Insights into the Interaction Mechanism of DTP3 with MKK7 by Using STD-NMR and Computational Approaches
Abstract
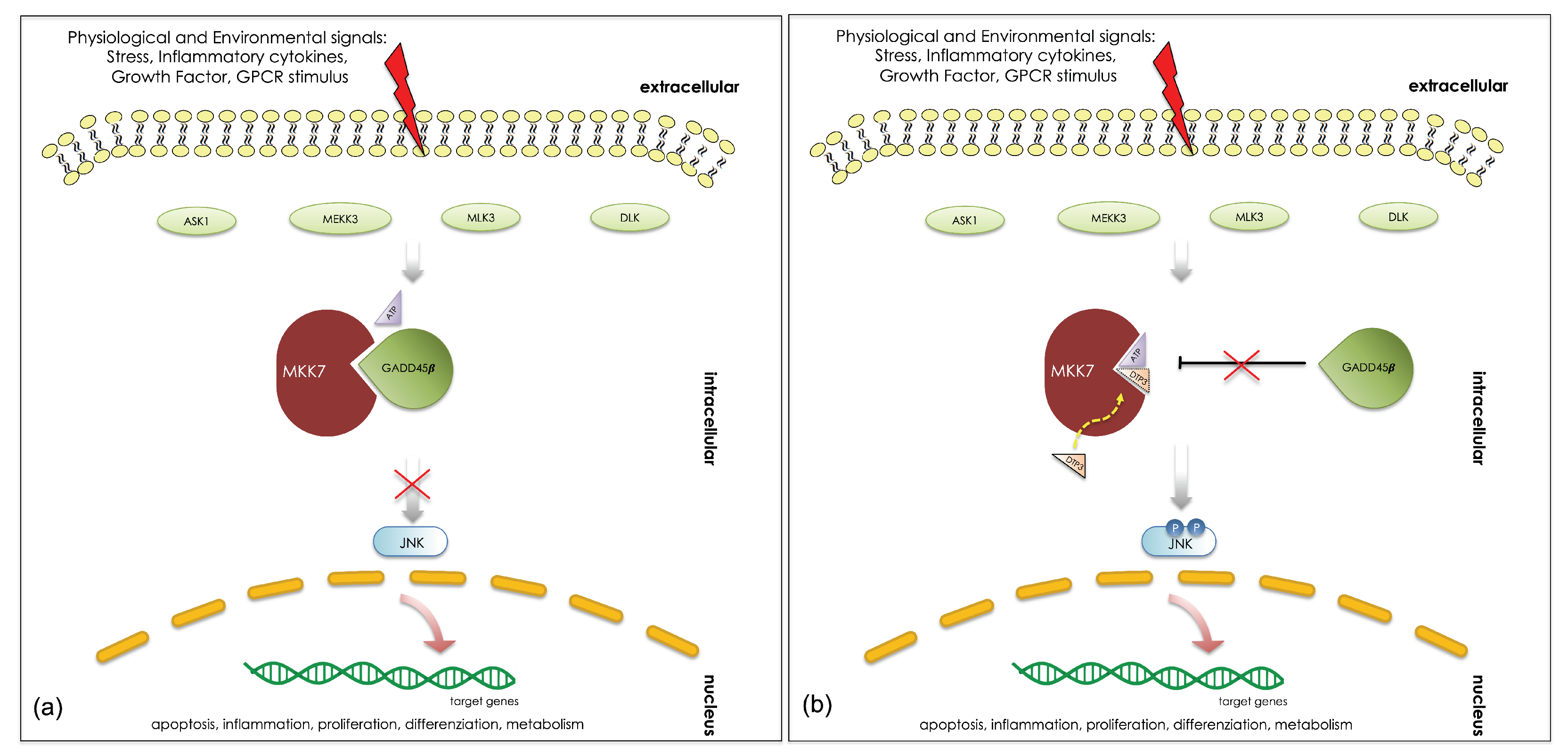
1. Introduction
2. Materials and Methods
2.1. Synthesis and Purification of FITC--DTP3 and FITC--SCRB Peptides
2.2. Fluorescence Quenching Ligand Binding Assay
2.3. NMR Analyses
2.3.1. NMR Spectroscopy of Free Molecules
2.3.2. NMR Spectroscopy of Tripeptide–Protein Interactions
2.4. Computational Studies
2.4.1. Molecular Dynamics Simulations
2.4.2. Zernike Descriptors
2.4.3. Binding Sites Detection and Molecular Docking
3. Results
3.1. Peptide Synthesis
3.2. Fluorescence Quenching Ligand Binding Assay
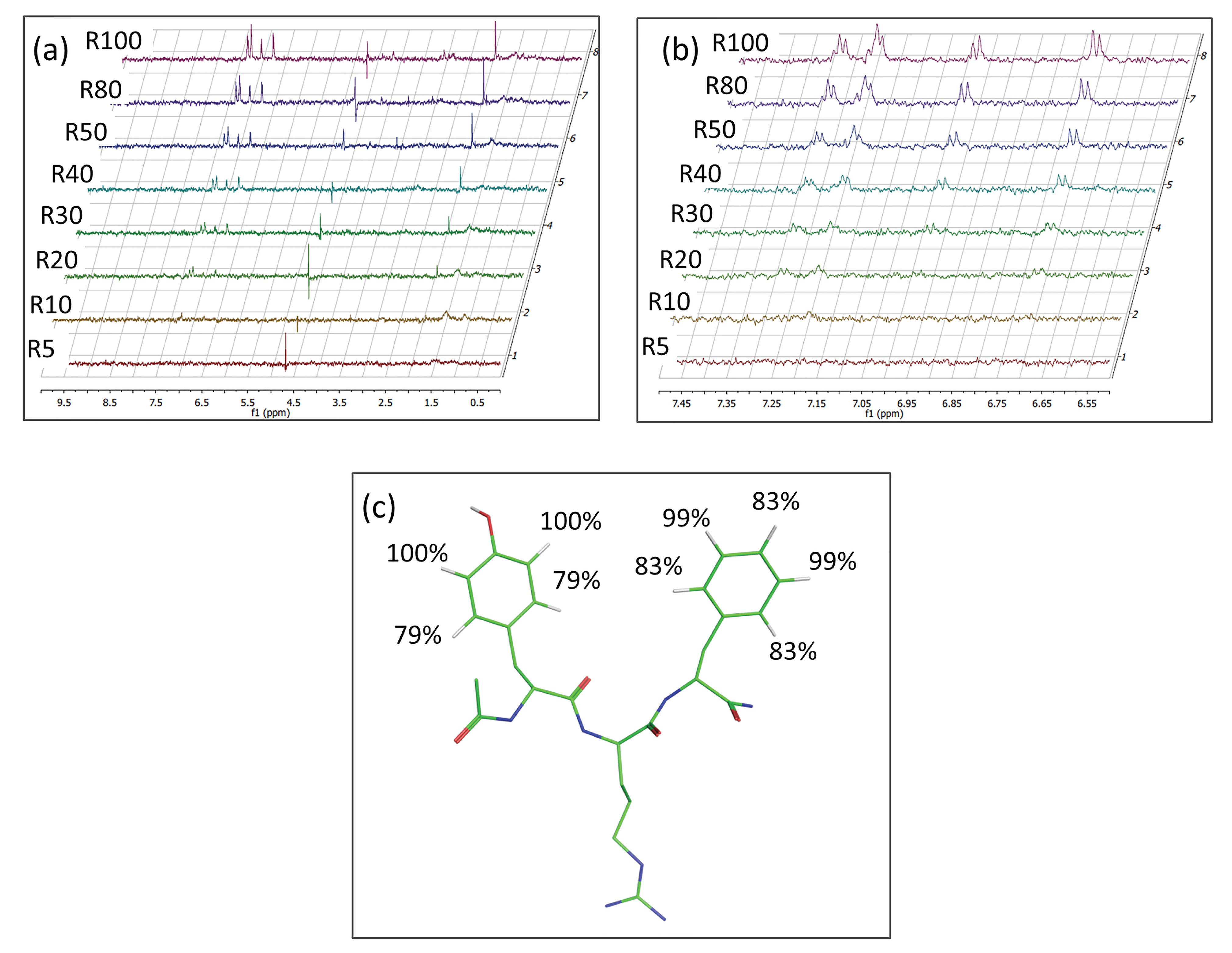
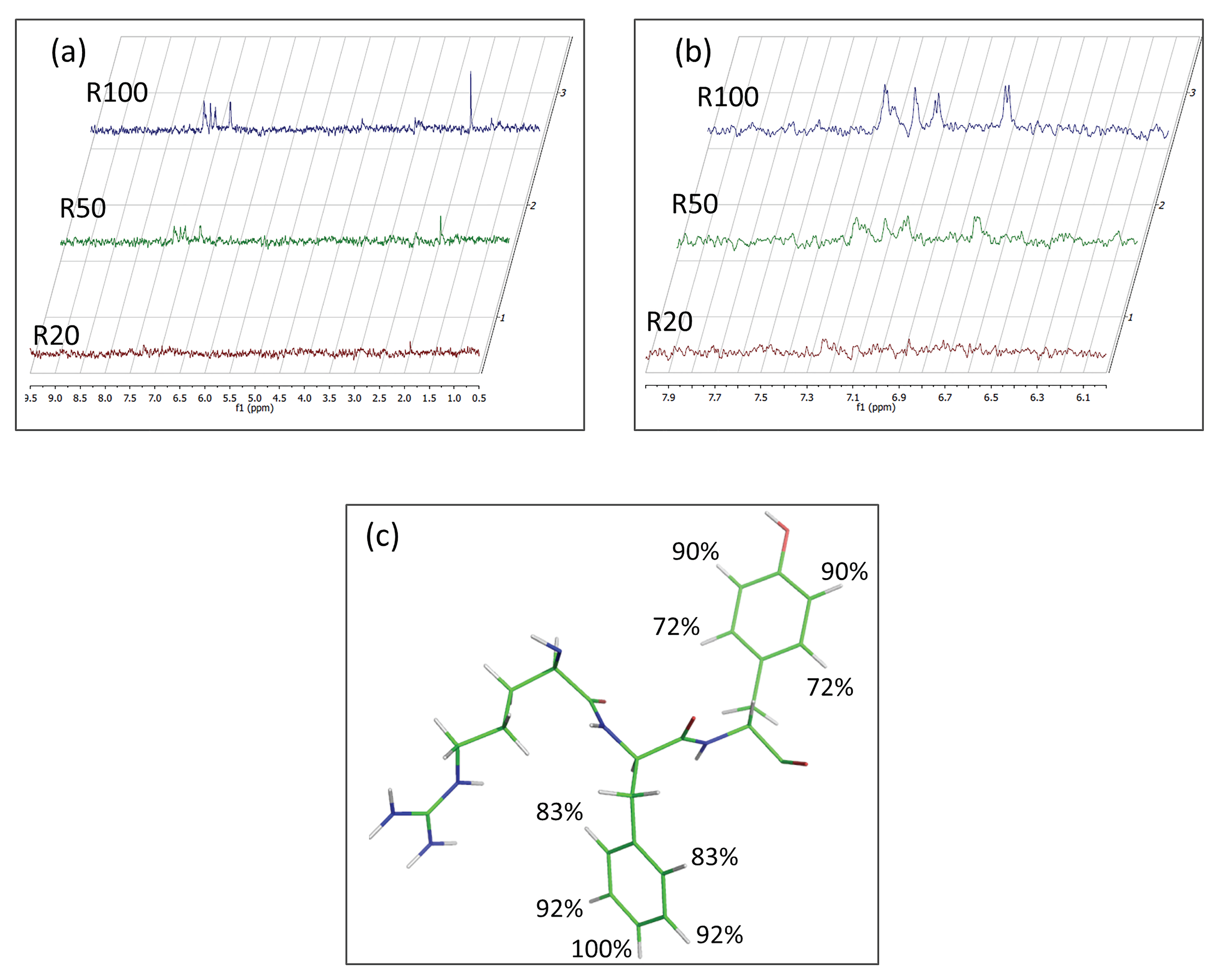
3.3. Interaction studies of DTP3 and SCRB with MKK7-KD by STD-NMR
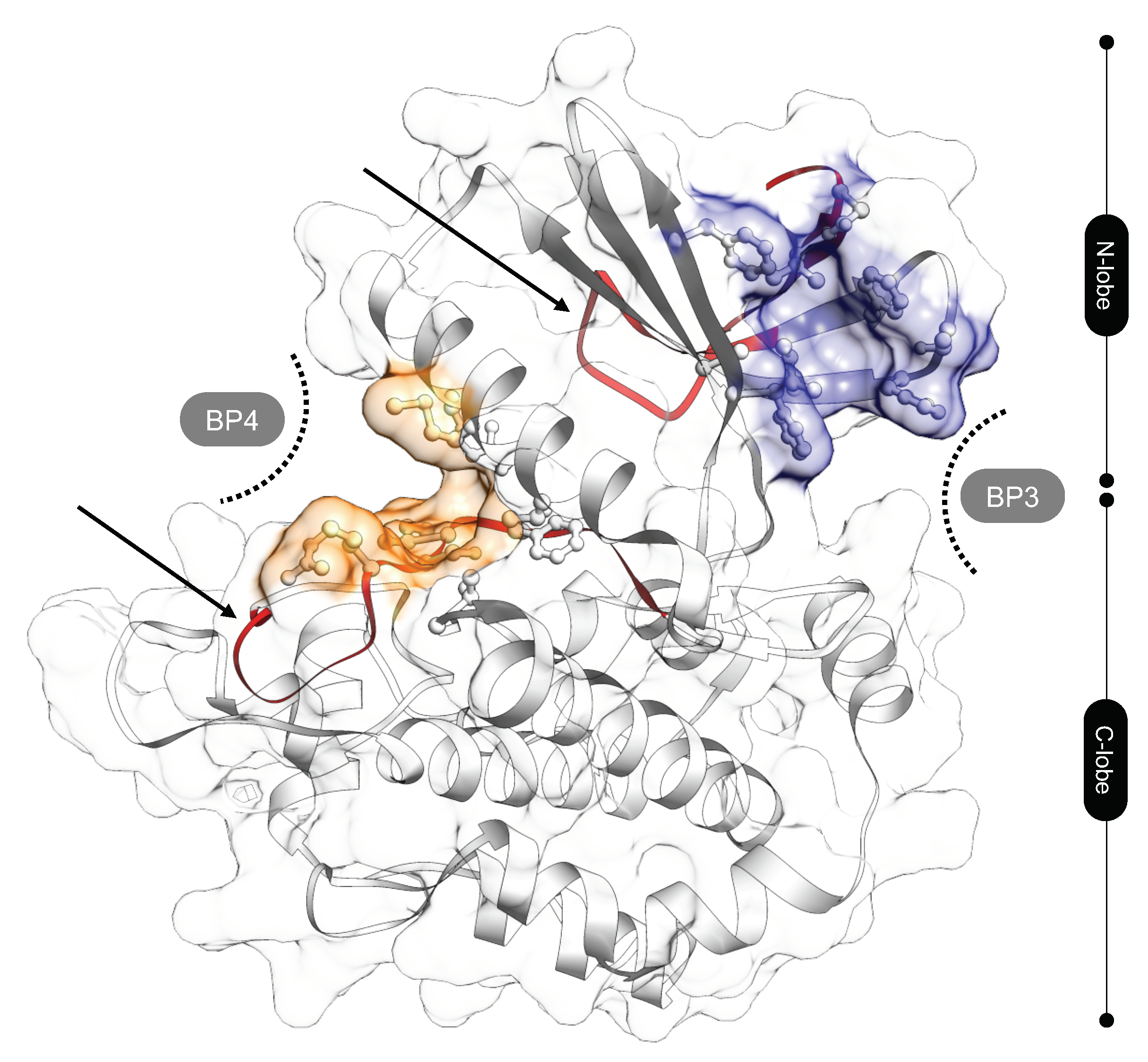
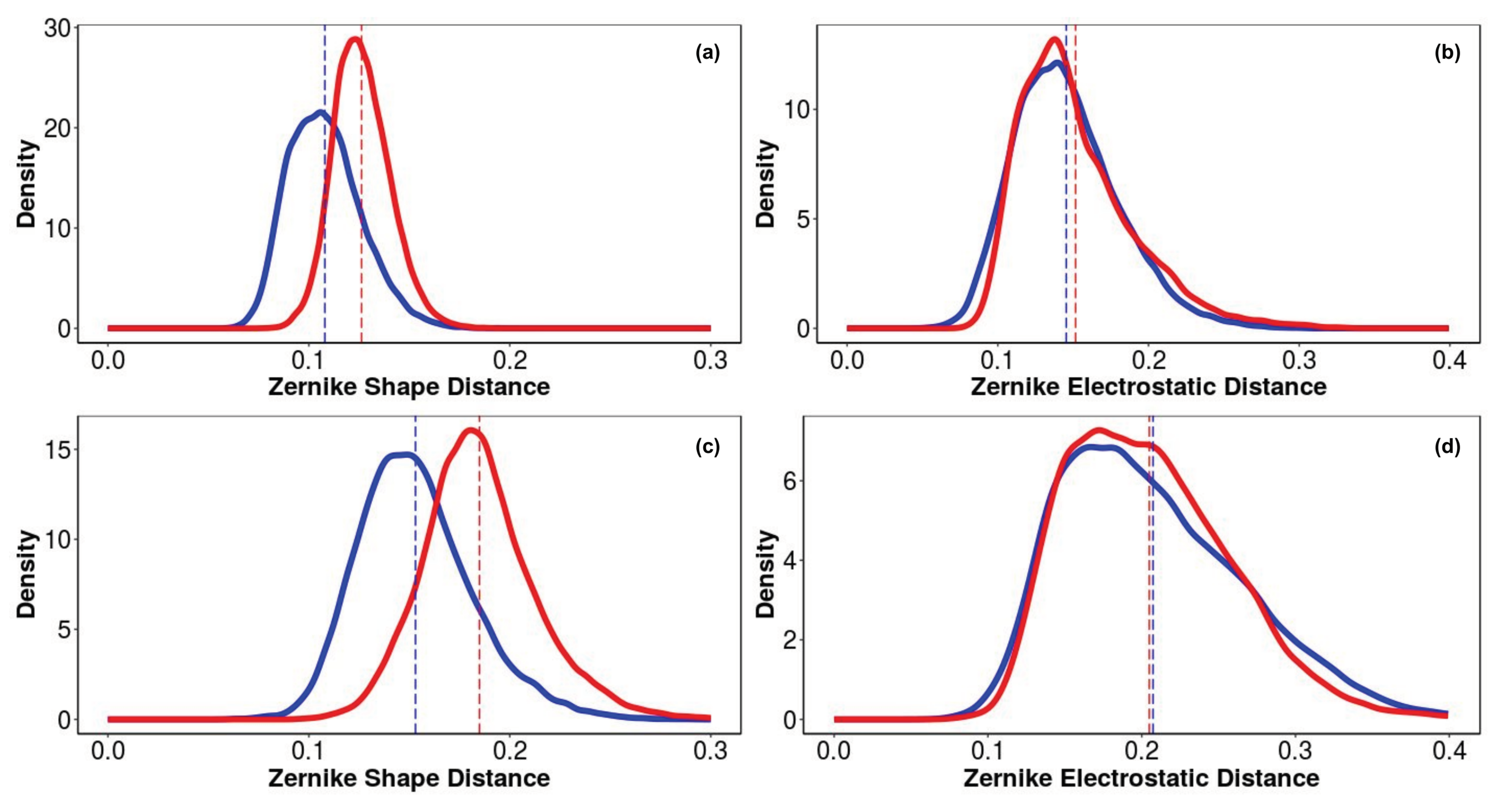
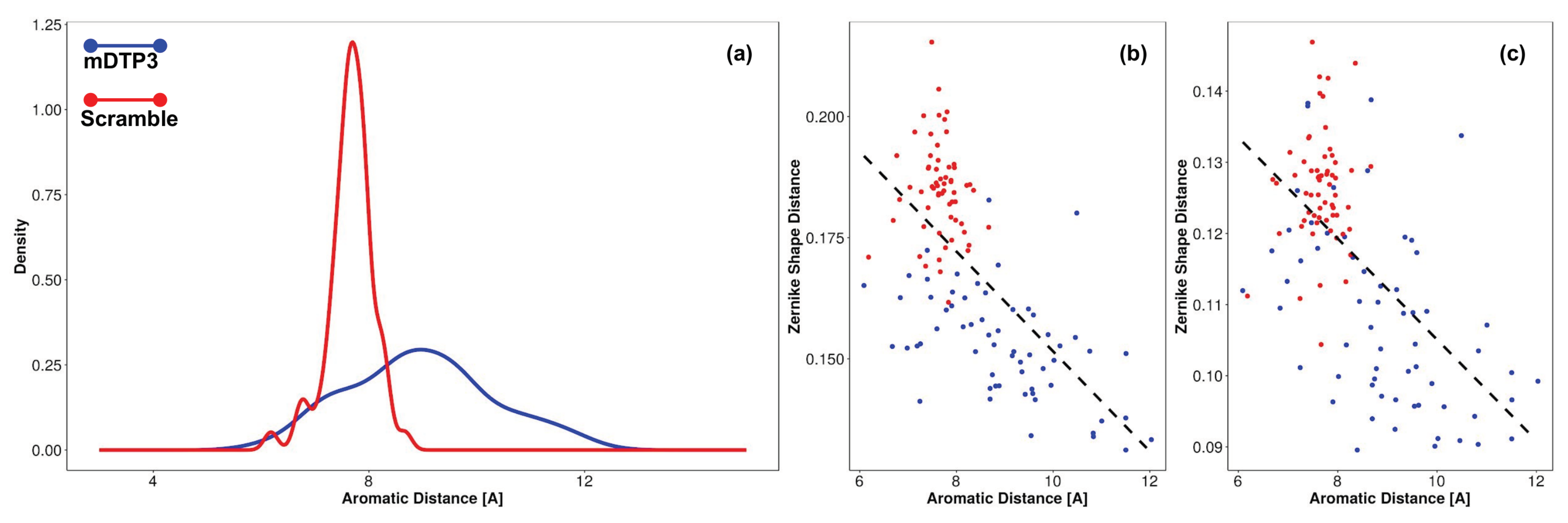
3.4. Identification of the DTP3 Binding Sites on MKK7-KD Surface and Their Characterization by Using Zernike Polynomials
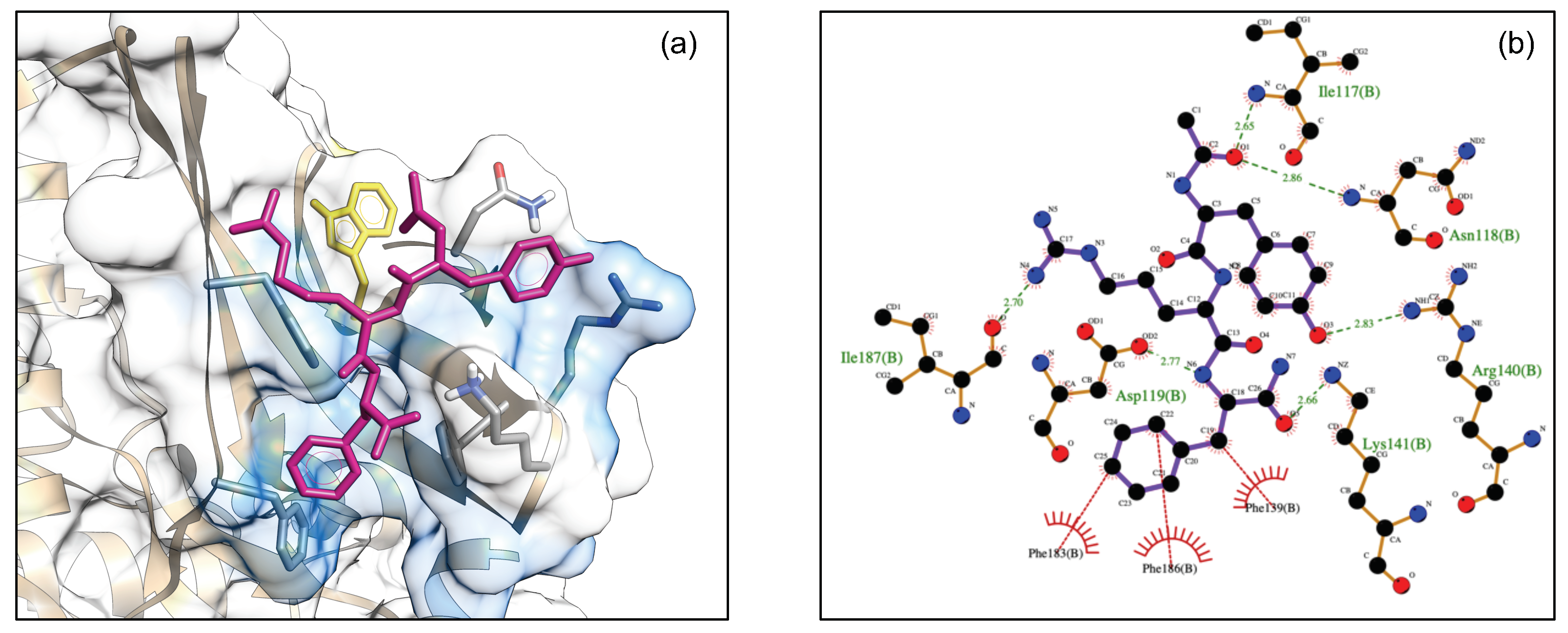
