Novel 1,3,4-oxadiazole Targets STAT3 Signaling to Induce Antitumor Effect in Lung Cancer
Abstract
1. Introduction
2. Results
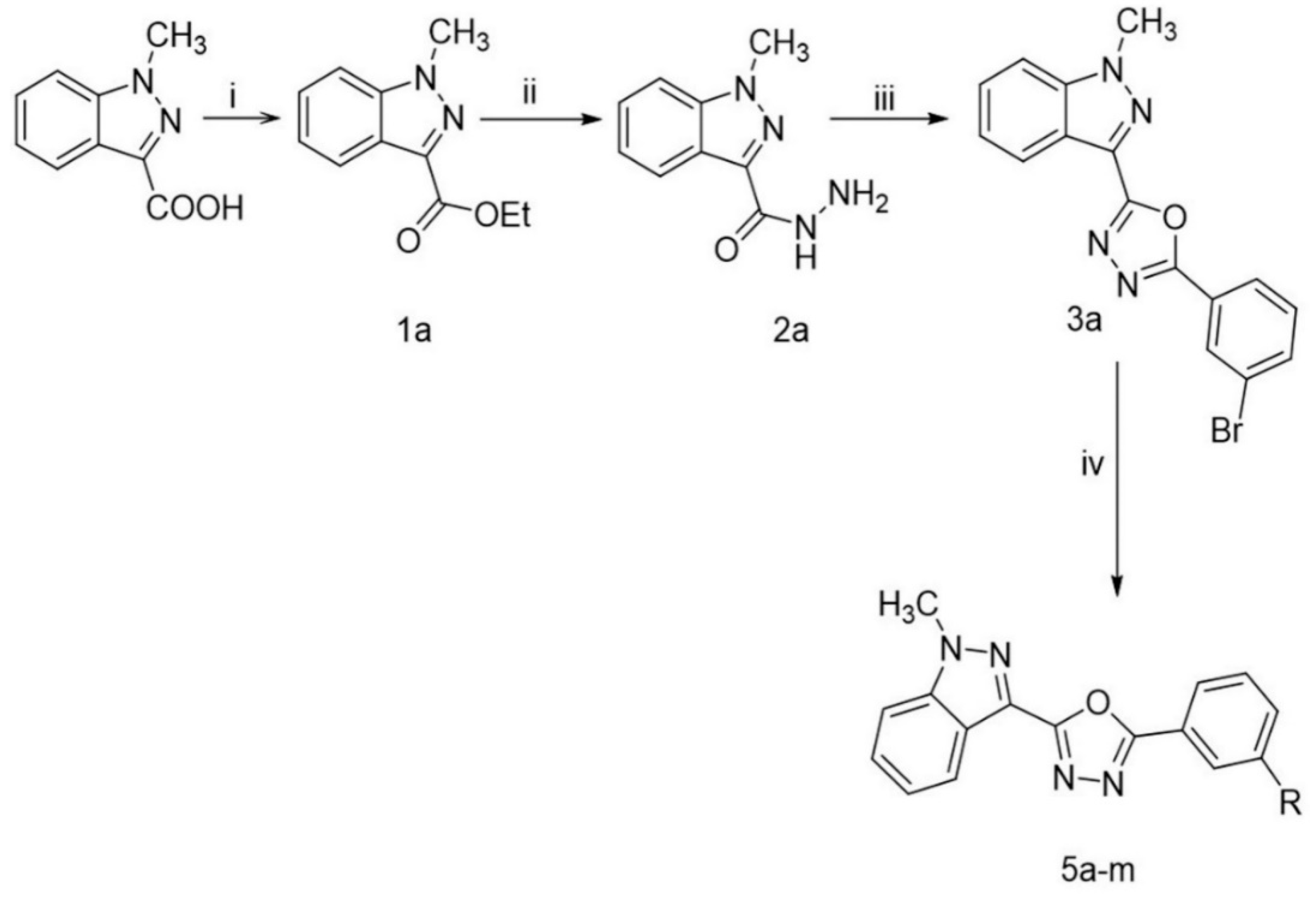
2.1. Chemistry
2.2. Biology
2.2.1. CHK9 Induces Cytotoxicity in Lung Cancer Cells
2.2.2. CHK9 Triggers Apoptosis in Lung Cancer Cells
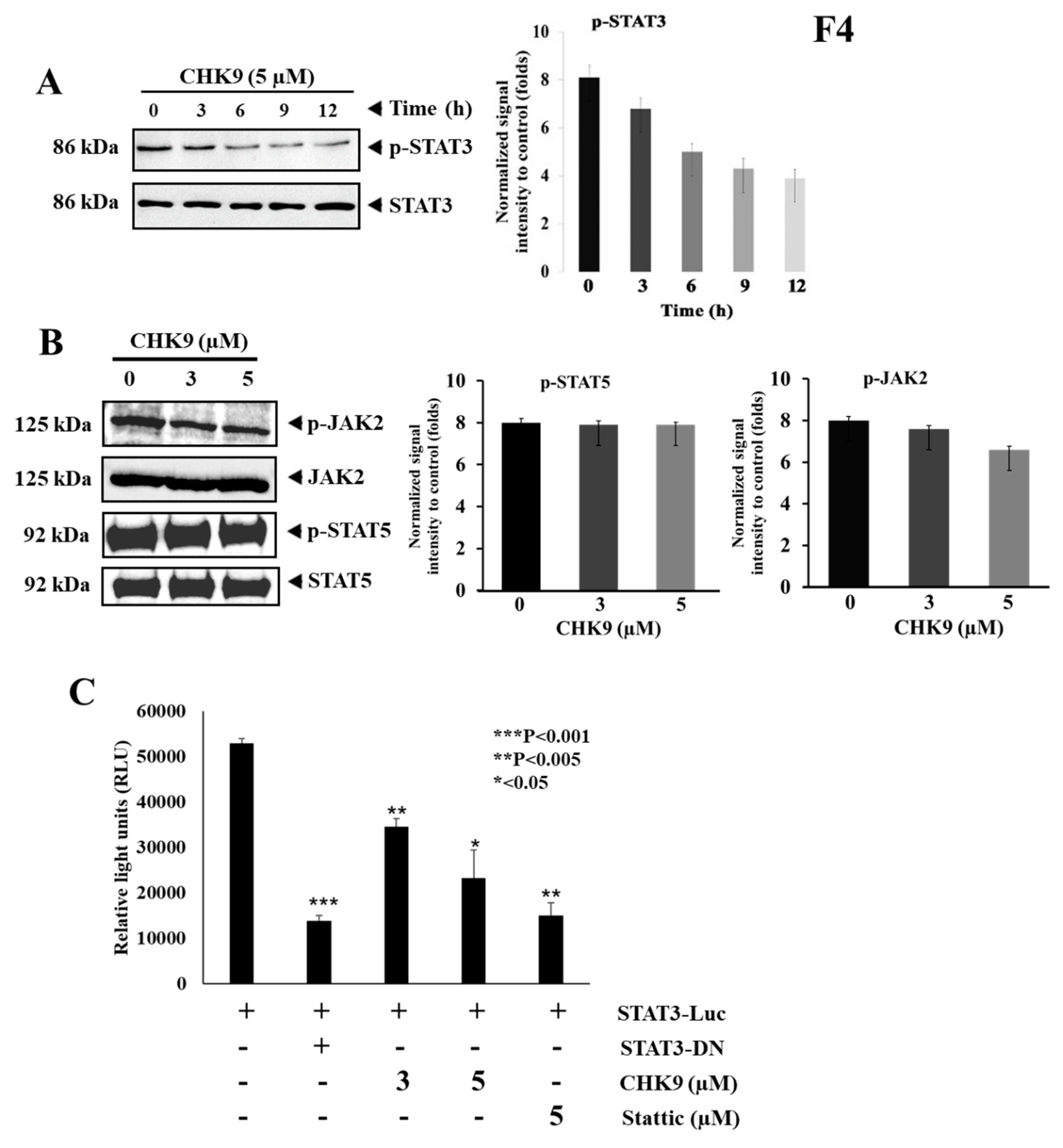
2.2.3. CHK9 Specifically Inhibits STAT3 Activation without Modulating the Activity of JAK2 in Lung Cancer Cells
2.2.4. CHK9 Mitigates STAT3-Dependent Gene Expression in Lung Cancer Cells
2.2.5. CHK9 Decreases the Expression of STAT3-Targeted Genes and p53 in Lung Cancer Cells
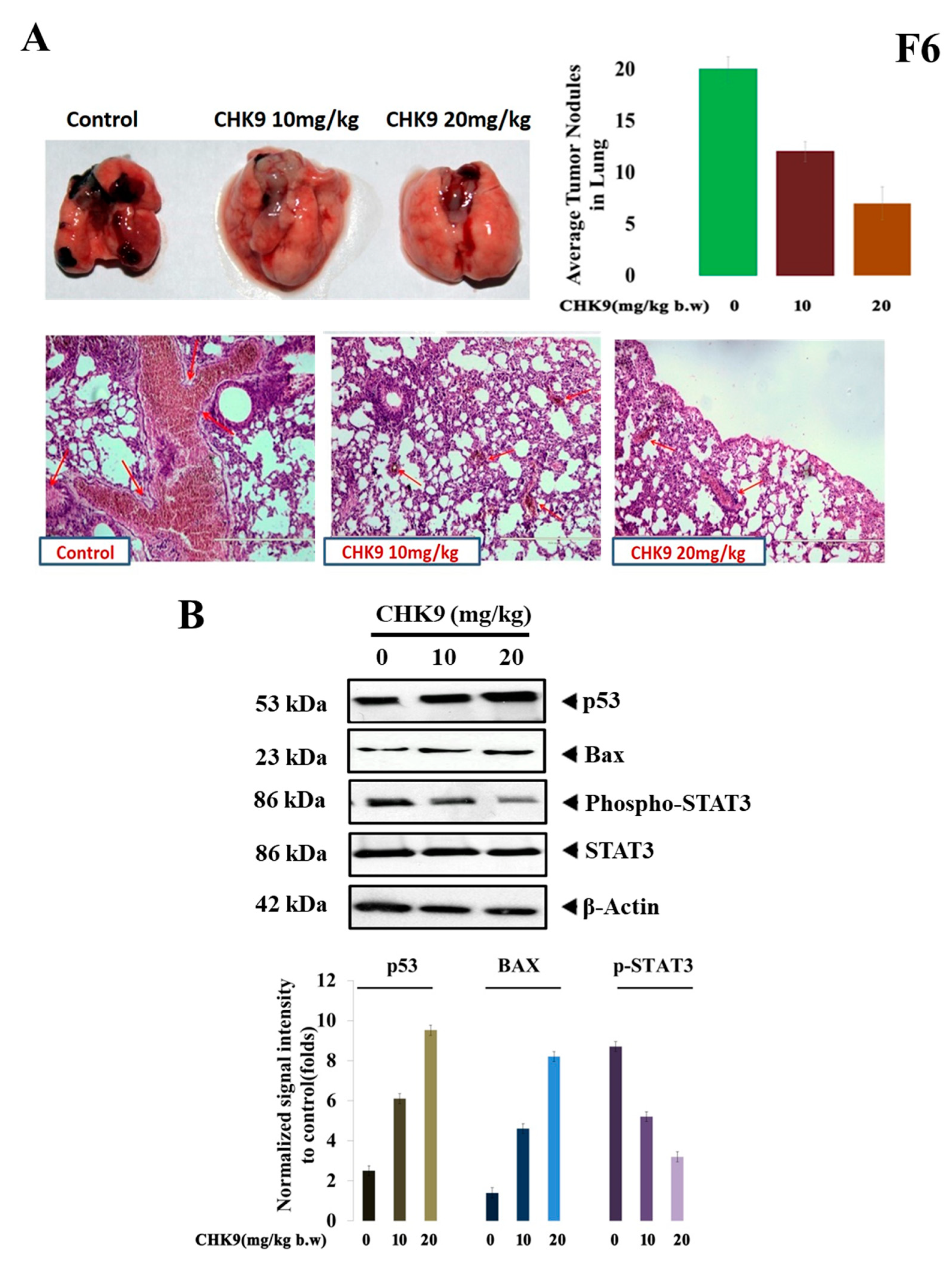
2.2.6. CHK9 Regress In Vivo Tumor Growth by Inhibiting STAT3 Signaling Cascade
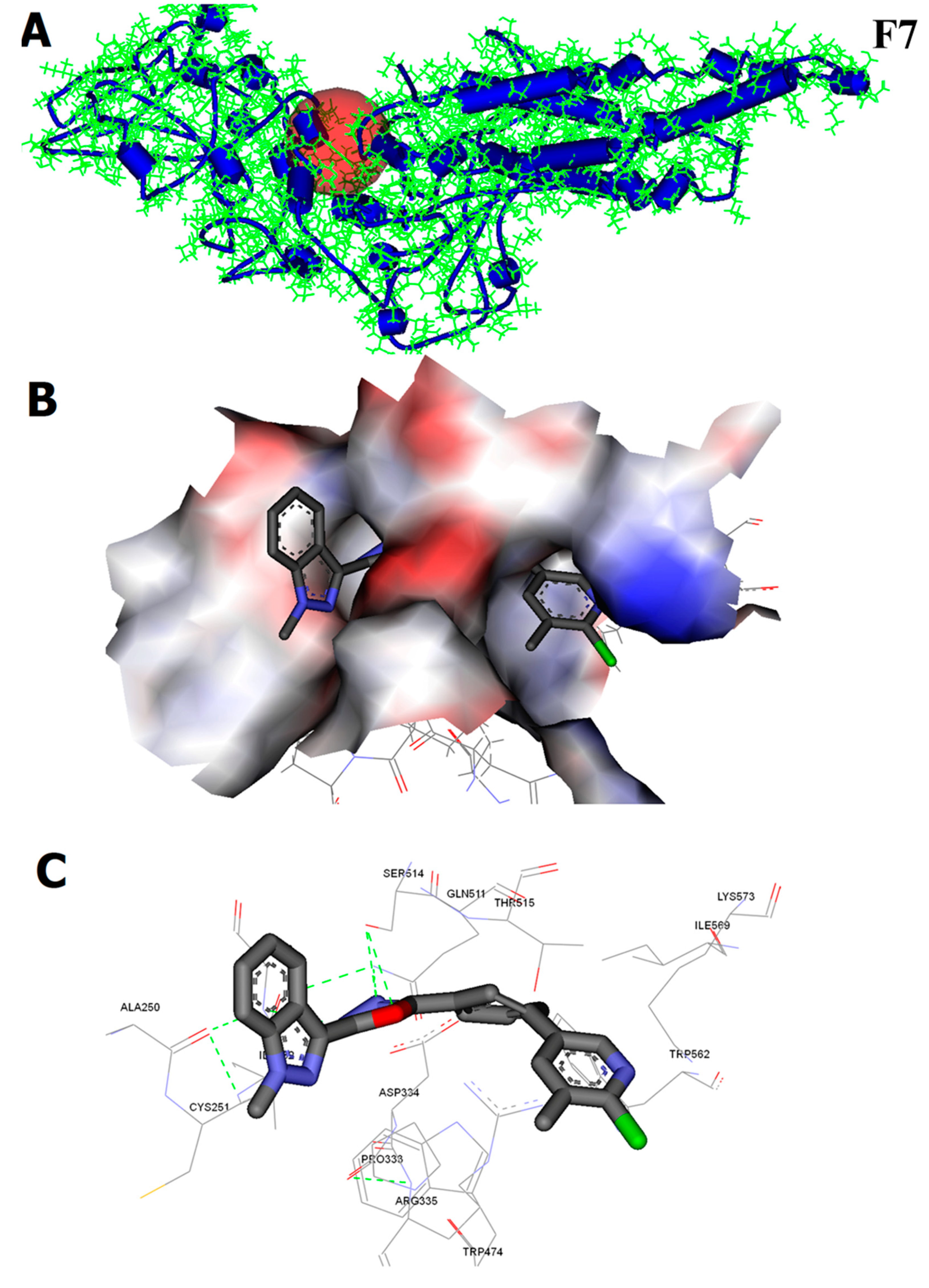
2.2.7. CHK9 Interacts with the SH2 Domain of STAT3
3. Discussion
4. Materials and Methods
4.1. Chemistry
4.1.1. Chemicals, Reagents, and Instruments
4.1.2. Synthesis of ethyl 1-methyl-1H-indazole-3-carboxylate (1a)
