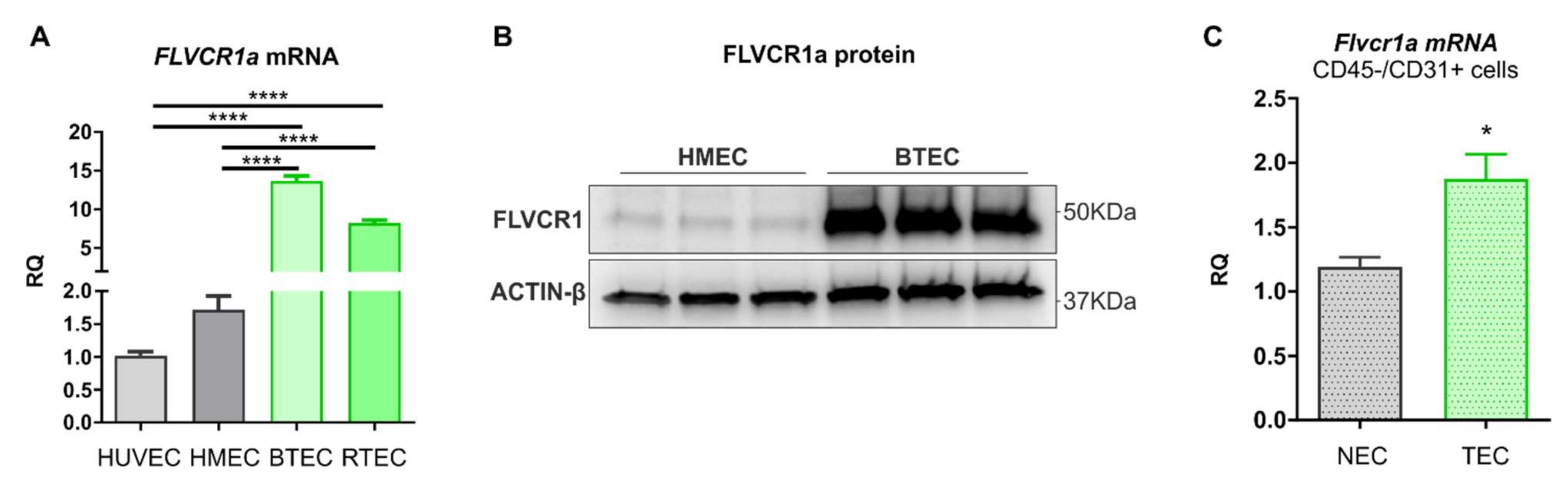
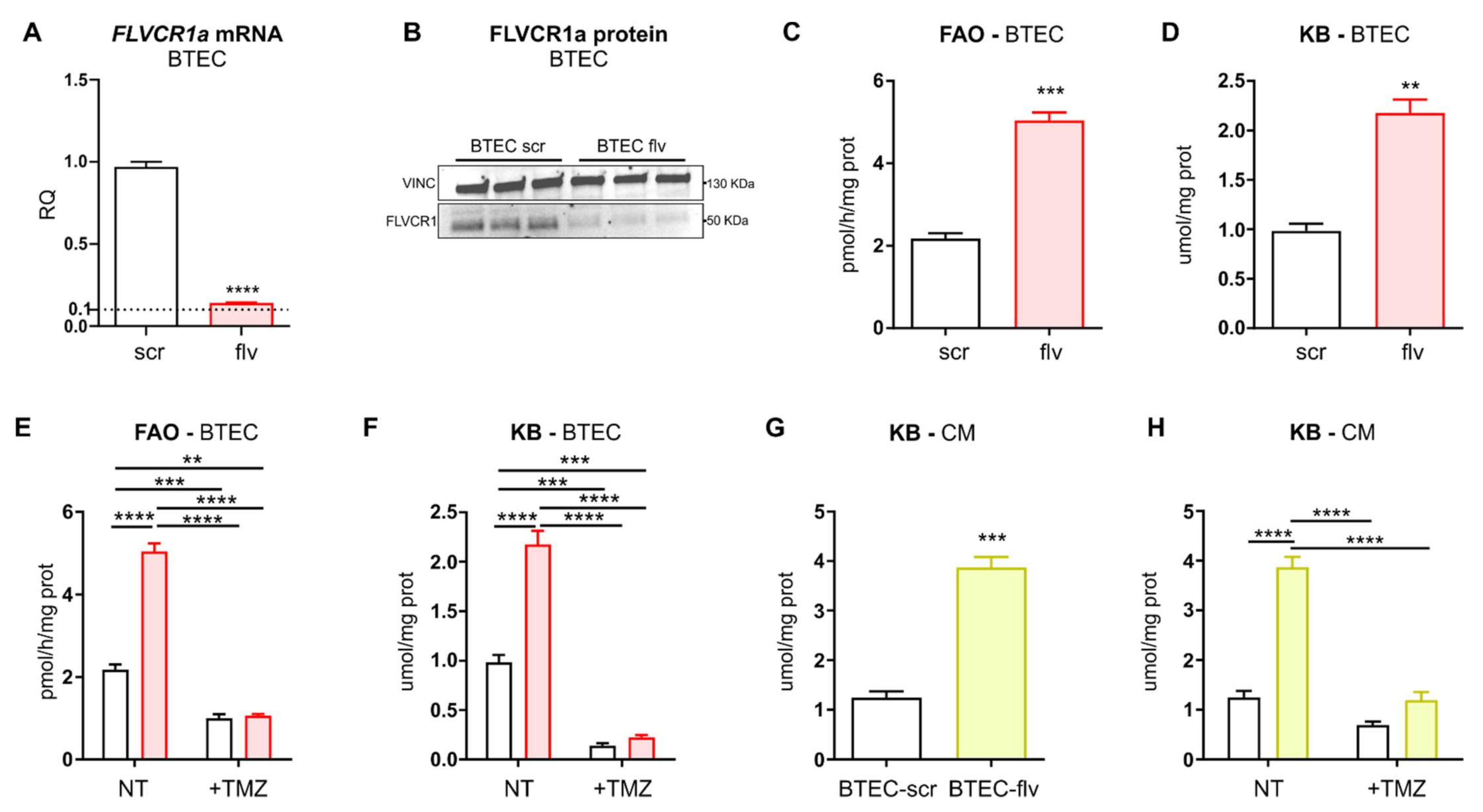
Here, we show that FLVCR1a is overexpressed in TECs, both in vitro and in vivo. Heme metabolism disruption in TEC by targeting FLVCR1a leads to enhanced fatty acid oxidation (FAO) and accumulation of ketone bodies (KBs), a likely side-product of fatty acids (FAs) breakdown. These metabolic alterations, in turn, modify the pattern of EC-derived metabolites, with a high release of KBs in the extracellular environment. Finally, our data indicate that elevated levels of KBs within the TME are sufficient to promote oxidative metabolism in the CC.
