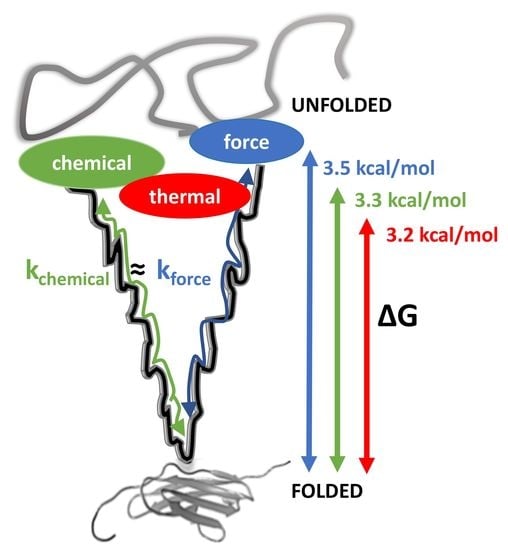
Protein Unfolding: Denaturant vs. Force
Abstract
1. Introduction
2. Materials and Methods
2.1. Expression and Purification of the Individual I83 Domain and Halo-(I83)6 Polymer
2.2. Chemical Stability
2.3. Thermal Stability
2.4. Chemical Folding Kinetics
2.5. Flow Cell Preparation
2.6. Single-Molecule Measurements
2.7. Force-Induced Folding and Refolding Kinetics
2.8. Comparison of Mechanical and Chemical Stability and Kinetics
3. Results
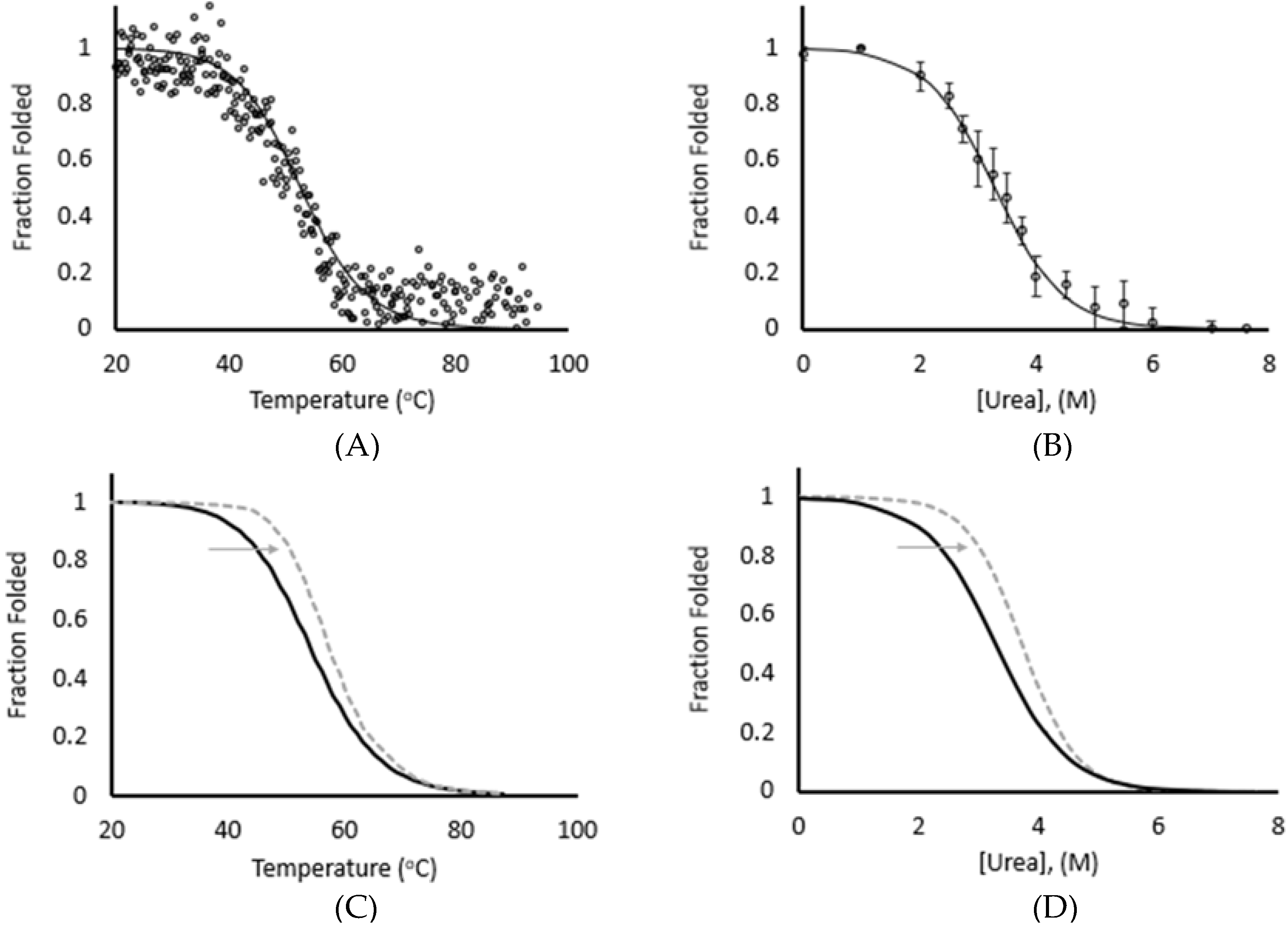
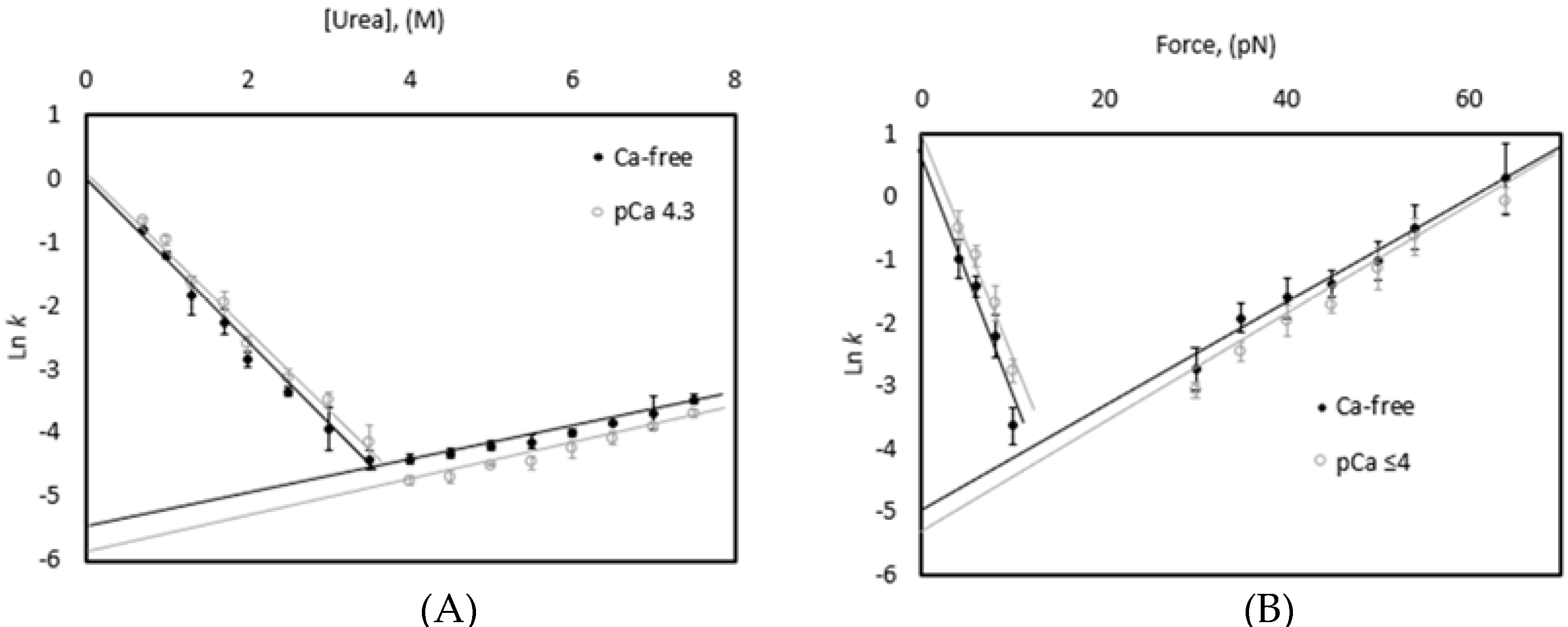
3.1. Thermal and Chemical Equilibrium Studies Yield Similar Stabilities
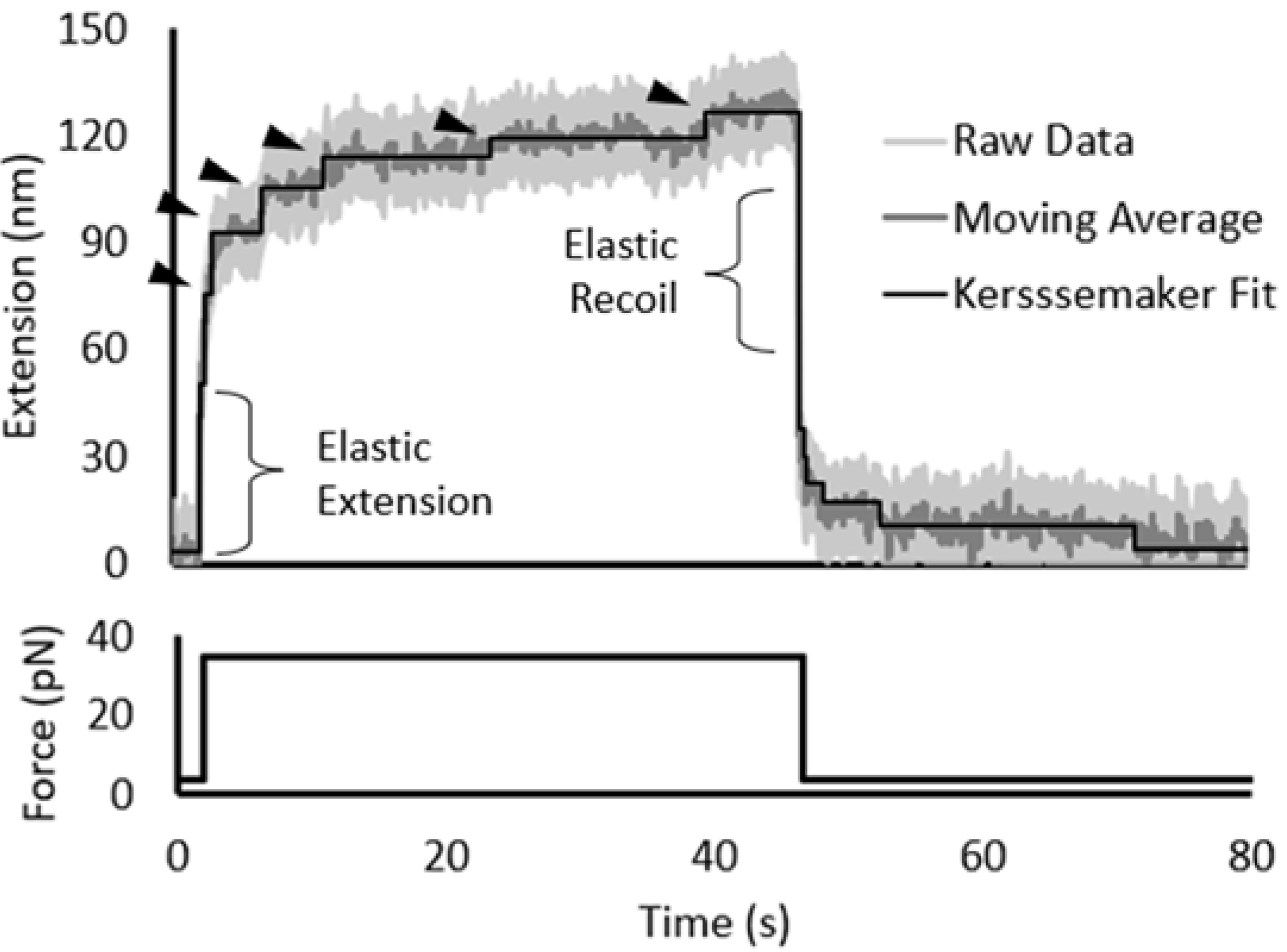
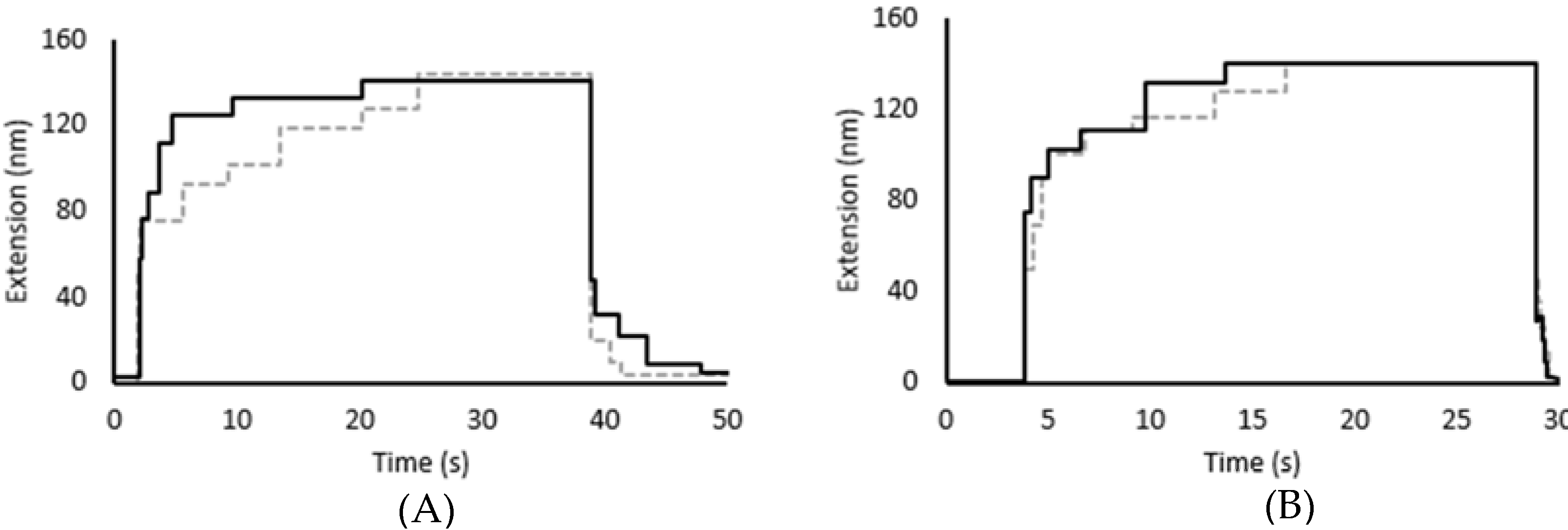
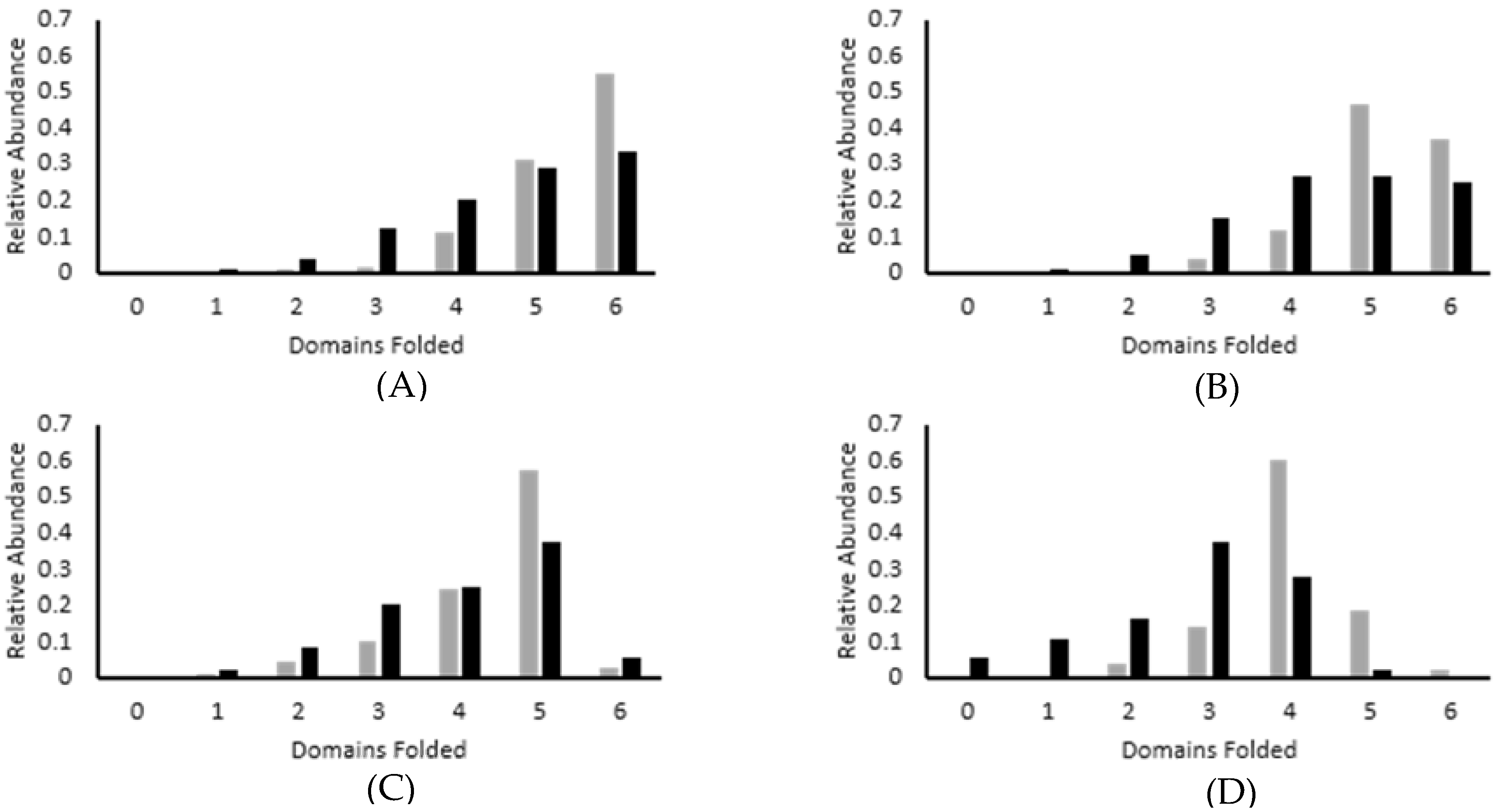
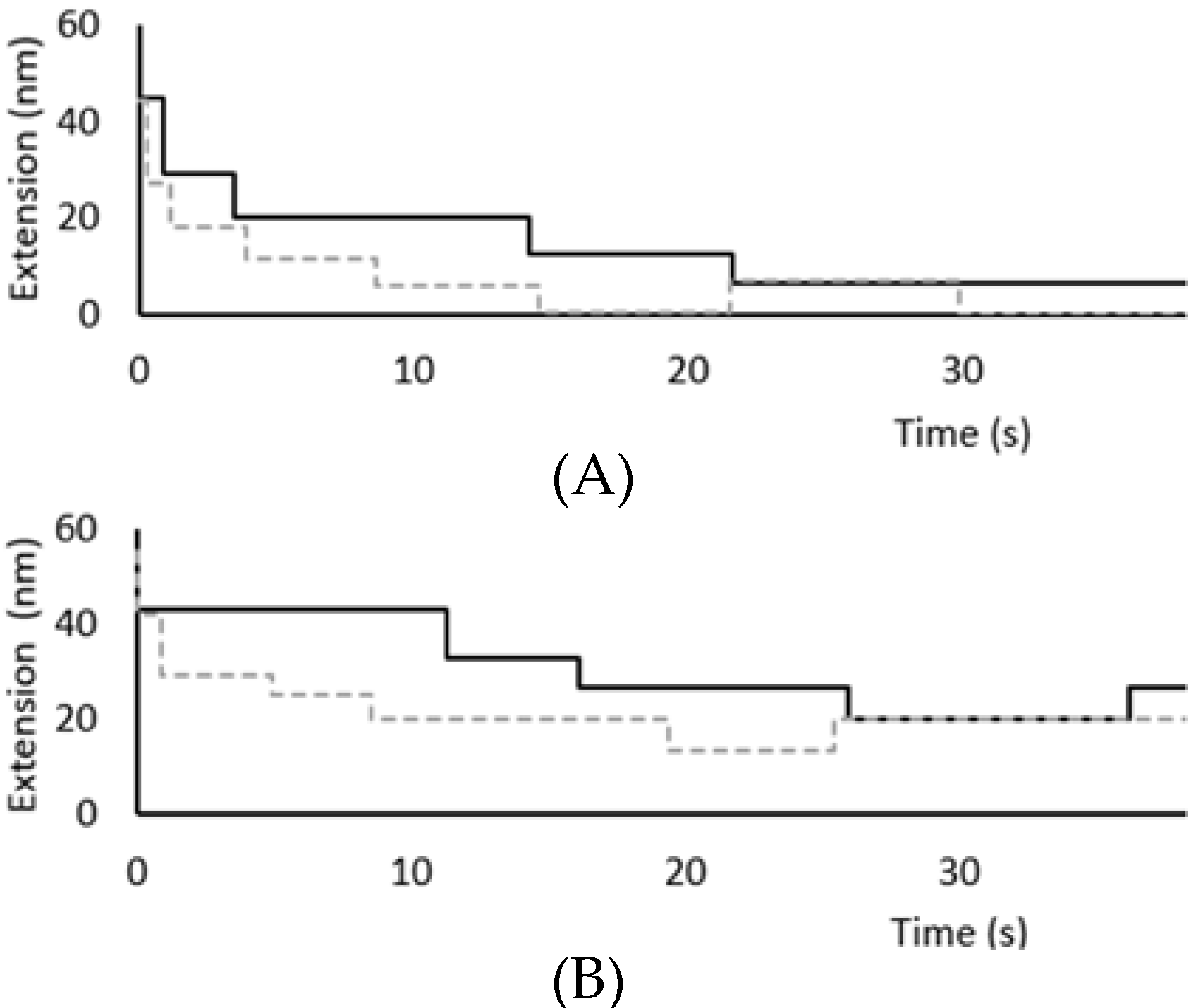
3.2. Force Unfolding and Refolding of Halo-(I83)6 Shows Stabilization in Calcium
3.3. Chemical and Mechanical Folding Yield Similar Rate Constants and Free Energy Values
4. Discussion
4.1. Correlations Observed across Methods
4.2. Mechanistic Role for Calcium Binding to I83 Domain
4.3. Force Unfolding of Physiological Proteins including Ig-like Domains in Titin
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Anfinsen, C.B. Principles that govern the folding of protein chains. Science 1973, 181, 223–230. [Google Scholar] [CrossRef]
- Levinthal, C. Are there pathways for protein folding? J. Chim. Phys. 1968, 65, 44–45. [Google Scholar] [CrossRef]
- Eftink, M.R. The use of fluorescence methods to monitor unfolding transitions in proteins. Biophys. J. 1994, 66, 482–501. [Google Scholar] [CrossRef]
- Kelly, C.; Pace, N.; Gage, M.; Pfuhl, M. Solution NMR Structure of Titin N2A Region Ig Domain I83 and Its Interaction with Metal Ions. J. Mol. Biol. 2021, 433, 166977. [Google Scholar] [CrossRef]
- Royer, C.A. Probing Protein Folding and Conformational Transitions with Fluorescence. Chem. Rev. 2006, 106, 1769–1784. [Google Scholar] [CrossRef]
- Cassaignau, A.M.E.; Cabrita, L.D.; Christodoulou, J. How Does the Ribosome Fold the Proteome? Annu. Rev. Biochem. 2020, 89, 389–415. [Google Scholar] [CrossRef]
- Maciuba, K.; Rajasekaran, N.; Chen, X.; Kaiser, C.M. Co-translational folding of nascent polypeptides: Multi-layered mechanisms for the efficient biogenesis of functional proteins. BioEssays News Rev. Mol. Cell. Dev. Biol. 2021, 43, e2100042. [Google Scholar] [CrossRef]
