Inhaled Corticosteroids and the Lung Microbiome in COPD
Abstract
:1. Introduction
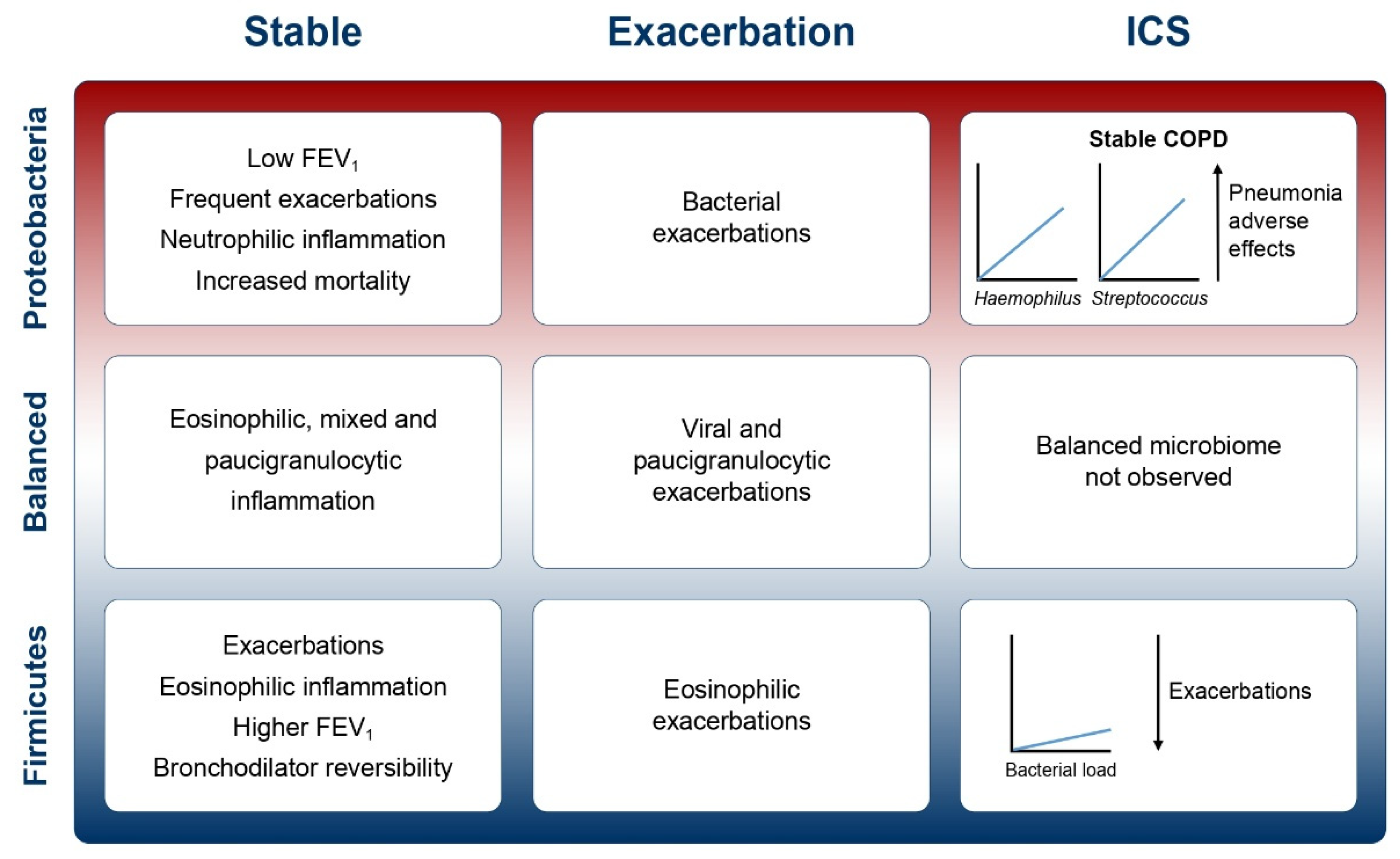
2. What Do We Know about the Microbiome in COPD?
2.1. The Microbiome of the COPD Lung versus the Healthy Lung
2.2. The Microbiome in Stable COPD
2.3. The Microbiome at Exacerbation
3. Interactions between Inflammation, Clinical Characteristics and the Microbiome
3.1. Neutrophilic and Eosinophilic Inflammation
3.2. Pneumonia
4. ICS—A Help or a Hinderance? What Effect Does ICS Use Have on the Microbiome?
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Agusti, A.; Bel, E.; Thomas, M.; Vogelmeier, C.; Brusselle, G.; Holgate, S.; Humbert, M.; Jones, P.; Gibson, P.G.; Vestbo, J.; et al. Treatable traits: Toward precision medicine of chronic airway diseases. Eur. Respir. J. 2016, 47, 410–419. [Google Scholar] [CrossRef] [Green Version]
- Anderson, G.P. Endotyping asthma: New insights into key pathogenic mechanisms in a complex, heterogeneous disease. Lancet 2008, 372, 1107–1119. [Google Scholar] [CrossRef]
- Han, M.K.; Agusti, A.; Calverley, P.M.; Celli, B.R.; Criner, G.; Curtis, J.L.; Fabbri, L.M.; Goldin, J.G.; Jones, P.W.; Macnee, W.; et al. Chronic obstructive pulmonary disease phenotypes: The future of COPD. Am. J. Respir. Crit. Care Med. 2010, 182, 598–604. [Google Scholar] [CrossRef] [Green Version]
- Agusti, A.; Bafadhel, M.; Beasley, R.; Bel, E.H.; Faner, R.; Gibson, P.G.; Louis, R.; McDonald, V.M.; Sterk, P.J.; Thomas, M.; et al. Precision medicine in airway diseases: Moving to clinical practice. Eur. Respir. J. 2017, 50, 1701655. [Google Scholar] [CrossRef]
- Vestbo, J.; Hurd, S.S.; Agusti, A.G.; Jones, P.W.; Vogelmeier, C.; Anzueto, A.; Barnes, P.J.; Fabbri, L.M.; Martinez, F.J.; Nishimura, M.; et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am. J. Respir. Crit. Care Med. 2013, 187, 347–365. [Google Scholar] [CrossRef] [PubMed]
- Global Initiative for Chronic Obstructive Lung Disease. Global Strategy for the Diagnosis, Management, and Prevention of Chronic Obstructive Pulmonary Disease (2021 report). Available online: https://goldcopd.org/wp-content/uploads/2020/11/GOLD-REPORT-2021-v1.1-25Nov20_WMV.pdf (accessed on 14 April 2021).