4.1.3. Synthesis of 1-methyl-1H-indazole-3-carbohydrazide (2a)
4.1.4. Synthesis 2-(3-bromophenyl)-5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazole (3a)
4.1.5. General Procedure for the Synthesis of Compounds (5a–m)
4.1.6. 2-(1-methyl-1H-indazol-3-yl)-5-(3-(pyridin-3-yl)phenyl)-1,3,4-oxadiazoles (5a)
4.1.7. 2-(3’-methoxy-[1,1’-biphenyl]-3-yl)-5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazole (5b)
4.1.8. 2-(4’-chloro-[1,1’-biphenyl]-3-yl)-5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazole (5c)
4.1.9. 2-(1-methyl-1H-indazol-3-yl)-5-(3-(naphthalen-1-yl)phenyl)-1,3,4-oxadiazole (5d)
4.1.10. 2-(1-methyl-1H-indazol-3-yl)-5-(2’-methyl-[1,1’-biphenyl]-3-yl)-1,3,4-oxadiazole (5e)
4.1.11. 2-(1-methyl-1H-indazol-3-yl)-5-(3-(6-methylpyridin-3-yl)phenyl)-1,3,4-oxadiazole (5f)
4.1.12. 2-(1-methyl-1H-indazol-3-yl)-5-(3-(pyrimidin-5-yl)phenyl)-1,3,4-oxadiazole(5g)
4.1.13. 2-(3-(5,6-dimethylpyridin-3-yl)phenyl)-5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazole (5h)