- Powers, E.T.; Gierasch, L.M. The Proteome Folding Problem and Cellular Proteostasis. J. Mol. Biol. 2021, 167197. [Google Scholar] [CrossRef]
- Aguirre, C.; Goto, Y.; Costas, M. Thermal and chemical unfolding pathways of PaSdsA1 sulfatase, a homo-dimer with topologically interlinked chains. FEBS Lett. 2016, 590, 202–214. [Google Scholar] [CrossRef]
- Saha, P.; Manna, C.; Chakrabarti, J.; Ghosh, M. Reversible thermal unfolding of a yfdX protein with chaperone-like activity. Sci. Rep. 2016, 6, 29541. [Google Scholar] [CrossRef]
- Mahapa, A.; Mandal, S.; Biswas, A.; Jana, B.; Polley, S.; Sau, S.; Sau, K. Chemical and thermal unfolding of a global staphylococcal virulence regulator with a flexible C-terminal end. PLoS ONE 2015, 10, e0122168. [Google Scholar] [CrossRef]
- Candotti, M.; Esteban-Martín, S.; Salvatella, X.; Orozco, M. Toward an atomistic description of the urea-denatured state of proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 5933–5938. [Google Scholar] [CrossRef]
- Baldwin, R.L. Temperature dependence of the hydrophobic interaction in protein folding. Proc. Natl. Acad. Sci. USA 1986, 83, 8069–8072. [Google Scholar] [CrossRef]
- Bryngelson, J.D.; Onuchic, J.N.; Socci, N.D.; Wolynes, P.G. Funnels, pathways, and the energy landscape of protein folding: A synthesis. Proteins 1995, 21, 167–195. [Google Scholar] [CrossRef]
- Alonso-Caballero, A.; Tapia-Rojo, R.; Badilla, C.L.; Fernandez, J.M. Magnetic tweezers meets AFM: Ultra-stable protein dynamics across the force spectrum. bioRxiv 2021. [Google Scholar] [CrossRef]
- Haldar, S.; Tapia-Rojo, R.; Eckels, E.C.; Valle-Orero, J.; Fernandez, J.M. Trigger factor chaperone acts as a mechanical foldase. Nat. Commun. 2017, 8, 668. [Google Scholar] [CrossRef]
- Popa, I.; Rivas-Pardo, J.A.; Eckels, E.C.; Echelman, D.J.; Badilla, C.L.; Valle-Orero, J.; Fernandez, J.M. A HaloTag Anchored Ruler for Week-Long Studies of Protein Dynamics. J. Am. Chem. Soc. 2016, 138, 10546–10553. [Google Scholar] [CrossRef]
- Rief, M.; Gautel, M.; Oesterhelt, F.; Fernandez, J.M.; Gaub, H.E. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science 1997, 276, 1109–1112. [Google Scholar] [CrossRef]
- Jagannathan, B.; Marqusee, S. Protein folding and unfolding under force. Biopolymers 2013, 99, 860–869. [Google Scholar] [CrossRef]