- White, P.; Thornton, H.; Pinnock, H.; Georgopoulou, S.; Booth, H.P. Overtreatment of COPD with inhaled corticosteroids—Implications for safety and costs: Cross-sectional observational study. PLoS ONE 2013, 8, e75221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savran, O.; Godtfredsen, N.; Sorensen, T.; Jensen, C.; Ulrik, C.S. COPD patients prescribed inhaled corticosteroid in general practice: Based on disease characteristics according to guidelines? Chron. Respir. Dis. 2019, 16, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas, A.E.M.; Smeenk, F.W.J.M.; Smeele, I.J.; Van Schayck, C.P. Overtreatment with inhaled corticosteroids and diagnostic problems in primary care patients, an exploratory study. Fam. Pract. 2008, 25, 86–91. [Google Scholar] [CrossRef] [Green Version]
- Calverley, P.M.; Anderson, J.A.; Celli, B.; Ferguson, G.T.; Jenkins, C.; Jones, P.W.; Yates, J.C.; Vestbo, J.; Torch Investigators. Salmeterol and fluticasone propionate and survival in chronic obstructive pulmonary disease. N. Engl. J. Med. 2007, 356, 775–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dransfield, M.T.; Bourbeau, J.; Jones, P.W.; Hanania, N.A.; Mahler, D.A.; Vestbo, J.; Wachtel, A.; Martinez, F.J.; Barnhart, F.; Sanford, L.; et al. Once-daily inhaled fluticasone furoate and vilanterol versus vilanterol only for prevention of exacerbations of COPD: Two replicate double-blind, parallel-group, randomised controlled trials. Lancet Respir. Med. 2013, 1, 210–223. [Google Scholar] [CrossRef]
- Drummond, M.B.; Dasenbrook, E.C.; Pitz, M.W.; Murphy, D.J.; Fan, E. Inhaled corticosteroids in patients with stable chronic obstructive pulmonary disease: A systematic review and meta-analysis. JAMA 2008, 300, 2407–2416. [Google Scholar] [CrossRef]
- Singh, S.; Amin, A.V.; Loke, Y.K. Long-term use of inhaled corticosteroids and the risk of pneumonia in chronic obstructive pulmonary disease: A meta-analysis. Arch. Intern. Med. 2009, 169, 219–229. [Google Scholar] [CrossRef]
- Miravitlles, M.; Auladell-Rispau, A.; Monteagudo, M.; Vázquez-Niebla, J.C.; Mohammed, J.; Nuñez, A.; Urrútia, G. Systematic review on long-term adverse effects of inhaled corticosteroids in the treatment of COPD. Eur. Respir. Rev. 2021, 30. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, R.; Breen, D.; Wilson, S.; Djukanovic, R. Inflammatory cells in the airways in COPD. Thorax 2006, 61, 448–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moermans, C.; Heinen, V.; Nguyen, M.; Henket, M.; Sele, J.; Manise, M.; Corhay, J.L.; Louis, R. Local and systemic cellular inflammation and cytokine release in chronic obstructive pulmonary disease. Cytokine 2011, 56, 298–304. [Google Scholar] [CrossRef]
- Lonergan, M.; Dicker, A.J.; Crichton, M.L.; Keir, H.R.; Van Dyke, M.K.; Mullerova, H.; Miller, B.E.; Tal-Singer, R.; Chalmers, J.D. Blood neutrophil counts are associated with exacerbation frequency and mortality in COPD. Respir. Res. 2020, 21, 166. [Google Scholar] [CrossRef]
- Singh, D.; Wedzicha, J.A.; Siddiqui, S.; de la Hoz, A.; Xue, W.; Magnussen, H.; Miravitlles, M.; Chalmers, J.D.; Calverley, P.M.A. Blood eosinophils as a biomarker of future COPD exacerbation risk: Pooled data from 11 clinical trials. Respir. Res. 2020, 21, 240. [Google Scholar] [CrossRef] [PubMed]
- Cox, G. Glucocorticoid treatment inhibits apoptosis in human neutrophils. Separation of survival and activation outcomes. J. Immunol. 1995, 154, 4719–4725. [Google Scholar]
- Bentley, A.M.; Hamid, Q.; Robinson, D.S.; Schotman, E.; Meng, Q.; Assoufi, B.; Kay, A.B.; Durham, S.R. Prednisolone treatment in asthma. Reduction in the numbers of eosinophils, T cells, tryptase-only positive mast cells, and modulation of IL-4, IL-5, and interferon-gamma cytokine gene expression within the bronchial mucosa. Am. J. Respir. Crit. Care Med. 1996, 153, 551–556. [Google Scholar] [CrossRef]
- Stolberg, V.R.; McCubbrey, A.L.; Freeman, C.M.; Brown, J.P.; Crudgington, S.W.; Taitano, S.H.; Saxton, B.L.; Mancuso, P.; Curtis, J.L. Glucocorticoid-Augmented Efferocytosis Inhibits Pulmonary Pneumococcal Clearance in Mice by Reducing Alveolar Macrophage Bactericidal Function. J. Immunol. 2015, 195, 174–184. [Google Scholar] [CrossRef] [Green Version]