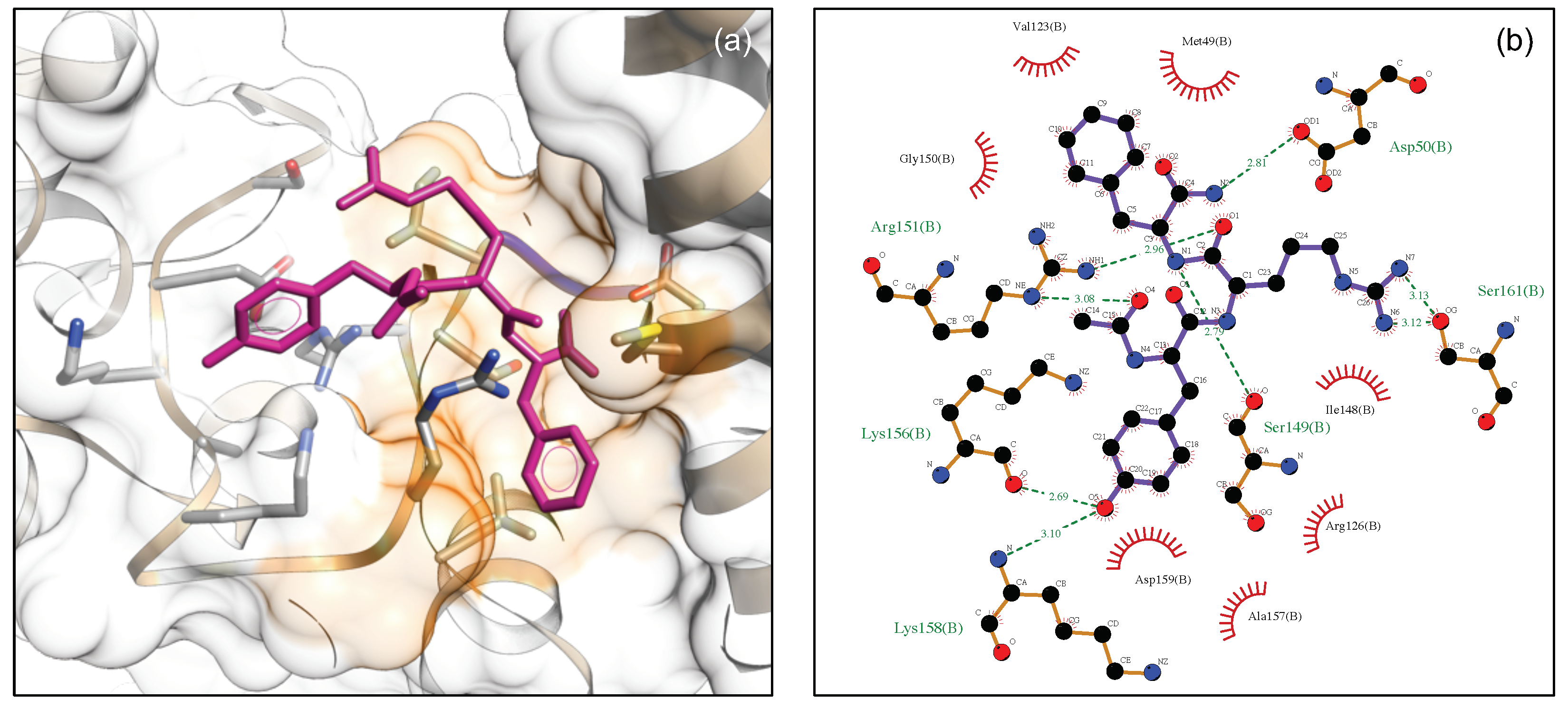
3.5. MKK7-DTP3 Molecular Docking Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, L.; Karin, M. Mammalian MAP kinase signalling cascades. Nature 2001, 410, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Cuevas, B.D.; Abell, A.N.; Johnson, G.L. Role of mitogen-activated protein kinase kinase kinases in signal integration. Oncogene 2007, 26, 3159–3171. [Google Scholar] [CrossRef] [PubMed]
- Park, J.G.; Aziz, N.; Cho, J.Y. MKK7, the essential regulator of JNK signaling involved in cancer cell survival: A newly emerging anticancer therapeutic target. Ther. Adv. Med Oncol. 2019, 11, 1758835919875574. [Google Scholar] [CrossRef] [PubMed]
- Haeusgen, W.; Herdegen, T.; Waetzig, V. The bottleneck of JNK signaling: Molecular and functional characteristics of MKK4 and MKK7. Eur. J. Cell Biol. 2011, 90, 536–544. [Google Scholar] [CrossRef] [PubMed]
- Tornatore, L.; Sandomenico, A.; Raimondo, D.; Low, C.; Rocci, A.; Tralau-Stewart, C.; Capece, D.; D’Andrea, D.; Bua, M.; Boyle, E.; et al. Cancer-Selective Targeting of the Nf-KB Survival Pathway With Gadd45B/Mkk7 Inhibitors. Cancer Cell 2014, 26, 495–508. [Google Scholar] [CrossRef]