- Martonfalvi, Z.; Bianco, P.; Linari, M.; Caremani, M.; Nagy, A.; Lombardi, V.; Kellermayer, M. Low-force transitions in single titin molecules reflect a memory of contractile history. J. Cell Sci. 2014, 127, 858–870. [Google Scholar] [CrossRef]
- Carrion-Vazquez, M.; Marszalek, P.E.; Oberhauser, A.F.; Fernandez, J.M. Atomic force microscopy captures length phenotypes in single proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 11288–11292. [Google Scholar] [CrossRef]
- Ionescu-Zanetti, C.; Khurana, R.; Gillespie, J.R.; Petrick, J.S.; Trabachino, L.C.; Minert, L.J.; Carter, S.A.; Fink, A.L. Monitoring the assembly of Ig light-chain amyloid fibrils by atomic force microscopy. Proc. Natl. Acad. Sci. USA 1999, 96, 13175–13179. [Google Scholar] [CrossRef]
- Rief, M.; Gautel, M.; Gaub, H.E. Unfolding forces of titin and fibronectin domains directly measured by AFM. Adv. Exp. Med. Biol. 2000, 481, 129–136, discussion 137–141. [Google Scholar] [CrossRef]
- Zhang, B.; Xu, G.; Evans, J.S. A kinetic molecular model of the reversible unfolding and refolding of titin under force extension. Biophys. J. 1999, 77, 1306–1315. [Google Scholar] [CrossRef][Green Version]
- Botello, E.; Harris, N.C.; Sargent, J.; Chen, W.H.; Lin, K.J.; Kiang, C.H. Temperature and chemical denaturant dependence of forced unfolding of titin I27. J. Phys. Chem. B 2009, 113, 10845–10848. [Google Scholar] [CrossRef]
- Carrion-Vazquez, M.; Oberhauser, A.F.; Fowler, S.B.; Marszalek, P.E.; Broedel, S.E.; Clarke, J.; Fernandez, J.M. Mechanical and chemical unfolding of a single protein: A comparison. Proc. Natl. Acad. Sci. USA 1999, 96, 3694–3699. [Google Scholar] [CrossRef]
- Kelly, C.M.; Manukian, S.; Kim, E.; Gage, M.J. Differences in stability and calcium sensitivity of the Ig domains in titin’s N2A region. Protein Sci. 2020, 29, 1160–1171. [Google Scholar] [CrossRef] [PubMed]
- Neuman, K.C.; Nagy, A. Single-molecule force spectroscopy: Optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 2008, 5, 491–505. [Google Scholar] [CrossRef]
- Eckels, E.C.; Haldar, S.; Tapia-Rojo, R.; Rivas-Pardo, J.A.; Fernández, J.M. The Mechanical Power of Titin Folding. Cell Rep. 2019, 27, 1836–1847.e1834. [Google Scholar] [CrossRef]