- Patterson, C.M.; Morrison, R.L.; D’Souza, A.; Teng, X.S.; Happel, K.I. Inhaled fluticasone propionate impairs pulmonary clearance of Klebsiella pneumoniae in mice. Respir. Res. 2012, 13, 40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van de Garde, M.D.; Martinez, F.O.; Melgert, B.N.; Hylkema, M.N.; Jonkers, R.E.; Hamann, J. Chronic exposure to glucocorticoids shapes gene expression and modulates innate and adaptive activation pathways in macrophages with distinct changes in leukocyte attraction. J. Immunol. 2014, 192, 1196–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Machin, M.; Russell, K.E.; Pavlidis, S.; Zhu, J.; Barnes, P.J.; Chung, K.F.; Adcock, I.M.; Durham, A.L. Corticosteroid modulation of immunoglobulin expression and B-cell function in COPD. FASEB J. 2016, 30, 2014–2026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, P.J. Inhaled corticosteroids. Pharmaceuticals 2010, 3, 514–540. [Google Scholar] [CrossRef] [Green Version]
- Belvisi, M.G.; Bottomley, K.M. The role of matrix metalloproteinases (MMPs) in the pathophysiology of chronic obstructive pulmonary disease (COPD): A therapeutic role for inhibitors of MMPs? Inflamm. Res. Off. J. Eur. Histamine Res. Soc. 2003, 52, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Belchamber, K.B.R.; Singh, R.; Batista, C.M.; Whyte, M.K.; Dockrell, D.H.; Kilty, I.; Robinson, M.J.; Wedzicha, J.A.; Barnes, P.J.; Donnelly, L.E. Defective bacterial phagocytosis is associated with dysfunctional mitochondria in COPD macrophages. Eur. Respir. J. 2019, 54, 1802244. [Google Scholar] [CrossRef]
- Culpitt, S.V.; Rogers, D.F.; Shah, P.; De Matos, C.; Russell, R.E.; Donnelly, L.E.; Barnes, P.J. Impaired inhibition by dexamethasone of cytokine release by alveolar macrophages from patients with chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2003, 167, 24–31. [Google Scholar] [CrossRef]
- Russell, R.E.; Culpitt, S.V.; DeMatos, C.; Donnelly, L.; Smith, M.; Wiggins, J.; Barnes, P.J. Release and activity of matrix metalloproteinase-9 and tissue inhibitor of metalloproteinase-1 by alveolar macrophages from patients with chronic obstructive pulmonary disease. Am. J. Respir. Cell Mol. Biol. 2002, 26, 602–609. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.; Belchamber, K.B.R.; Fenwick, P.S.; Chana, K.; Donaldson, G.; Wedzicha, J.A.; Barnes, P.J.; Donnelly, L.E. Defective monocyte-derived macrophage phagocytosis is associated with exacerbation frequency in COPD. Respir. Res. 2021, 22, 113. [Google Scholar] [CrossRef]
- Belchamber, K.B.; Thomas, C.M.; Dunne, A.E.; Barnes, P.J.; Donnelly, L.E. Comparison of fluticasone propionate and budesonide on COPD macrophage and neutrophil function. Int. J. Chron. Obstruct. Pulmon. Dis 2018, 13, 2883–2897. [Google Scholar] [CrossRef] [Green Version]
- Mammen, M.J.; Sethi, S. COPD and the microbiome. Respirology 2016, 21, 590–599. [Google Scholar] [CrossRef] [Green Version]
- Papi, A.; Bellettato, C.M.; Braccioni, F.; Romagnoli, M.; Casolari, P.; Caramori, G.; Fabbri, L.M.; Johnston, S.L. Infections and airway inflammation in chronic obstructive pulmonary disease severe exacerbations. Am. J. Respir. Crit. Care Med. 2006, 173, 1114–1121. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.; Murphy, T.F. Infection in the pathogenesis and course of chronic obstructive pulmonary disease. N. Engl. J. Med. 2008, 359, 2355–2365. [Google Scholar] [CrossRef]
- Dicker, A.J.; Huang, J.T.J.; Lonergan, M.; Keir, H.R.; Fong, C.J.; Tan, B.; Cassidy, A.J.; Finch, S.; Mullerova, H.; Miller, B.E.; et al. The sputum microbiome, airway inflammation, and mortality in chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2021, 147, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Turturice, B.A.; McGee, H.S.; Oliver, B.; Baraket, M.; Nguyen, B.T.; Ascoli, C.; Ranjan, R.; Rani, A.; Perkins, D.L.; Finn, P.W. Atopic asthmatic immune phenotypes associated with airway microbiota and airway obstruction. PLoS ONE 2017, 12, e0184566. [Google Scholar] [CrossRef] [Green Version]
- Ramakrishnan, V.R.; Holt, J.; Nelson, L.F.; Ir, D.; Robertson, C.E.; Frank, D.N. Determinants of the Nasal Microbiome: Pilot Study of Effects of Intranasal Medication Use. Allergy Rhinol. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baumgardner, D.J. Oral fungal microbiota: To thrush and beyond. J. Patient Cent. Res. Rev. 2019, 6, 252–261. [Google Scholar] [CrossRef] [PubMed]
- Charlson, E.S.; Bittinger, K.; Haas, A.R.; Fitzgerald, A.S.; Frank, I.; Yadav, A.; Bushman, F.D.; Collman, R.G. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 2011, 184, 957–963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dickson, R.P.; Erb-Downward, J.R.; Freeman, C.M.; McCloskey, L.; Beck, J.M.; Huffnagle, G.B.; Curtis, J.L. Spatial Variation in the Healthy Human Lung Microbiome and the Adapted Island Model of Lung Biogeography. Ann. Am. Thorac. Soc. 2015, 12, 821–830. [Google Scholar] [CrossRef]