- Shraga, A.; Olshvang, E.; Davidzohn, N.; Khoshkenar, P.; Germain, N.; Shurrush, K.; Carvalho, S.; Avram, L.; Albeck, S.; Unger, T.; et al. Covalent Docking Identifies a Potent and Selective MKK7 Inhibitor. Cell Chem. Biol. 2019, 26, 98–108.e5. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef]
- Tornatore, L.; Capece, D.; D’Andrea, D.; Begalli, F.; Verzella, D.; Bennett, J.; Acton, G.; Campbell, E.A.; Kelly, J.; Tarbit, M.; et al. Preclinical toxicology and safety pharmacology of the first-in-class GADD45β/MKK7 inhibitor and clinical candidate, DTP3. Toxicol. Rep. 2019, 6, 369–379. [Google Scholar] [CrossRef]
- Tornatore, L.; Capece, D.; D’Andrea, D.; Begalli, F.; Verzella, D.; Bennett, J.; Acton, G.; Campbell, E.A.; Kelly, J.; Tarbit, M.; et al. Clinical proof of concept for a safe and effective NF-κB-targeting strategy in multiple myeloma. Br. J. Haematol. 2019, 185, 588. [Google Scholar] [CrossRef]
- Tornatore, L.; Marasco, D.; Dathan, N.; Vitale, R.M.; Benedetti, E.; Papa, S.; Franzoso, G.; Ruvo, M.; Monti, S.M. Gadd45β forms a Homodimeric Complex that Binds Tightly to MKK7. J. Mol. Biol. 2008, 378, 97–111. [Google Scholar] [CrossRef]
- Rega, C.; Russo, R.; Focà, A.; Sandomenico, A.; Iaccarino, E.; Raimondo, D.; Milanetti, E.; Tornatore, L.; Franzoso, G.; Pedone, P.V.; et al. Probing the interaction interface of the GADD45β/MKK7 and MKK7/DTP3 complexes by chemical cross-linking mass spectrometry. Int. J. Biol. Macromol. 2018, 114, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Schröder, M.; Tan, L.; Wang, J.; Liang, Y.; Gray, N.S.; Knapp, S.; Chaikuad, A. Catalytic Domain Plasticity of MKK7 Reveals Structural Mechanisms of Allosteric Activation and Diverse Targeting Opportunities. Cell Chem. Biol. 2020, 27, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Mayer, M.; Meyer, B. Group epitope mapping by saturation transfer difference NMR to identify segments of a ligand in direct contact with a protein receptor. J. Am. Chem. Soc. 2001, 123, 6108–6117. [Google Scholar] [CrossRef] [PubMed]