4.1.14. 2-(3-(6-chloro-5-methylpyridin-3-yl)phenyl)-5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazole (5i, CHK9)
4.1.15. 2-(3-(6-fluoro-5-methylpyridin-3-yl)phenyl)-5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazole (5j)
4.1.16. 3’-(5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazol-2-yl)-[1,1’-biphenyl]-3-carbaldehyde (5k)
4.1.17. N-cyclopentyl-3’-(5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazol-2-yl)-[1,1’-biphenyl]-3-carboxamide (5l)
4.1.18. 2-(4’-fluoro-[1,1’-biphenyl]-3-yl)-5-(1-methyl-1H-indazol-3-yl)-1,3,4-oxadiazole (5m)
4.2. Biology
4.2.1. Animal Cell Culture and Growth Inhibition Studies
4.2.2. Flow Cytometric Analysis
4.2.3. Annexin V Staining
4.2.4. TUNEL Assay
4.2.5. DAPI Staining
4.2.6. Preparation of Cell Lysate
4.2.7. STAT3 Luciferase Reporter Assay
4.2.8. Immunoblot Analysis
4.2.9. In silico Studies
4.2.10. In Vivo Experiments
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Statistical Analysis
Ethics Statement
References
- Esposito, L.; Conti, D.; Ailavajhala, R.; Khalil, N.; Giordano, A. Lung Cancer: Are we up to the Challenge? Curr. Genom. 2010, 11, 513–518. [Google Scholar] [CrossRef]
- Yang, M.H.; Lee, J.H.; Ko, J.-H.; Jung, S.H.; Sethi, G.; Ahn, K.S. Brassinin Represses Invasive Potential of Lung Carcinoma Cells through Deactivation of PI3K/Akt/mTOR Signaling Cascade. Molecules 2019, 24, 1584. [Google Scholar] [CrossRef]
- Lee, J.H.; Mohan, C.D.; Basappa, S.; Rangappa, S.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Kumar, A.P.; Sethi, G.; Ahn, K.S.; et al. The IκB Kinase Inhibitor ACHP Targets the STAT3 Signaling Pathway in Human Non-Small Cell Lung Carcinoma Cells. Biomolecules 2019, 9, 875. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Lee, S.-G.; Sethi, G.; Ahn, K.S. Ophiopogonin D, a Steroidal Glycoside Abrogates STAT3 Signaling Cascade and Exhibits Anti-Cancer Activity by Causing GSH/GSSG Imbalance in Lung Carcinoma. Cancers 2018, 10, 427. [Google Scholar] [CrossRef]