- Cui, L.; Morris, A.; Ghedin, E. The human mycobiome in health and disease. Genome Med. 2013, 5, 63. [Google Scholar] [CrossRef] [Green Version]
- Sethi, S.; Maloney, J.; Grove, L.; Wrona, C.; Berenson, C.S. Airway inflammation and bronchial bacterial colonization in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 2006, 173, 991–998. [Google Scholar] [CrossRef]
- Soler, N.; Ewig, S.; Torres, A.; Filella, X.; Gonzalez, J.; Zaubet, A. Airway inflammation and bronchial microbial patterns in patients with stable chronic obstructive pulmonary disease. Eur. Respir. J. 1999, 14, 1015–1022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangelov, K.; Sethi, S. Role of infections. Clin. Chest Med. 2014, 35, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Ali, N.; Mac Aogain, M.; Morales, R.F.; Tiew, P.Y.; Chotirmall, S.H. Optimisation and benchmarking of targeted amplicon sequencing for mycobiome analysis of respiratory specimens. Int. J. Mol. Sci. 2019, 20, 4991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fricker, A.M.; Podlesny, D.; Fricke, W.F. What is new and relevant for sequencing-based microbiome research? A mini-review. J. Adv. Res. 2019, 19, 105–112. [Google Scholar] [CrossRef] [PubMed]
- Morgan, X.C.; Huttenhower, C. Chapter 12: Human microbiome analysis. PLoS Comput. Biol. 2012, 8, e1002808. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.J.; Erb-Downward, J.R.; Dickson, R.P.; Curtis, J.L.; Huffnagle, G.B.; Han, M.K. Understanding the role of the microbiome in chronic obstructive pulmonary disease: Principles, challenges, and future directions. Transl. Res. 2017, 179, 71–83. [Google Scholar] [CrossRef] [Green Version]
- Huffnagle, G.B.; Dickson, R.P.; Lukacs, N.W. The respiratory tract microbiome and lung inflammation: A two-way street. Mucosal. Immunol. 2017, 10, 299–306. [Google Scholar] [CrossRef] [Green Version]
- Sommariva, M.; Le Noci, V.; Bianchi, F.; Camelliti, S.; Balsari, A.; Tagliabue, E.; Sfondrini, L. The lung microbiota: Role in maintaining pulmonary immune homeostasis and its implications in cancer development and therapy. Cell Mol. Life Sci. 2020, 77, 2739–2749. [Google Scholar] [CrossRef] [Green Version]
- Abdel-Aziz, M.I.; Brinkman, P.; Vijverberg, S.J.H.; Neerincx, A.H.; Riley, J.H.; Bates, S.; Hashimoto, S.; Kermani, N.Z.; Chung, K.F.; Djukanovic, R.; et al. Sputum microbiome profiles identify severe asthma phenotypes of relative stability at 12 to 18 months. J. Allergy Clin. Immunol. 2021, 147, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Taylor, S.L.; Leong, L.E.X.; Choo, J.M.; Wesselingh, S.; Yang, I.A.; Upham, J.W.; Reynolds, P.N.; Hodge, S.; James, A.L.; Jenkins, C.; et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J. Allergy Clin. Immunol. 2018, 141, 94–103.e115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.; Lee, S.; Go, M.J.; Lee, S.Y.; Kim, S.C.; Lee, C.H.; Cho, B.K. Analysis of the mouse gut microbiome using full-length 16S rRNA amplicon sequencing. Sci. Rep. 2016, 6, 29681. [Google Scholar] [CrossRef] [Green Version]
- Benitez-Paez, A.; Sanz, Y. Multi-locus and long amplicon sequencing approach to study microbial diversity at species level using the MinION portable nanopore sequencer. Gigascience 2017, 6, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kai, S.; Matsuo, Y.; Nakagawa, S.; Kryukov, K.; Matsukawa, S.; Tanaka, H.; Iwai, T.; Imanishi, T.; Hirota, K. Rapid bacterial identification by direct PCR amplification of 16S rRNA genes using the MinION nanopore sequencer. FEBS Open Bio 2019, 9, 548–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuo, Y.; Komiya, S.; Yasumizu, Y.; Yasuoka, Y.; Mizushima, K.; Takagi, T.; Kryukov, K.; Fukuda, A.; Morimoto, Y.; Naito, Y.; et al. Full-length 16S rRNA gene amplicon analysis of human gut microbiota using MinION nanopore sequencing confers species-level resolution. BMC Microbiol. 2021, 21, 35. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.R.; Pop, M.; Deboy, R.T.; Eckburg, P.B.; Turnbaugh, P.J.; Samuel, B.S.; Gordon, J.I.; Relman, D.A.; Fraser-Liggett, C.M.; Nelson, K.E. Metagenomic analysis of the human distal gut microbiome. Science 2006, 312, 1355–1359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Einarsson, G.G.; Comer, D.M.; McIlreavey, L.; Parkhill, J.; Ennis, M.; Tunney, M.M.; Elborn, J.S. Community dynamics and the lower airway microbiota in stable chronic obstructive pulmonary disease, smokers and healthy non-smokers. Thorax 2016, 71, 795–803. [Google Scholar] [CrossRef] [Green Version]