- Zoete, V.; Cuendet, M.A.; Grosdidier, A.; Michielin, O. SwissParam: A fast force field generation tool for small organic molecules. J. Comput. Chem. 2011, 32, 2359–2368. [Google Scholar] [CrossRef] [PubMed]
- Mark, P.; Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 2001, 105, 9954–9960. [Google Scholar] [CrossRef]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 014101. [Google Scholar] [CrossRef]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindah, E. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef]
- Fogolari, F.; Corazza, A.; Yarra, V.; Jalaru, A.; Viglino, P.; Esposito, G. Bluues: A program for the analysis of the electrostatic properties of proteins based on generalized Born radii. BMC Bioinform. 2012, 13, S18. [Google Scholar] [CrossRef]
- Dolinsky, T.J.; Nielsen, J.E.; McCammon, J.A.; Baker, N.A. PDB2PQR: An automated pipeline for the setup of Poisson-Boltzmann electrostatics calculations. Nucleic Acids Res. 2004, 32, W665. [Google Scholar] [CrossRef]
- Venkatraman, V.; Sael, L.; Kihara, D. Potential for protein surface shape analysis using spherical harmonics and 3d zernike descriptors. Cell Biochem. Biophys. 2009, 54, 23–32. [Google Scholar] [CrossRef]