- Eckels, E.C.; Tapia-Rojo, R.; Rivas-Pardo, J.A.; Fernandez, J.M. The Work of Titin Protein Folding as a Major Driver in Muscle Contraction. Annu. Rev. Physiol. 2018, 80, 327–351. [Google Scholar] [CrossRef]
- Rivas-Pardo, J.A.; Eckels, E.C.; Popa, I.; Kosuri, P.; Linke, W.A.; Fernandez, J.M. Work Done by Titin Protein Folding Assists Muscle Contraction. Cell Rep. 2016, 14, 1339–1347. [Google Scholar] [CrossRef]
- Pace, C.N.; Shaw, K.L. Linear extrapolation method of analyzing solvent denaturation curves. Proteins 2000, 41, 1–7. [Google Scholar] [CrossRef]
- von Helmholtz, H.; Hittorf, J.W.; Waals, J.D. Physical Memoirs Selected and Translated from Foreign Sources, under the Direction of the Physical Society of London; Taylor and Francis: London, UK, 1891; Volume 1, p. 510. [Google Scholar]
- Loeff, L.; Kerssemakers, J.W.J.; Joo, C.; Dekker, C. AutoStepfinder: A fast and automated step detection method for single-molecule analysis. Patterns 2021, 2, 100256. [Google Scholar] [CrossRef]
- Ferris, M.M.; Habbersett, R.C.; Wolinsky, M.; Jett, J.H.; Yoshida, T.M.; Keller, R.A. Statistics of single-molecule measurements: Applications in flow-cytometry sizing of DNA fragments. Cytometry. Part A J. Int. Soc. Anal. Cytol. 2004, 60, 41–52. [Google Scholar] [CrossRef]
- Rabbani, G.; Ahmad, E.; Zaidi, N.; Fatima, S.; Khan, R.H. pH-Induced molten globule state of Rhizopus niveus lipase is more resistant against thermal and chemical denaturation than its native state. Cell Biochem. Biophys. 2012, 62, 487–499. [Google Scholar] [CrossRef]
- Rabbani, G.; Ahmad, E.; Zaidi, N.; Khan, R.H. pH-dependent conformational transitions in conalbumin (ovotransferrin), a metalloproteinase from hen egg white. Cell Biochem. Biophys. 2011, 61, 551–560. [Google Scholar] [CrossRef]
- Lapidus, L.J. Protein unfolding mechanisms and their effects on folding experiments. F1000Research 2017, 6, 1723. [Google Scholar] [CrossRef]
- Stirnemann, G.; Kang, S.G.; Zhou, R.; Berne, B.J. How force unfolding differs from chemical denaturation. Proc. Natl. Acad. Sci. USA 2014, 111, 3413–3418. [Google Scholar] [CrossRef]
- Greenfield, N.J. Using circular dichroism collected as a function of temperature to determine the thermodynamics of protein unfolding and binding interactions. Nat. Protoc. 2006, 1, 2527–2535. [Google Scholar] [CrossRef]