- Haldar, K.; George, L.; Wang, Z.; Mistry, V.; Ramsheh, M.Y.; Free, R.C.; John, C.; Reeve, N.F.; Miller, B.E.; Tal-Singer, R.; et al. The sputum microbiome is distinct between COPD and health, independent of smoking history. Respir. Res. 2020, 21, 183. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, Y.; Yan, Z.; Liu, H.; Chen, B.; Liang, Z.; Wang, F.; Miller, B.E.; Tal-Singer, R.; Yi, X.; et al. Multi-omic meta-analysis identifies functional signatures of airway microbiome in chronic obstructive pulmonary disease. ISME J. 2020, 14, 2748–2765. [Google Scholar] [CrossRef]
- Zhou, B.R.; Zhang, J.A.; Zhang, Q.; Permatasari, F.; Xu, Y.; Wu, D.; Yin, Z.Q.; Luo, D. Palmitic acid induces production of proinflammatory cytokines interleukin-6, interleukin-1beta, and tumor necrosis factor-alpha via a NF-kappaB-dependent mechanism in HaCaT keratinocytes. Mediat. Inflamm. 2013, 2013, 530429. [Google Scholar] [CrossRef] [Green Version]
- Sato, Y.; Fujimoto, S.; Mukai, E.; Sato, H.; Tahara, Y.; Ogura, K.; Yamano, G.; Ogura, M.; Nagashima, K.; Inagaki, N. Palmitate induces reactive oxygen species production and beta-cell dysfunction by activating nicotinamide adenine dinucleotide phosphate oxidase through Src signaling. J. Diabetes Investig. 2014, 5, 19–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segal, L.N.; Clemente, J.C.; Tsay, J.C.; Koralov, S.B.; Keller, B.C.; Wu, B.G.; Li, Y.; Shen, N.; Ghedin, E.; Morris, A.; et al. Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 2016, 1, 16031. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaeloudes, C.; Kuo, C.H.; Haji, G.; Finch, D.K.; Halayko, A.J.; Kirkham, P.; Chung, K.F.; Adcock, I.M. Metabolic re-patterning in COPD airway smooth muscle cells. Eur. Respir. J. 2017, 50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Locantore, N.; Haldar, K.; Ramsheh, M.Y.; Beech, A.S.; Ma, W.; Brown, J.R.; Tal-Singer, R.; Barer, M.R.; Bafadhel, M.; et al. Inflammatory Endotype-associated Airway Microbiome in Chronic Obstructive Pulmonary Disease Clinical Stability and Exacerbations: A Multicohort Longitudinal Analysis. Am. J. Respir. Crit. Care Med. 2021, 203, 1488–1502. [Google Scholar] [CrossRef]
- Opron, K.; Begley, L.A.; Erb-Downward, J.R.; Freeman, C.; Madapoosi, S.; Alexis, N.E.; Barjaktarevic, I.; Graham Barr, R.; Bleecker, E.R.; Bowler, R.P.; et al. Lung microbiota associations with clinical features of COPD in the SPIROMICS cohort. NPJ Biofilms Microbiomes 2021, 7, 14. [Google Scholar] [CrossRef]
- Wang, Z.; Bafadhel, M.; Haldar, K.; Spivak, A.; Mayhew, D.; Miller, B.E.; Tal-Singer, R.; Johnston, S.L.; Ramsheh, M.Y.; Barer, M.R.; et al. Lung microbiome dynamics in COPD exacerbations. Eur. Respir. J. 2016, 47, 1082–1092. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Singh, R.; Miller, B.E.; Tal-Singer, R.; Van Horn, S.; Tomsho, L.; Mackay, A.; Allinson, J.P.; Webb, A.J.; Brookes, A.J.; et al. Sputum microbiome temporal variability and dysbiosis in chronic obstructive pulmonary disease exacerbations: An analysis of the COPDMAP study. Thorax 2018, 73, 331–338. [Google Scholar] [CrossRef] [Green Version]
- Mayhew, D.; Devos, N.; Lambert, C.; Brown, J.R.; Clarke, S.C.; Kim, V.L.; Magid-Slav, M.; Miller, B.E.; Ostridge, K.K.; Patel, R.; et al. Longitudinal profiling of the lung microbiome in the AERIS study demonstrates repeatability of bacterial and eosinophilic COPD exacerbations. Thorax 2018, 73, 422–430. [Google Scholar] [CrossRef] [Green Version]
- Keir, H.R.; Dicker, A.; Lonergan, M.; Crichton, M.; Miller, B.E.; Tal-Singer, R.; Chalmers, J.D. Clinical endotypes of exacerbation are associated with differences in microbial composition and diversity in COPD. Eur. Respir. J. 2020, 56. [Google Scholar] [CrossRef]
- Leitao Filho, F.S.; Alotaibi, N.M.; Ngan, D.; Tam, S.; Yang, J.; Hollander, Z.; Chen, V.; FitzGerald, J.M.; Nislow, C.; Leung, J.M.; et al. Sputum Microbiome Is Associated with 1-Year Mortality after Chronic Obstructive Pulmonary Disease Hospitalizations. Am. J. Respir. Crit. Care Med. 2019, 199, 1205–1213. [Google Scholar] [CrossRef]
- Bafadhel, M.; McKenna, S.; Terry, S.; Mistry, V.; Reid, C.; Haldar, P.; McCormick, M.; Haldar, K.; Kebadze, T.; Duvoix, A.; et al. Acute exacerbations of chronic obstructive pulmonary disease: Identification of biologic clusters and their biomarkers. Am. J. Respir. Crit. Care Med. 2011, 184, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Ghebre, M.A.; Pang, P.H.; Diver, S.; Desai, D.; Bafadhel, M.; Haldar, K.; Kebadze, T.; Cohen, S.; Newbold, P.; Rapley, L.; et al. Biological exacerbation clusters demonstrate asthma and chronic obstructive pulmonary disease overlap with distinct mediator and microbiome profiles. J. Allergy Clin. Immunol. 2018, 141, 2027–2036.e2012. [Google Scholar] [CrossRef] [Green Version]