2. Materials and Methods
2.1. Cell Cultures
Human umbilical endothelial cells (HUVECs) were propagated in M199 medium (Invitrogen) with 20% heat-inactivated low-endotoxin fetal bovine serum (FBS; Gibco by Thermo Fisher Scientific, Waltham, MA, USA, catalog n° 10270106), 100 U/mL penicillin, 100 μg/mL streptomycin, 20 U/mL Heparin sodium salt from porcine intestinal mucosa (Sigma-Aldrich, St. Louis, MO, USA), and 10 ng/mL recombinant human FGF—basic (PeproTech, Hong Kong, China). HUVECs were used up to passage 6. Human adult dermal microvascular endothelial cells (HMECs) were purchased by Lonza and propagated in EndoGRO-MV-VEGF Complete Culture Media Kit (SCME003 Merck Millipore, Burlington, MA, USA) and used up to passage 12. Breast-tumor-derived endothelial cells (BTEC) and renal-tumor-derived endothelial cells (RTEC) from human breast lobular-infiltrating carcinoma biopsy and renal carcinoma, respectively, were isolated and characterized in the laboratory of Professor Benedetta Bussolati, Department of Molecular Biotechnology and Health Sciences, University of Torino, Torino, Italy [
17,
18]. BTECs and RTECs were maintained in EndoGRO-MV-VEGF Complete Culture Media Kit (SCME003 Merck Millipore, Burlington, MA, USA). Lewis lung carcinoma LL/2 (LLC1) cells (ATCC: CRL-1642) were cultured in Dulbecco’s modified Eagle’s medium (DMEM, high glucose, GlutaMAX supplement; Gibco by Thermo Fisher Scientific, Waltham, MA, USA, catalog n° 61965059) supplemented with 10% heat-inactivated low-endotoxin fetal bovine serum (FBS; Gibco by Thermo Fisher Scientific, Waltham, MA, USA, catalog n° 10270106). All cell media were ordinarily supplemented with antibiotics (100 μg/mL penicillin and 100 ug/mL streptomycin; Gibco by Thermo Fisher Scientific, Waltham, MA, USA, catalog n° 15140122). Cells were maintained in a 37 °C and 5% CO
2 air incubator and routinely screened for absence of mycoplasma contamination.
2.2. Reagents and Materials
BTECs were treated with 500 uM trimetazidine (1-(2,3,4-Trimethoxybenzyl) piperazine dihydrochloride, 653322 Sigma-Aldrich, St. Louis, MO, USA) dissolved in Phosphate-Buffered Saline (PBS) 1× solution for 72 h. Afterward, the conditioned medium (CM) and cell lysate were collected and used for metabolic analysis or in LLC cells treatment. LLC cells were treated with increasing concentrations (i.e., 1, 5, 10 mM) of β-hydroxybutyrate (H6501 Sigma-Aldrich, St. Louis, MO, USA) dissolved in PBS 1× solution.
2.3. FLVCR1a Gene Silencing
FLVCR1a silencing was performed as described in [
19]. Briefly, an shRNA targeting the sequence correspondent to the first exon of the human FLVCR1 gene was used to specifically down-regulate FLVCR1a (TRC Lentiviral pLKO.1 Human FLVCR1 shRNA set RHS4533-EG28982, clone TRCN0000059599; Dharmacon, Lafayette, CO, USA) in BTECs. For control cells, a pLKO.1 lentiviral vector expressing a scramble (scr) shRNA was used. The lentiviruses were produced in HEK293FT cells. Cells were infected with the lentiviruses in the presence of Sequabrene™ (S2667 Sigma-Aldrich, St. Louis, MO, USA). Following lentiviral transduction, cells were maintained in selective medium containing 0.002 mg/mL puromycin (puromycin dihydrochloride from Streptomyces alboniger, Sigma-Aldrich, St. Louis, MO, USA, catalog n° P8833).
2.4. Conditioned Medium (CM) Preparation
FLVCR1a-silenced and control BTECs (4–5 × 105) were seeded in 10 mm tissue-culture-treated dishes in the presence of 6 mL of EndoGRO-MV-VEGF Complete Culture Media. After 72 h, BTEC-derived CM was collected and centrifugated at 300× g at 4 °C to eliminate cell debris. Afterward, CM was diluted 1:3 with complete DMEM culture medium. The obtained diluted CM was used to treat LLC cells for 24 h.