- Rabbani, G.; Ahmad, E.; Khan, M.V.; Ashraf, M.T.; Bhat, R.; Khan, R.H. Impact of structural stability of cold adapted Candida antarctica lipase B (CaLB): In relation to pH, chemical and thermal denaturation. RSC Adv. 2015, 5, 20115–20131. [Google Scholar] [CrossRef]
- Rabbani, G.; Kaur, J.; Ahmad, E.; Khan, R.H.; Jain, S.K. Structural characteristics of thermostable immunogenic outer membrane protein from Salmonella enterica serovar Typhi. Appl. Microbiol. Biotechnol. 2014, 98, 2533–2543. [Google Scholar] [CrossRef]
- Valle-Orero, J.; Rivas-Pardo, J.A.; Popa, I. Multidomain proteins under force. Nanotechnology 2017, 28, 174003. [Google Scholar] [CrossRef][Green Version]
- Borgia, A.; Williams, P.M.; Clarke, J. Single-molecule studies of protein folding. Annu. Rev. Biochem. 2008, 77, 101–125. [Google Scholar] [CrossRef]
- Chen, Y.; Ding, F.; Nie, H.; Serohijos, A.W.; Sharma, S.; Wilcox, K.C.; Yin, S.; Dokholyan, N.V. Protein folding: Then and now. Arch. Biochem. Biophys. 2008, 469, 4–19. [Google Scholar] [CrossRef]
- Kellermayer, M.S.; Smith, S.B.; Granzier, H.L.; Bustamante, C. Folding-unfolding transitions in single titin molecules characterized with laser tweezers. Science 1997, 276, 1112–1116. [Google Scholar] [CrossRef]
- Crick, A.J.; Theron, M.; Tiffert, T.; Lew, V.L.; Cicuta, P.; Rayner, J.C. Quantitation of malaria parasite-erythrocyte cell-cell interactions using optical tweezers. Biophys. J. 2014, 107, 846–853. [Google Scholar] [CrossRef]
- Le, S.; Liu, R.; Lim, C.T.; Yan, J. Uncovering mechanosensing mechanisms at the single protein level using magnetic tweezers. Methods 2016, 94, 13–18. [Google Scholar] [CrossRef]
- Bianco, P.; Mártonfalvi, Z.; Naftz, K.; Kőszegi, D.; Kellermayer, M. Titin domains progressively unfolded by force are homogenously distributed along the molecule. Biophys. J. 2015, 109, 340–345. [Google Scholar] [CrossRef]
- Mártonfalvi, Z.; Bianco, P.; Naftz, K.; Ferenczy, G.G.; Kellermayer, M. Force generation by titin folding. Protein Sci. A Publ. Protein Soc. 2017, 26, 1380–1390. [Google Scholar] [CrossRef]
- Chen, H.; Yuan, G.; Winardhi, R.S.; Yao, M.; Popa, I.; Fernandez, J.M.; Yan, J. Dynamics of equilibrium folding and unfolding transitions of titin immunoglobulin domain under constant forces. J. Am. Chem. Soc. 2015, 137, 3540–3546. [Google Scholar] [CrossRef]