- Brinkmann, V.; Reichard, U.; Goosmann, C.; Fauler, B.; Uhlemann, Y.; Weiss, D.S.; Weinrauch, Y.; Zychlinsky, A. Neutrophil extracellular traps kill bacteria. Science 2004, 303, 1532–1535. [Google Scholar] [CrossRef] [PubMed]
- Grabcanovic-Musija, F.; Obermayer, A.; Stoiber, W.; Krautgartner, W.D.; Steinbacher, P.; Winterberg, N.; Bathke, A.C.; Klappacher, M.; Studnicka, M. Neutrophil extracellular trap (NET) formation characterises stable and exacerbated COPD and correlates with airflow limitation. Respir. Res. 2015, 16, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obermayer, A.; Stoiber, W.; Krautgartner, W.D.; Klappacher, M.; Kofler, B.; Steinbacher, P.; Vitkov, L.; Grabcanovic-Musija, F.; Studnicka, M. New aspects on the structure of neutrophil extracellular traps from chronic obstructive pulmonary disease and in vitro generation. PLoS ONE 2014, 9, e97784. [Google Scholar] [CrossRef]
- Pedersen, F.; Marwitz, S.; Holz, O.; Kirsten, A.; Bahmer, T.; Waschki, B.; Magnussen, H.; Rabe, K.F.; Goldmann, T.; Uddin, M.; et al. Neutrophil extracellular trap formation and extracellular DNA in sputum of stable COPD patients. Respir. Med. 2015, 109, 1360–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dicker, A.J.; Crichton, M.L.; Pumphrey, E.G.; Cassidy, A.J.; Suarez-Cuartin, G.; Sibila, O.; Furrie, E.; Fong, C.J.; Ibrahim, W.; Brady, G.; et al. Neutrophil extracellular traps are associated with disease severity and microbiota diversity in patients with chronic obstructive pulmonary disease. J. Allergy Clin. Immunol. 2018, 141, 117–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agusti, A.; Fabbri, L.M.; Singh, D.; Vestbo, J.; Celli, B.; Franssen, F.M.E.; Rabe, K.F.; Papi, A. Inhaled corticosteroids in COPD: Friend or foe? Eur. Respir. J. 2018, 52, 1801219. [Google Scholar] [CrossRef] [PubMed]
- Kolsum, U.; Donaldson, G.C.; Singh, R.; Barker, B.L.; Gupta, V.; George, L.; Webb, A.J.; Thurston, S.; Brookes, A.J.; McHugh, T.D.; et al. Blood and sputum eosinophils in COPD; relationship with bacterial load. Respir. Res. 2017, 18, 88. [Google Scholar] [CrossRef]
- Zhou, W.; Tan, J. The expression and the clinical significance of eosinophils, PCT and CRP in patients with acute exacerbation of chronic obstructive pulmonary disease complicated with pulmonary infection. Am. J. Transl. Res. 2021, 13, 3451–3458. [Google Scholar]
- Pavord, I.D.; Lettis, S.; Anzueto, A.; Barnes, N. Blood eosinophil count and pneumonia risk in patients with chronic obstructive pulmonary disease: A patient-level meta-analysis. Lancet Respir. Med. 2016, 4, 731–741. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.A.; Faner, R.; Oscullo, G.; de la Rosa, D.; Soler-Cataluna, J.J.; Ballester, M.; Agusti, A. Inhaled Steroids, Circulating Eosinophils, Chronic Airway Infection, and Pneumonia Risk in Chronic Obstructive Pulmonary Disease. A Network Analysis. Am. J. Respir. Crit. Care Med. 2020, 201, 1078–1085. [Google Scholar] [CrossRef]
- Crim, C.; Calverley, P.M.; Anderson, J.A.; Celli, B.; Ferguson, G.T.; Jenkins, C.; Jones, P.W.; Willits, L.R.; Yates, J.C.; Vestbo, J. Pneumonia risk in COPD patients receiving inhaled corticosteroids alone or in combination: TORCH study results. Eur. Respir. J. 2009, 34, 641–647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crim, C.; Dransfield, M.T.; Bourbeau, J.; Jones, P.W.; Hanania, N.A.; Mahler, D.A.; Vestbo, J.; Wachtel, A.; Martinez, F.J.; Barnhart, F.; et al. Pneumonia risk with inhaled fluticasone furoate and vilanterol compared with vilanterol alone in patients with COPD. Ann. Am. Thorac. Soc. 2015, 12, 27–34. [Google Scholar] [CrossRef]
- 87. Rabe, K.F.; Martinez, F.J.; Ferguson, G.T.; Wang, C.; Singh, D.; Wedzicha, J.A.; Trivedi, R.; St. Rose, E.; Ballal, S.; McLaren, J.; et al. Triple inhaled therapy at two glucocorticoid doses in moderate-to-very-severe COPD. N. Engl. J. Med. 2020, 383, 35–48. [Google Scholar] [CrossRef]
- Lipson, D.A.; Barnhart, F.; Brealey, N.; Brooks, J.; Criner, G.J.; Day, N.C.; Dransfield, M.T.; Halpin, D.M.G.; Han, M.K.; Jones, C.E.; et al. Once-daily single-inhaler triple versus dual therapy in patients with COPD. N. Engl. J. Med. 2018, 378, 1671–1680. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Sun, J.; Huang, Q.; Liu, Y.; Yuan, M.; Ma, C.; Yan, H. Inhaled Corticosteroids and the Pneumonia Risk in Patients With Chronic Obstructive Pulmonary Disease: A Meta-analysis of Randomized Controlled Trials. Front. Pharmacol. 2021, 12, 691621. [Google Scholar] [CrossRef] [PubMed]