- Krivák, R.; Hoksza, D. P2Rank: Machine learning based tool for rapid and accurate prediction of ligand binding sites from protein structure. J. Cheminform. 2018, 10, 39. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2009, 31, 455–461. [Google Scholar] [CrossRef] [PubMed]
- Feinstein, W.P.; Brylinski, M. Calculating an optimal box size for ligand docking and virtual screening against experimental and predicted binding pockets. J. Cheminform. 2015, 7, 18. [Google Scholar] [CrossRef]
- Laskowski, R.A.; Swindells, M.B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011, 51, 2778–2786. [Google Scholar] [CrossRef]
- Sandomenico, A.; Russo, A.; Palmieri, G.; Bergamo, P.; Gogliettino, M.; Falcigno, L.; Ruvo, M. Small peptide inhibitors of acetyl-peptide hydrolase having an uncommon mechanism of inhibition and a stable bent conformation. J. Med. Chem. 2012, 55, 2102–2111. [Google Scholar] [CrossRef] [PubMed]
- Di Rienzo, L.; Milanetti, E.; Lepore, R.; Olimpieri, P.P.; Tramontano, A. Superposition-free comparison and clustering of antibody binding sites: Implications for the prediction of the nature of their antigen. Sci. Rep. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Kihara, D.; Sael, L.; Chikhi, R.; Esquivel-Rodriguez, J. Molecular Surface Representation Using 3D Zernike Descriptors for Protein Shape Comparison and Docking. Curr. Protein Pept. Sci. 2011, 12, 520–530. [Google Scholar] [CrossRef]
- Di Rienzo, L.; Milanetti, E.; Alba, J.; D’Abramo, M. Quantitative Characterization of Binding Pockets and Binding Complementarity by Means of Zernike Descriptors. J. Chem. Inf. Model. 2020, 2020, 1390–1398. [Google Scholar] [CrossRef]
- Daberdaku, S.; Ferrari, C. Exploring the potential of 3D Zernike descriptors and SVM for protein-protein interface prediction. BMC Bioinform. 2018, 19, 35. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sandomenico, A.; Di Rienzo, L.; Calvanese, L.; Iaccarino, E.; D’Auria, G.; Falcigno, L.; Chambery, A.; Russo, R.; Franzoso, G.; Tornatore, L.; et al. Insights into the Interaction Mechanism of DTP3 with MKK7 by Using STD-NMR and Computational Approaches. Biomedicines 2021, 9, 20. https://doi.org/10.3390/biomedicines9010020