- Dutta, S.; Tsiros, C.; Sundar, S.L.; Athar, H.; Moore, J.; Nelson, B.; Gage, M.J.; Nishikawa, K. Calcium increases titin N2A binding to F-actin and regulated thin filaments. Sci. Rep. 2018, 8, 14575. [Google Scholar] [CrossRef]
- Hayashi, C.; Ono, Y.; Doi, N.; Kitamura, F.; Tagami, M.; Mineki, R.; Arai, T.; Taguchi, H.; Yanagida, M.; Hirner, S.; et al. Multiple molecular interactions implicate the connectin/titin N2A region as a modulating scaffold for p94/calpain 3 activity in skeletal muscle. J. Biol. Chem. 2008, 283, 14801–14814. [Google Scholar] [CrossRef]
- Huebsch, K.A.; Kudryashova, E.; Wooley, C.M.; Sher, R.B.; Seburn, K.L.; Spencer, M.J.; Cox, G.A. Mdm muscular dystrophy: Interactions with calpain 3 and a novel functional role for titin’s N2A domain. Hum. Mol. Genet. 2005, 14, 2801–2811. [Google Scholar] [CrossRef][Green Version]
- Nishikawa, K.; Lindstedt, S.L.; Hessel, A.; Mishra, D. N2A Titin: Signaling Hub and Mechanical Switch in Skeletal Muscle. Int. J. Mol. Sci. 2020, 21, 3974. [Google Scholar] [CrossRef]
- Zhou, T.; Fleming, J.R.; Franke, B.; Bogomolovas, J.; Barsukov, I.; Rigden, D.J.; Labeit, S.; Mayans, O. CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex. FEBS Lett. 2016, 590, 3098–3110. [Google Scholar] [CrossRef]
- Zhou, T.; Fleming, J.R.; Lange, S.; Hessel, A.L.; Bogomolovas, J.; Stronczek, C.; Grundei, D.; Ghassemian, M.; Biju, A.; Börgeson, E.; et al. Molecular Characterisation of Titin N2A and Its Binding of CARP Reveals a Titin/Actin Cross-linking Mechanism. J. Mol. Biol. 2021, 433, 166901. [Google Scholar] [CrossRef]
- Ono, Y.; Ojima, K.; Shinkai-Ouchi, F.; Hata, S.; Sorimachi, H. An eccentric calpain, CAPN3/p94/calpain-3. Biochimie 2016, 122, 169–187. [Google Scholar] [CrossRef]
- Garvey, S.M.; Rajan, C.; Lerner, A.P.; Frankel, W.N.; Cox, G.A. The muscular dystrophy with myositis (mdm) mouse mutation disrupts a skeletal muscle-specific domain of titin. Genomics 2002, 79, 146–149. [Google Scholar] [CrossRef]
- Lanzicher, T.; Zhou, T.; Saripalli, C.; Keschrumrus, V.; Smith III, J.E.; Mayans, O.; Sbaizero, O.; Granzier, H. Single-Molecule Force Spectroscopy on the N2A Element of Titin: Effects of Phosphorylation and CARP. Front. Physiol. 2020, 11, 173. [Google Scholar] [CrossRef]