- Kamal, F.; Glanville, N.; Xia, W.; Bakhsoliani, E.; Aniscenko, J.; Bartlett, N.W.; Edwards, M.R.; Johnston, S.L.; Singanayagam, A. Beclomethasone Has Lesser Suppressive Effects on Inflammation and Antibacterial Immunity Than Fluticasone or Budesonide in Experimental Infection Models. Chest 2020, 158, 947–951. [Google Scholar] [CrossRef] [PubMed]
- Heijink, I.H.; Jonker, M.R.; de Vries, M.; van Oosterhout, A.J.; Telenga, E.; Ten Hacken, N.H.; Postma, D.S.; van den Berge, M. Budesonide and fluticasone propionate differentially affect the airway epithelial barrier. Respir. Res. 2016, 17, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Provost, K.A.; Smith, M.; Miller-Larsson, A.; Gudleski, G.D.; Sethi, S. Bacterial regulation of macrophage bacterial recognition receptors in COPD are differentially modified by budesonide and fluticasone propionate. PLoS ONE 2019, 14, e0207675. [Google Scholar] [CrossRef]
- Contoli, M.; Pauletti, A.; Rossi, M.R.; Spanevello, A.; Casolari, P.; Marcellini, A.; Forini, G.; Gnesini, G.; Marku, B.; Barnes, N.; et al. Long-term effects of inhaled corticosteroids on sputum bacterial and viral loads in COPD. Eur. Respir. J. 2017, 50. [Google Scholar] [CrossRef] [Green Version]
- Leitao Filho, F.S.; Takiguchi, H.; Akata, K.; Ra, S.W.; Moon, J.Y.; Kim, H.K.; Cho, Y.; Yamasaki, K.; Milne, S.; Yang, J.; et al. Effects of Inhaled Corticosteroid/long-acting beta-2 Agonist Combination on the Airway Microbiome of Patients with COPD: A Randomized Controlled Trial (DISARM). Am. J. Respir. Crit. Care Med. 2021. [Google Scholar] [CrossRef]
- Singanayagam, A.; Glanville, N.; Cuthbertson, L.; Bartlett, N.W.; Finney, L.J.; Turek, E.; Bakhsoliani, E.; Calderazzo, M.A.; Trujillo-Torralbo, M.-B.; Footitt, J.; et al. Inhaled corticosteroid suppression of cathelicidin drives dysbiosis and bacterial infection in chronic obstructive pulmonary disease. Sci. Transl. Med. 2019, 11, eaav3879. [Google Scholar] [CrossRef]
- Garcha, D.S.; Thurston, S.J.; Patel, A.R.; Mackay, A.J.; Goldring, J.J.; Donaldson, G.C.; McHugh, T.D.; Wedzicha, J.A. Changes in prevalence and load of airway bacteria using quantitative PCR in stable and exacerbated COPD. Thorax 2012, 67, 1075–1080. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.J.; Sethi, S.; Murphy, T.; Nariya, S.; Boushey, H.A.; Lynch, S.V. Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J. Clin. Microbiol. 2014, 52, 2813–2823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barbier, M.; Agustí, A.; Albertí, S. Fluticasone propionate reduces bacterial airway epithelial invasion. Eur. Respir. J. 2008, 32, 1283–1288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, C.; Goldmann, T.; Rohmann, K.; Rupp, J.; Marwitz, S.; Rotta Detto Loria, J.; Limmer, S.; Zabel, P.; Dalhoff, K.; Drömann, D. Budesonide Inhibits Intracellular Infection with Non-Typeable Haemophilus influenzae Despite Its Anti-Inflammatory Effects in Respiratory Cells and Human Lung Tissue: A Role for p38 MAP Kinase. Respiration 2015, 90, 416–425. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Wang, X.; Yang, X.; Liu, Z.; Wu, M.; Li, G. Budesonide suppresses pulmonary antibacterial host defense by down-regulating cathelicidin-related antimicrobial peptide in allergic inflammation mice and in lung epithelial cells. BMC Immunol. 2013, 14, 7. [Google Scholar] [CrossRef] [Green Version]
- Branzk, N.; Lubojemska, A.; Hardison, S.E.; Wang, Q.; Gutierrez, M.G.; Brown, G.D.; Papayannopoulos, V. Neutrophils sense microbe size and selectively release neutrophil extracellular traps in response to large pathogens. Nat. Immunol. 2014, 15, 1017–1025. [Google Scholar] [CrossRef] [Green Version]
- Menegazzi, R.; Decleva, E.; Dri, P. Killing by neutrophil extracellular traps: Fact or folklore? Blood 2012, 119, 1214–1216. [Google Scholar] [CrossRef] [Green Version]
- Scott, J.; Harris, G.J.; Pinder, E.M.; Macfarlane, J.G.; Hellyer, T.P.; Rostron, A.J.; Conway Morris, A.; Thickett, D.R.; Perkins, G.D.; McAuley, D.F.; et al. Exchange protein directly activated by cyclic AMP (EPAC) activation reverses neutrophil dysfunction induced by beta2-agonists, corticosteroids, and critical illness. J. Allergy Clin. Immunol. 2016, 137, 535–544. [Google Scholar] [CrossRef]
| Authors | Year | Study Population | Treatment | Endpoints | Observations |
|---|---|---|---|---|---|
| Turturice [36] | 2017 | Adult atopic asthma, intermittent or mild/moderate persistent symptoms (n = 13); age-matched non-asthmatic controls (n = 6) | 6 weeks’ FP vs. before treatment | Inflammatory markers (cytokine + chemokine panel) and metagenomic sequencing of lower airway microbiome in BAL | Reductions in S. pneumoniae, N. meningitis, E. faecium, E. faecalis. Associated with decreased MIP-1β, increased IL-2 |