Sandomenico A, Di Rienzo L, Calvanese L, Iaccarino E, D’Auria G, Falcigno L, Chambery A, Russo R, Franzoso G, Tornatore L, et al. Insights into the Interaction Mechanism of DTP3 with MKK7 by Using STD-NMR and Computational Approaches. Biomedicines. 2021; 9(1):20. https://doi.org/10.3390/biomedicines9010020
Chicago/Turabian StyleSandomenico, Annamaria, Lorenzo Di Rienzo, Luisa Calvanese, Emanuela Iaccarino, Gabriella D’Auria, Lucia Falcigno, Angela Chambery, Rosita Russo, Guido Franzoso, Laura Tornatore, and et al. 2021. "Insights into the Interaction Mechanism of DTP3 with MKK7 by Using STD-NMR and Computational Approaches" Biomedicines 9, no. 1: 20. https://doi.org/10.3390/biomedicines9010020
APA StyleSandomenico, A., Di Rienzo, L., Calvanese, L., Iaccarino, E., D’Auria, G., Falcigno, L., Chambery, A., Russo, R., Franzoso, G., Tornatore, L., D’Abramo, M., Ruvo, M., Milanetti, E., & Raimondo, D. (2021). Insights into the Interaction Mechanism of DTP3 with MKK7 by Using STD-NMR and Computational Approaches. Biomedicines, 9(1), 20. https://doi.org/10.3390/biomedicines9010020