- Brynnel, A.; Hernandez, Y.; Kiss, B.; Lindqvist, J.; Adler, M.; Kolb, J.; van der Pijl, R.; Gohlke, J.; Strom, J.; Smith, J.; et al. Downsizing the molecular spring of the giant protein titin reveals that skeletal muscle titin determines passive stiffness and drives longitudinal hypertrophy. Elife 2018, 7, e40532. [Google Scholar] [CrossRef]
- Lewit-Bentley, A.; Réty, S. EF-hand calcium-binding proteins. Curr. Opin. Struct. Biol. 2000, 10, 637–643. [Google Scholar] [CrossRef]
- Tan, Y.; Schneider, T.; Leong, M.; Aravind, L.; Zhang, D.; Zhulin, I.B. Novel Immunoglobulin Domain Proteins Provide Insights into Evolution and Pathogenesis of SARS-CoV-2-Related Viruses. ASM J. 2020, 11, e00760-20. [Google Scholar] [CrossRef]
- Buck, C.A. Immunoglobulin superfamily: Structure, function and relationship to other receptor molecules. Semin. Cell Biol. 1992, 3, 179–188. [Google Scholar] [CrossRef]
- Mendelsohn, C.L.; Wimmer, E.; Racaniello, V.R. Cellular receptor for poliovirus: Molecular cloning, nucleotide sequence, and expression of a new member of the immunoglobulin superfamily. Cell 1989, 56, 855–865. [Google Scholar] [CrossRef]
- Improta, S.; Politou, A.S.; Pastore, A. Immunoglobulin-like modules from titin I-band: Extensible components of muscle elasticity. Structure 1996, 4, 323–337. [Google Scholar] [CrossRef]
- Fürst, D.O.; Gautel, M. The anatomy of a molecular giant: How the sarcomere cytoskeleton is assembled from immunoglobulin superfamily molecules. J. Mol. Cell. Cardiol. 1995, 27, 951–959. [Google Scholar] [CrossRef]
- Labeit, S.; Kolmerer, B.; Linke, W.A. The giant protein titin. Emerging roles in physiology and pathophysiology. Circ. Res. 1997, 80, 290–294. [Google Scholar] [CrossRef]
- Marino, M.; Svergun, D.I.; Kreplak, L.; Konarev, P.V.; Maco, B.; Labeit, D.; Mayans, O. Poly-Ig tandems from I-band titin share extended domain arrangements irrespective of the distinct features of their modular constituents. J. Muscle Res. Cell Motil. 2005, 26, 355–365. [Google Scholar] [CrossRef]
- Watanabe, K.; Nair, P.; Labeit, D.; Kellermayer, M.S.; Greaser, M.; Labeit, S.; Granzier, H. Molecular mechanics of cardiac titin’s PEVK and N2B spring elements. J. Biol. Chem. 2002, 277, 11549–11558. [Google Scholar] [CrossRef]
- Nagy, A.; Grama, L.; Huber, T.; Bianco, P.; Trombitás, K.; Granzier, H.L.; Kellermayer, M.S. Hierarchical extensibility in the PEVK domain of skeletal-muscle titin. Biophys. J. 2005, 89, 329–336. [Google Scholar] [CrossRef]