2.5. Animals
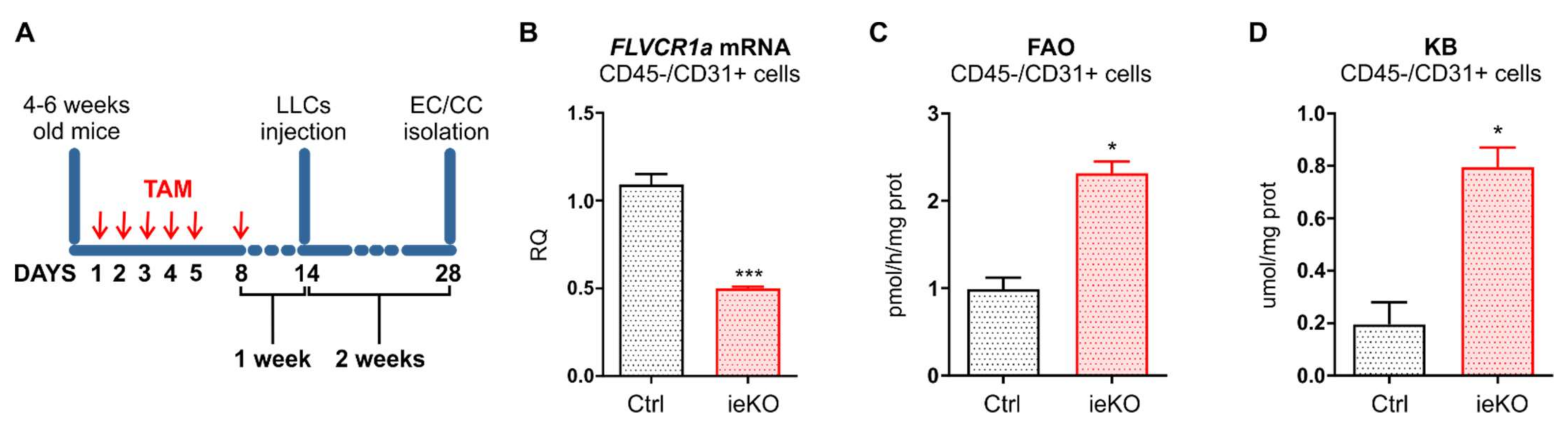
Tamoxifen-inducible, endothelial-specific Flvcr1a-null mice (Flvcr1aiEC-KO) were generated in our laboratory. Briefly, previously generated Flvcr1aflox/flox mice were crossed with Cdh5(PAC)-CreERT2 mice (Tg (Cdh5-cre/ERT2)1Rha, kindly provided by Ralf H. Adams) on a C57BL/6 background. Mice were genotyped by polymerase chain reaction (PCR) analyses on genomic DNA from tail biopsies. To detect the Cdh5-Cre allele, primers Cre-Fw (5′-ACACCTGCTACCATATCATCCTAC-3′) and Cre-Rev (5′-CATCGACCGGTAATGCAG-3′) were used. To analyze the LoxP sites on Flvcr1 gene, primers ILox-Fw (5′-TCTAAGGCCCAGTAGGACCC-3′) and ILox-Rev (5′-GAAAGCATTTCCGTCCGCCC-3′) were used, given a 280 bp band for the floxed allele and a 242 bp band for the wild-type allele. To inactivate Flvcr1a selectively in endothelial cells, 4–6 weeks old Flvcr1aflox/flox; Cdh5-CreERT2 mice were treated intraperitoneally with 1 mg/day tamoxifen (Sigma-Aldrich, St. Louis, MO, USA, catalog n° T5648) for 5 consecutive days, followed by 1 additional day after a 2-day treatment-free interval. To detect the Flvcr1a-null allele resulting from Cdh5-Cre activity, primers ILox-Fw (5′-TCTAAGGCCCAGTAGGACCC-3′) and IILox-Rev (5′-AGAGGGCAACCTCGGTGTCC-3′) were used, given a 320 bp fragment. Tamoxifen-treated Flvcr1aflox/flox mice were used as controls. All the mice were provided with food and water ad libitum. All experiments with animals were approved by the Italian Ministry of Health (562/2018-PR, 20 July 2018).
2.6. Xenograft Tumor Model
For the LLC xenograft model, 5 × 105 LL/2 (LLC1) murine cells suspended in 100 μL PBS were injected subcutaneously into the flanks of immunocompetent syngeneic C57BL/6 mice. For tumor induction in Flvcr1aiEC-KO mice and controls, mice were treated intraperitoneally with tamoxifen (Sigma-Aldrich, St. Louis, MO, USA; 1 mg/day for 5 consecutive days and 1 additional day after a 2-day treatment-free interval) one week before LLC cells injection.
2.7. Isolation of TEC and CC from LLC-Xenografts
Tumor-associated endothelial cells (TECs) and cancer cells (CCs) were isolated from LLC subcutaneous tumors developed in tamoxifen-inducible, endothelial-specific Flvcr1a-null mice (Flvcr1aiEC-KO) and controls. Briefly, tumors were dissected and minced into 1–2 mm fragments with a scalpel. Tissue pieces were incubated at 37 °C for 60 min in 10 mL of pre-warmed Dulbecco’s Phosphate Buffered Saline (DPBS) with calcium and magnesium (Lonza Pharma & Biotech, Basel, Switzerland, catalog n. BE17-513F) and 2 mg/mL collagenase (collagenase from Clostridium histolyticum, Type I, Sigma-Aldrich, St. Louis, MO, USA, catalog n. C0130), with regular shacking until a single cell suspension was obtained. During this incubation, the cells were mechanically dissociated at 10 min intervals by pipetting. To stop the collagenase activity, DMEM (GIBCO by Thermo Fisher Scientific, Waltham, MA, USA, catalog n. 61965059) containing 10% FBS (GIBCO by Thermo Fisher Scientific, Waltham, MA, USA, catalog n. 10270106) was added to the cell suspension, gently pelleted, and rinsed with PBS. The cells in PBS were then filtered through a 40 mm cell strainer (Corning Life Sciences, Corning, NY, USA, catalog n. 352340). Single-cell suspension was centrifuged at 300× g for 10 min, and TECs/CCs were isolated through MACS Technology by using nano-sized MicroBeads, following the manufacturer’s instructions. Particularly, a negative selection was performed using CD45 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany, catalog n. 130-052-301). CD45-negative cell fraction was then pelleted and incubated with CD31 MicroBeads (Miltenyi Biotec, Bergisch Gladbach, Germany, catalog n. 130-097-418) to isolate TECs (CD45−/CD31+ cell fraction). CCs resulted from the CD45−/CD31− cell fraction.