| Ramakrishnan [37] | 2018 | Adult chronic non-infectious rhinitis males (n = 4); healthy female (n = 1) | Non-infectious rhinitis males: mometasone furoate nasal spray QD for 1 month healthy female: BID topical mupirocin decolonisation | Serial nasal cavity swab microbiome analysis over 8 weeks (16S rRNA gene) | Increased abundance of staphylococci; reduced abundance of Moraxella and streptococci. Increase in diversity in 2/4 subjects |
| Contoli [93] | 2017 | Steroid-naïve, stable moderate COPD (n = 60) on treatment with SAL | 1:1 SAL/FP:SAL BID 12 months | Sputum bacterial load, microbiome composition (16S rRNA gene) | Increased bacterial load, an increased proportion of S. pneumoniae, H. influenzae with low blood/sputum eosinophils. Increase in diversity, an increased proportion of Firmicutes, Candida; the reduced proportion of Proteobacteria |
| Leitao Filho [94] | 2021 | Adults (n = 63) with stable moderate-to-severe COPD, 4-week ICS washout, 4-week run-in with FORM | BUD/FORM or FP/SAL vs. FORM | Bronchoscopy bacterial load, microbiome composition, microbiome changes vs. clinical parameters | In FP/Sal group, reduction in diversity, greater number of changes in microbiome from BL, decreased abundance of Haemophilus, decreased Proteobacteria:Firmicutes ratio |
| Singanayagam [95] | 2019 | Stable mild-moderate COPD: current ICS use (n = 10); non-use of ICS (muscarinic antagonist; SABA/LABA; n = 13) | FP, BUD, BD | Sputum bacterial load, microbiome composition (16S rRNA gene) | Increase in abundance of Streptococcus; increased bacterial load and diversity |
| Patients reporting exacerbations: current ICS use (n = 11); non-use of ICS (n = 16) | FP, BUD, BD | Sputum hCAP18, BAL cathepsin D concentration | Suppressed hCAP18 concentrations; increased cathepsin D concentrations | ||
| Mouse model of COPD; WT mice | FP vs. pre-FP | S. pneumoniae load, cathelicidin-related AMP concentration, cathepsin D concentration in BAL, whole lung, blood | Increased S. pneumoniae load; reduced concentrations of cathelicidin-related AMP; increased cathepsin D concentrations | ||
| Cathelicidin knock-out mouse | FP vs. control | Bacterial load; S. pneumoniae load in BAL | No effect of FP on cathelicidin knockouts | ||
| BEAS2B bronchial epithelial cells | FP vs. control | hCAP18 concentration; cathepsin D concentration | Suppressed hCAP18 concentrations; augmented cathepsin D induction | ||
| COPD primary bronchial epithelial cells | FP vs. control | hCAP18 concentration | Suppressed hCAP18 concentrations | ||
| Garcha [96] | 2012 | Stable COPD GOLD stages II–IV (n = 134) | n = 47 using ICS (median [IQR] beclomethasone-equivalent dosage 2000 (640–2000) mg daily) | Sputum bacterial load; severe airflow limitation | Higher airway bacterial load associated with higher ICS usage and more severe airflow limitation |
| Huang [97] | 2014 | COPD patients with bacterial infection (n = 12) | Antibiotics only vs. oral corticosteroids only vs. both | Sputum microbiome composition (16S rRNA gene) | Oral corticosteroids alone: increased proportion of Proteobacteria, Bacteroidetes and Firmicutes, particularly Enterobacteriaceae, Lachnospiraceae, Burkholderiaceae, Neisseriaceae Oral corticosteroids plus antibiotics: increase in Proteobacteria |
| Wang [68] | 2016 | Stable and exacerbative COPD patients (n = 87) | Antibiotics only vs. oral corticosteroids only vs. both | Sputum microbiome composition (16S rRNA gene) | Oral corticosteroids alone: decreased diversity; increased proportion of Proteobacteria; decrease in Streptococcus, increase in Haemophilus and Moraxella |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Keir, H.R.; Contoli, M.; Chalmers, J.D. Inhaled Corticosteroids and the Lung Microbiome in COPD. Biomedicines 2021, 9, 1312. https://doi.org/10.3390/biomedicines9101312
Keir HR, Contoli M, Chalmers JD. Inhaled Corticosteroids and the Lung Microbiome in COPD. Biomedicines. 2021; 9(10):1312. https://doi.org/10.3390/biomedicines9101312
Chicago/Turabian StyleKeir, Holly R., Marco Contoli, and James D. Chalmers. 2021. "Inhaled Corticosteroids and the Lung Microbiome in COPD" Biomedicines 9, no. 10: 1312. https://doi.org/10.3390/biomedicines9101312
APA StyleKeir, H. R., Contoli, M., & Chalmers, J. D. (2021). Inhaled Corticosteroids and the Lung Microbiome in COPD. Biomedicines, 9(10), 1312. https://doi.org/10.3390/biomedicines9101312