- Radke, M.H.; Polack, C.; Methawasin, M.; Fink, C.; Granzier, H.L.; Gotthardt, M. Deleting Full Length Titin Versus the Titin M-Band Region Leads to Differential Mechanosignaling and Cardiac Phenotypes. Circulation 2019, 139, 1813–1827. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Evans, J.S. Modeling AFM-induced PEVK extension and the reversible unfolding of Ig/FNIII domains in single and multiple titin molecules. Biophys. J. 2001, 80, 597–605. [Google Scholar] [CrossRef][Green Version]
- Yu, M.; Lu, J.H.; Le, S.; Yan, J. Unexpected Low Mechanical Stability of Titin I27 Domain at Physiologically Relevant Temperature. J. Phys. Chem. Lett. 2021, 12, 7914–7920. [Google Scholar] [CrossRef]
- Jacobson, G.N.; Clark, P.L. Quality over quantity: Optimizing co-translational protein folding with non-’optimal’ synonymous codons. Curr. Opin. Struct. Biol. 2016, 38, 102–110. [Google Scholar] [CrossRef]
| Thermal | Chemical | |
|---|---|---|
| ΔGunfolding (kcal/mol) Ca-free | 3.2 ± 0.1 | 3.3 ± 0.3 |
| ΔGrefolding (kcal/mol) Ca-free | 3.1 ± 0.2 | 3.4 ± 0.04 |
| ΔGunfolding (kcal/mol) pCa 4.3 | 4.2 ± 0.1 | 4.8 ± 0.1 |
| Chemical | Force | |||
|---|---|---|---|---|
| Ca-Free | pCa 4.3 | Ca-Free | pCa ≤ 4 | |
| Natural Log of Unfolding Rate Ln ku | −5.52 ± 0.10 | −6.04 ± 0.08 | −5.01 ± 0.34 | −5.68 ± 0.21 |
| Unfolding Rate ku (s−1) | 0.004 ± 0.0004 | 0.0024 ± 0.0002 | 0.0067 ± 0.0019 | 0.0034 ± 0.0006 |
| Natural Log of Refolding Rate Ln kf | −0.013 ± 0.15 | 0.13 ± 0.14 | 1.00 ± 0.28 | 1.20 ± 0.23 |
| Refolding Rate Kf (s−1) | 0.99 ± 0.13 | 1.14 ± 0.15 | 2.72 ± 0.66 | 3.32 ± 0.68 |
| ΔGunfolding Equation (7) (kcal/mol) | 3.3 ± 0.25 | 3.7 ± 0.22 | 3.5 ± 0.62 | 4.0 ± 0.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kelly, C.; Gage, M.J. Protein Unfolding: Denaturant vs. Force. Biomedicines 2021, 9, 1395. https://doi.org/10.3390/biomedicines9101395
Kelly C, Gage MJ. Protein Unfolding: Denaturant vs. Force. Biomedicines. 2021; 9(10):1395. https://doi.org/10.3390/biomedicines9101395
Chicago/Turabian StyleKelly, Colleen, and Matthew J. Gage. 2021. "Protein Unfolding: Denaturant vs. Force" Biomedicines 9, no. 10: 1395. https://doi.org/10.3390/biomedicines9101395
APA StyleKelly, C., & Gage, M. J. (2021). Protein Unfolding: Denaturant vs. Force. Biomedicines, 9(10), 1395. https://doi.org/10.3390/biomedicines9101395