2.8. RNA Extraction and Quantitative Real-Time PCR Analysis
RNA extraction and quantitative real-time PCR (qRT-PCR) analyses were performed as described previously [
19]. Briefly, total RNA was extracted using PureLink RNA Mini Kit (Thermo Fisher Scientific, Waltham, MA, USA), and 0.5–1 μg of total RNA was transcribed into complementary DNA (cDNA) by High-Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). qRT-PCR was performed using gene-specific TaqMan™ Gene Expression Assays (Thermo Fisher Scientific Waltham, MA, USA). To detect
FLVCR1a expression, specific primers and probes were designed using Primer Express Software Version 3.0 (Thermo Fisher Scientific, Waltham, MA, USA). qRT-PCR was performed on a 7900HT Fast or QuantStudio™ 6 Flex Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA), and the analyses were completed using RQ Manager or QuantStudio Real-Time PCR software. Transcript abundance, normalized to 18 s messenger ribonucleic acid (mRNA) expression, is expressed as a fold change over a calibrator sample.
2.9. Western Blot Analysis
To assess FLVCR1a expression, BTECs were lysed by rotation for 30 min at 4 °C in RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.5, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 1 mM EDTA). The buffer was freshly supplemented with 1 mM phosphatase inhibitor cocktail (Sigma-Aldrich, St. Louis, MO, USA, catalog n° P0044), 1 mM PMSF (Sigma-Aldrich, St. Louis, MO, USA, catalog n° 93482-50ML-F), and protease-inhibitor cocktail (La Roche, Basel, Switzerland, catalog n° 04693116001). The cell lysate was clarified by centrifugation for 10 min at 4 °C. Protein concentration in the supernatant was assessed by Bradford assay. For FLVCR1a protein detection, 10 μg of protein extracts were incubated 10 min at 37 °C with 1 μL of PNGase-F from Elizabethkingia meningoseptica (Sigma-Aldrich, St. Louis, MO, USA, catalog n° P-7367) to remove protein glycosylation. Before loading on 4–15% mini-PROTEAN TGX precast gel (Bio-Rad, Hercules, CA, USA, catalog n° 4568084), samples were incubated 5 min at 37 °C (FLVCR1) or 10 min at 95 °C (Vinculin, Actin-β) in 4× Laemmli buffer freshly supplemented with 8% 2-mercaptoethanol. The primary antibodies and dilutions are as follows: FLVCR1 (C-4) (Santa Cruz Biotechnology, Dallas, TX, USA, catalog n° sc-390100; 1:500), Vinculin (homemade, 1:8000), and Actin-β (Invitrogen, catalog n° MA5-32540; 1:1000). The revelation was assessed using the ChemiDoc Imaging System (Bio-Rad, Hercules, CA, USA).
2.10. Fatty Acids β-Oxidation Measurement
Cells were washed with fresh medium, detached with trypsin/EDTA, re-suspended at 1 × 10
5 cells/mL in 0.2 mL of 100 mM Tris 10 mM/EDTA, and 50 µL aliquots were sonicated and used for protein measurements and normalization using the BCA1 kit (Sigma-Aldrich, St. Louis, MO, USA). The remaining samples were centrifuged at 13,000×
g for 5 min at room temperature and re-suspended in 0.5 mL Hepes 20 mM (pH 7.4), containing 0.24 mM fatty acid-free BSA, 0.5 mM
l-carnitine, 2 µCi [1-
14C]palmitic acid (3.3 mCi/mmol, PerkinElmer, Waltham, MA, USA) and transferred into test tubes that were tightly sealed with rubber caps. In each experimental set, cells were pre-incubated for 30 min with the carnitine palmitoyltransferase inhibitor etomoxir (1 µM) or with the AMP-kinase activator 5-aminoimidazole-4-carboxamide ribonucleotide AICAR (1 mM), as negative and positive controls, respectively. After 2 h incubation at 37 °C, 0.3 mL of a 1:1
v/
v phenylethylamine/methanol solution was added to each sample using a syringe, followed by 0.3 mL 0.8 N HClO
4. Samples were incubated for a further 1 h at room temperature, then centrifuged at 13,000×
g for 10 min. Both the supernatants, containing
14CO
2, and the precipitates, containing
14C-acid-soluble metabolites (ASM), i.e., the main products of fatty acid β-oxidation, were collected. The radioactivity of each supernatant and precipitate was counted by liquid scintillation, according to [
20].
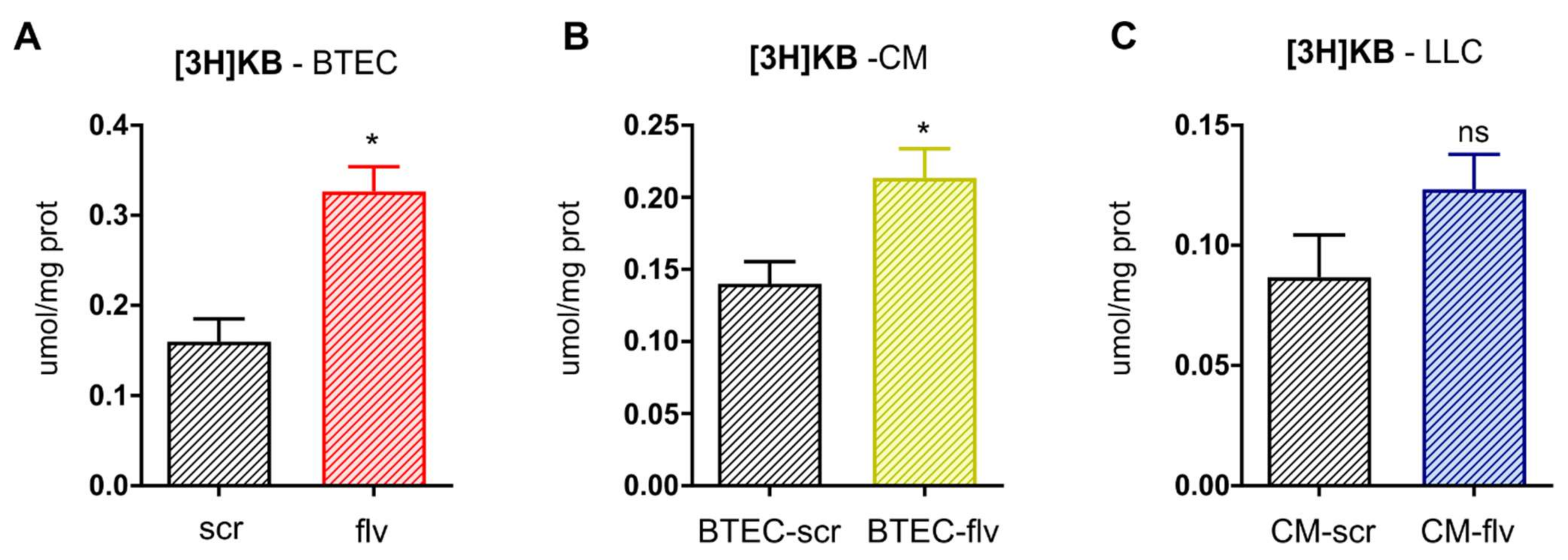
2.11. Ketone Bodies Measurement
The amount of β-hydroxybutyrate, taken as an index of ketone bodies (KBs) amount, was assessed in whole-cell lysate as well as in conditioned medium using the β-hydroxybutyrate Colorimetric Assay Kit (Cayman Chemical, Ann Arbor, MI, USA), as per manufacturer’s instructions. Results were expressed as µmol/mg cellular proteins.
2.12. Evaluation of Ketone Bodies Flux
BTEC cells were incubated 24 h with 1 µCi [
3H]-acetate (3.6 mCi/mmol, PerkinElmer), then washed five times with PBS, and let grow for additional 48 h in fresh medium. After 72 h of radiolabeling, both cells and medium were collected. One aliquot of medium was used to generate the BTEC CM and incubated 24 h on LLC cells; the remaining part was used for the measurement of acetoacetate, as reported [
21]. BTEC cells and LLC cells were collected by gentle scraping, re-suspended in PBS, and sonicated. A total of 100 µL cell lysates was used to measure the protein amount. A total of 300 µL of the lysates or of the medium were diluted 1:3 in the assay buffer (0.2 mM 3-(2-hydroxyphenyl) propionic acid, 10 mM
p-Nitrobenzene diazonium fluoroborate solution, 0.4 M citrate buffer, pH 3.5, at 1:1:2 volume). The samples were incubated 5 min at 37 °C degree and resolved by an HPLC system equipped with a UV detector (SPD-20A) (Shimadzu, Kyoto, Japan), using the elution conditions described in [
21]. The amount of [
3H]-acetoacetate, an index of KBs synthesized by [
3H]-acetate, was quantified according to calibration curves of serial dilutions of acetoacetate and expressed as µmol/mg cell proteins.
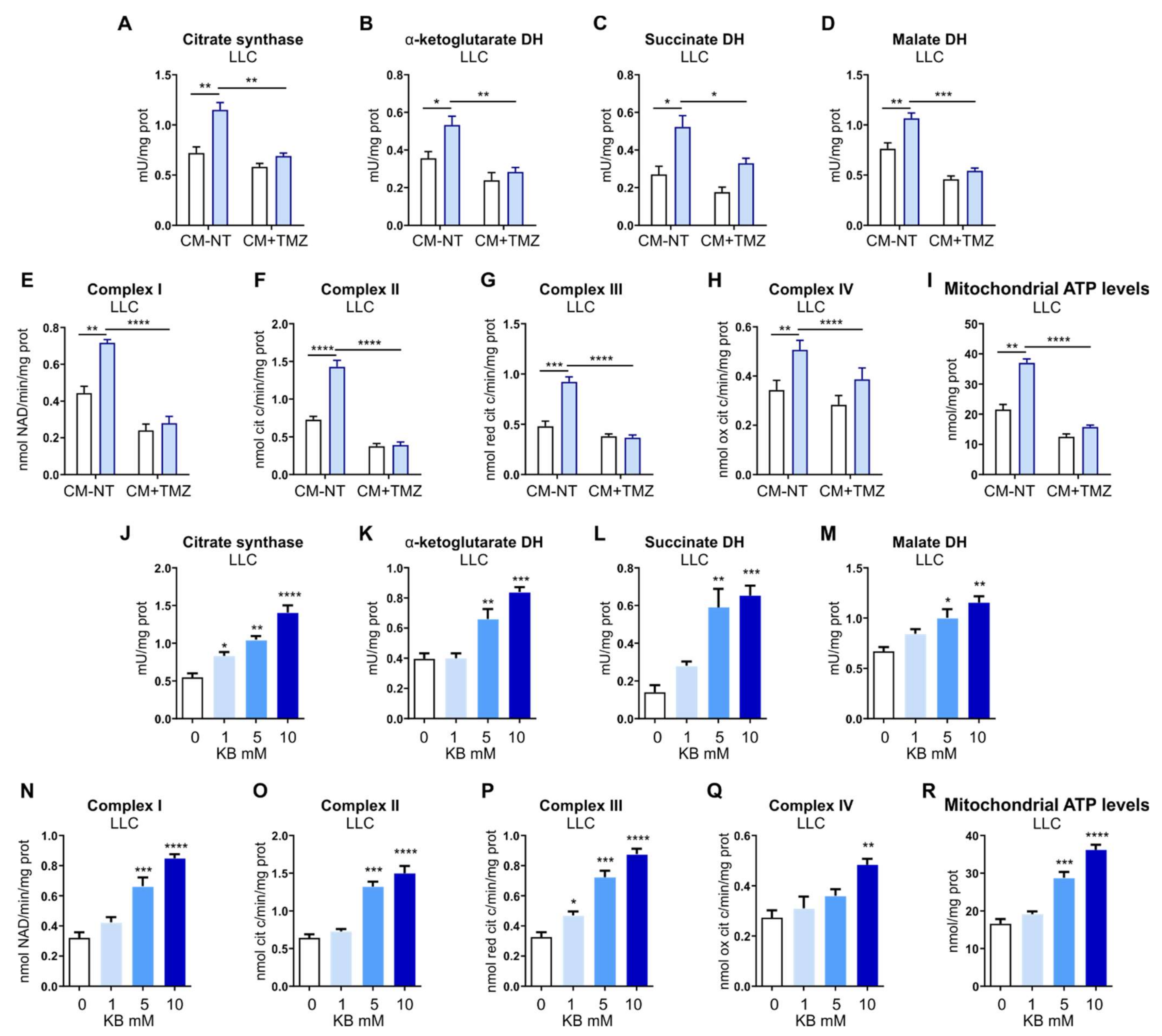
2.13. TCA Cycle Enzymes Activity
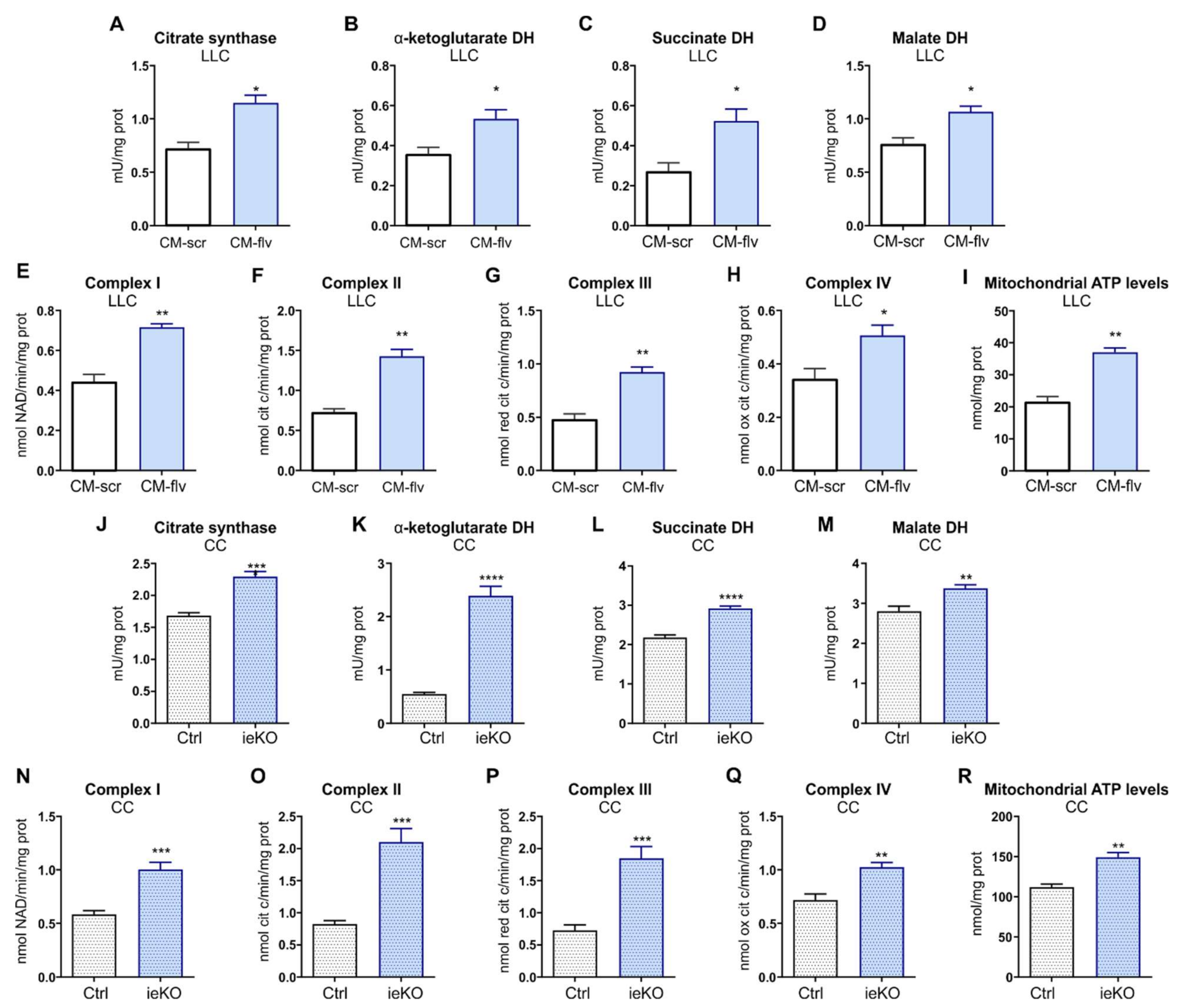
The enzymatic activities of citrate synthase, α-ketoglutarate dehydrogenase (DH), succinate dehydrogenase, and malate dehydrogenase were measured on 10 mg mitochondrial proteins using the Citrate Synthase Assay Kit (Sigma-Aldrich, St. Louis, MO, USA, catalog n° MAK193), Alpha Ketoglutarate (alpha KG) Assay Kit (Abcam, Cambridge, UK, catalog n° ab83431), Malate Dehydrogenase Assay Kit (Sigma-Aldrich, St. Louis, MO, USA, catalog n° MAK196), Succinate Dehydrogenase Activity Colorimetric Assay Kit (BioVision, Milpitas, CA, USA, catalog n° K660), as per manufacturer’s instructions. Results were expressed as mU/mg mitochondrial proteins.
2.14. The Activity of Mitochondrial ETC Complexes I–IV
According to [
22], cells were washed twice in ice-cold 0.1 M phosphate-buffered saline (PBS), then lysed in 0.5 mL buffer A (50 mmol/L Tris, 100 mmol/L KCl, 5 mmol/L MgCl
2, 1.8 mmol/L ATP, 1 mmol/L EDTA, pH 7.2), supplemented with protease-inhibitor cocktail III [100 mmol/L AEBSF, 80 mmol/L aprotinin, 5 mmol/L bestatin, 1.5 mmol/L E-64, 2 mmol/L leupeptin and 1 mmol/L pepstatin (Merck, Darmstadt, Germany), 1 mmol/L phenylmethylsulfonyl fluoride (PMSF), 250 mmol/L NaF. Samples were clarified by centrifuging at 650×
g for 3 min at 4 °C, and the supernatant was collected and centrifuged at 13,000×
g for 5 min at 4 °C. The new supernatant was discarded and the pellet containing mitochondria was washed in 0.5 mL buffer A and re-suspended in 0.25 mL buffer B (250 mmol/L sucrose, 15 μmol/L K
2HPO
4, 2 mmol/L MgCl
2, 0.5 mmol/L EDTA, 5%
w/
v bovine serum albumin). A 100 μL aliquot was sonicated and used for the measurement of protein content. The remaining not-sonicated part was used to measure the electron transport chain (ETC) complexes I–IV activities according to [
22]. Results were expressed as nmol NAD+/min/mg mitochondrial protein for complex I, nmol cyt c reduced/min/mg mitochondrial protein for complexes II–III, and nmoles cyt c oxidized/min/mg mitochondrial protein for complex IV.
2.15. ATP Levels in Mitochondria
The ATP levels in mitochondria extracts were measured with the ATP Bioluminescent Assay Kit (Sigma-Aldrich, St. Louis, MO, USA). ATP was quantified as relative light units (RLU) and converted into nmoles ATP/mg mitochondrial proteins, according to the calibration curve previously set.
2.16. Statistics
Statistical comparisons were conducted in Prism (GraphPad Software, Inc., La Jolla, CA, USA). Results are expressed as mean ± SEM.
4. Discussion
The results described here provide evidence of heme metabolism involvement in the crosstalk between tumor-associated endothelial cells (TECs) and cancer cells (CCs). The active role of the tumor microenvironment (TME) in cancer development and progression has become increasingly recognized [
23]. One of the most studied interplays within the TME involves endothelial cells (ECs) and CCs. Since blood vessels are required to supply oxygen and nutrients to the tumor, CCs stimulate angiogenesis by secreting growth factors, such as the Vascular-Endothelial Growth Factor (VEGF), which triggers EC re-activation. Nevertheless, the landscape is more complex than that, and metabolism plays a prominent role [
24]. CCs are indeed able to metabolically hijack and induce reprogramming in the surrounding stromal cells, including ECs. Consistently, TEC metabolism is profoundly perturbed in cancer [
25,
26]. For instance, the glycolytic rate is strongly increased in active angiogenic TECs compared to healthy ECs, thus sustaining the enhanced motility and active proliferation [
27]. Basically, TECs display a specific metabolic profile, which underlies a plethora of specialized endothelial functions that favor CC proliferation and support tumor growth. Moreover, the crosstalk is bidirectional, and increasing evidence highlights the importance of the metabolic cooperation that is established between CCs and surrounding stromal cells [
5].
Heme is a vital molecule involved in several biological processes, and recent findings pointed out its crucial role in ECs, specifically in the regulation of energy metabolism and angiogenesis [
16,
19,
28]. In particular, the loss of the cell membrane heme exporter FLVCR1a in ECs leads to embryonic death associated with impaired angiogenesis [
19]. Moreover, it has been recently demonstrated that the heme synthesis–export system is involved in the regulation of TEC energy metabolism. Indeed, FLVCR1a deficiency in active TECs results in increased TCA cycle flux and OXPHOS both in vitro and in vivo [
16]. Here, we found that FLVCR1a-deficient TECs enhanced FAO, which likely sustained the TCA cycle by providing acetyl-CoA (
Figure 7). Interestingly, FAs-derived acetyl-CoA can also be incorporated into KBs, small molecules that are produced through ketogenesis. KBs, namely, acetoacetate (AcAc), β-hydroxybutyrate (β-OHB), and acetone, represent an alternative energy source to glucose and are primarily synthetized in the liver under conditions of glucose shortage such as during fasting or prolonged exercise [
29]. Noteworthy, we observed an accumulation of KBs in FLVCR1a-deficient TECs (
Figure 7). This result agrees with our previous work showing higher FAO activity as well as increased β-OHB levels in
FLVCR1a-silenced colorectal cancer cells [
16]. Ketogenesis consists of multiple reactions occurring in mitochondria. First, two acetyl-CoA are condensed to form acetoacetyl-CoA (AcAc-CoA). A third acetyl-CoA is then added to produce β-hydroxy-β-methylglutaryl-CoA (HMG-CoA), a reaction catalyzed by the rate-limiting enzyme HMG-CoA synthase 2 (HMGCS2). Finally, the activity of β-hydroxymethyl-β-methylglutaryl-CoA lyase (HMGCL) generates AcAc that can spontaneously decarboxylate to acetone or be further converted into β-OHB by β-OHB dehydrogenase 1 (BDH1). While acetone can easily diffuse across cell membranes, AcAc and β-OHB are mainly transported via monocarboxylate transporters (in mammals, MCT1 and MCT2). As an alternative source of energy, circulating KBs are oxidized in extrahepatic tissues such as the heart, skeletal muscle, kidney, and brain to fuel mitochondrial metabolism in conditions of low glucose availability. KBs oxidation (ketolysis) requires the enzyme succinyl-CoA:3-oxoacid-CoA transferase (SCOT/OXCT1), which activates AcAc to AcAc-CoA through the exchange of a CoA moiety from succinyl-CoA [
29]. Importantly, succinyl-CoA is a hub metabolite shared across the TCA cycle, KBs oxidation, and heme biosynthesis. Hence, succinyl-CoA consumption through ketolysis might affect heme metabolism and vice versa. Finally, a reversible AcAc-CoA thiolase reaction yields two molecules of acetyl-CoA, which enter the TCA cycle and sustain oxidative metabolism.
The mechanism behind the KBs-mediated oxidative metabolic shift found in CCs conditioned by
FLVCR1a-deficient ECs is not fully elucidated. Tracing experiments with labeled acetate (i.e., [
3H]-acetate) supplementation in ECs clearly demonstrated that LLC cells are capable of importing KBs from the extracellular environment. In this context, monocarboxylate transporters (MCTs) may play a relevant role. Indeed, MCTs are involved in KBs transport across cell membranes and are generally dysregulated in cancer [
30]. Despite the enrichment of labeled KBs found in
FLVCR1a-silenced BTECs-derived CM following [
3H]-acetate supplementation, conditioned LLC cells showed only a slight increase in labeled KBs intracellular content. This result supports the hypothesis of rapid utilization of KBs to fuel the TCA cycle once taken up by the cancer cell.
Nevertheless, recent evidence also attributes to KBs a signaling activity. For instance, β-OHB is an endogenous inhibitor of class I nuclear histone deacetylase (HDACs) enzymes (i.e., HDAC1, HDAC3, HDAC4) in a dose-dependent manner (IC50 ~2–5 mM). Consistently, KBs enrichment in the extracellular milieu promotes histone hyperacetylation, resulting in a modified epigenetic landscape and subsequent rewired gene expression profile [
31,
32]. Although histones were the first identified targets, many non-histone proteins are subjected to HDACs-mediated deacetylation, including the proto-oncogene c-Myc and the tumor suppressor p53. Moreover, KBs may also indirectly contribute to protein hyperacetylation by increasing the intracellular pool of acetyl-CoA [
33].
β-OHB is the ligand for at least two G-protein-coupled receptors (GPRs or GPCRs) that bind short-chain fatty acids (FAs). GPR109A, even known as hydroxycarboxylic acid receptor 2 (HCAR2), is a Gi/o coupled GPCR showing an intermediate affinity for β-OHB (EC50 ~0.8 mM). Its activation reduces lipolysis in adipocytes, probably representing a feedback mechanism to regulate FA availability for KBs metabolism. Interestingly, GPR109A was recently identified as a tumor suppressor [
33]. Indeed, colon CCs express GPR109A at very low levels, whereas breast cancer cells completely abolish its expression. Noteworthy, GPR109A ectopic expression in colon CCs followed by GPR109A agonist exposure exerted anti-tumoral activity by increasing CC apoptosis [
34]. Furthermore, β-OHB is the antagonist of the GPR81
l-lactate receptor, also known as HCAR1. GPR81 is widely expressed in tissues, including ECs, and physiologically promotes angiogenesis upon exercise-associated increased circulating lactate. Consistent with the definition of lactate as an oncometabolite, GPR81 is overexpressed in many cancers, and in most cases, its expression levels positively correlate with tumor growth and metastasis. Indeed, lactate accumulates within the TME, promoting angiogenesis and shaping a pro-tumoral niche [
34,
35]. GPR81 activation results in decreased intracellular cyclic AMP (cAMP) and increased cellular Ca2+ levels, ultimately promoting CC growth. In such a context, β-OHB could antagonize the pro-tumorigenic effects of lactate by interacting with GPR81 [
34,
35]. Interestingly, a recent study points out a vasodilator effect of β-OHB on the vasculature, primarily mediated by potassium channels and independent of the traditional GPCRs activity [
36]. Hence, it would be interesting to understand whether in FLVCR1a deficiency models, KBs, besides altering the metabolism of surrounding CCs, might also impact the tumor vasculature architecture.
The connection between KBs and cancer is increasingly recognized. Nevertheless, current knowledge suggests a tumor-type-specific ability to use KBs as an alternative energetic fuel. Unlike normal cells, several CCs are not able to shift towards KB oxidation to sustain energetic metabolism. Consistently, ketolytic enzyme expression levels, such as BDH1 and SCOT, are low or undetectable in pancreatic cancer (PANC-1), lung cancer (H1299), and neuroblastoma (SK-N-AS) human cell lines as well as in human glioblastoma in vivo [
32,
37]. Consistently, KBs treatment inhibits neuroblastoma cell viability in vitro, whereas combination therapy of a ketogenic diet (KD) and antiangiogenic drug decreases tumor growth in a glioblastoma mouse model [
38,
39]. Furthermore, KD alone reduces tumor burden in a mouse model of metastatic brain cancer [
40]. Similarly, KD administration in mice significantly inhibits liver cancer cell growth [
41]. Finally, a recent study shows that β-OHB delays growth of melanoma orthotopic tumors by acting on the host immune system and displays synergistic antitumorigenic effects when given in combination with immunotherapy [
42]. Different outcomes come from studies on breast cancer. β-OHB administration in mice carrying MDA-MB-231 breast cancer cells xenografts results in a 2.5-fold increase in tumor volume [
43]. Moreover, ketogenic fibroblasts overexpressing HMGCS2 enhance the growth of co-injected MDA-MB-231 breast cancer cells in vivo [
44]. These data suggest that, differently from other cancer cell types, breast cancer cells can utilize fibroblast-derived KBs to fuel oxidative mitochondrial metabolism and benefit from this metabolic coupling. In spontaneous lung cancers, KBs are known sources of energy for cancer cells: interestingly, the switch from an anaerobic-glycolysis-dependent metabolism toward an FAO- and KB-oxidation-prevailing metabolism depends on the micro-environmental changes induced by treatment with antiangiogenic drugs [
45]. This metabolic rewiring allows sustained tumor growth over the long term despite exposure to antiangiogenic drugs but makes the tumor dependent on mitochondrial metabolism. In this scenario, the co-treatment with both antiangiogenics and mitochondrial metabolism inhibitors abrogates tumor growth owing to a metabolic synthetic lethality [
45]. Based on this evidence, FLVCR1a-targeting in tumor-associated ECs could unveil a metabolic vulnerability by making the tumor dependent on mitochondrial metabolism. In this scenario, combinatorial treatment with mitochondrial-respiration inhibitors and agents targeting heme metabolism might be a promising strategy for cancer treatment.