- Malhotra, J.; Malvezzi, M.; Negri, E.; La Vecchia, C.; Boffetta, P. Risk factors for lung cancer worldwide. Eur. Respir. J. 2016, 48, 889–902. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Shanmugam, M.K.; Narula, A.S.; Kim, C.; Lee, J.H.; Namjoshi, O.A.; Blough, B.E.; Sethi, G.; Ahn, K.S. Oxymatrine Attenuates Tumor Growth and Deactivates STAT5 Signaling in a Lung Cancer Xenograft Model. Cancers 2019, 11, 49. [Google Scholar] [CrossRef] [PubMed]
- Gadgeel, S.M.; Ramalingam, S.S.; Kalemkerian, G.P. Treatment of lung cancer. Radiol. Clin. 2012, 50, 961–974. [Google Scholar] [CrossRef] [PubMed]
- Howington, J.A.; Blum, M.G.; Chang, A.C.; Balekian, A.A.; Murthy, S.C. Treatment of stage I and II non-small cell lung cancer: Diagnosis and management of lung cancer: American College of Chest Physicians evidence-based clinical practice guidelines. Chest 2013, 143, e278S–e313S. [Google Scholar] [CrossRef] [PubMed]
- Available online: https://seer.cancer.gov/archive/csr/1975_2015/ (accessed on 11 March 2020).
- Mohan, C.D.; Bharathkumar, H.; Bulusu, K.C.; Pandey, V.; Rangappa, S.; Fuchs, J.E.; Shanmugam, M.K.; Dai, X.; Li, F.; Deivasigamani, A.; et al. Development of a novel azaspirane that targets the Janus kinase-signal transducer and activator of transcription (STAT) pathway in hepatocellular carcinoma in vitro and in vivo. J. Biol. Chem. 2014, 289, 34296–34307. [Google Scholar] [CrossRef]
- Baek, S.H.; Lee, J.H.; Kim, C.; Ko, J.H.; Ryu, S.H.; Lee, S.G.; Yang, W.M.; Um, J.Y.; Chinnathambi, A.; Alharbi, S.A.; et al. Ginkgolic Acid C 17:1, Derived from Ginkgo biloba Leaves, Suppresses Constitutive and Inducible STAT3 Activation through Induction of PTEN and SHP-1 Tyrosine Phosphatase. Molecules 2017, 22, 276. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Baek, S.H.; Ko, J.H.; Lee, S.G.; Yang, W.M.; Um, J.Y.; Sethi, G.; Ahn, K.S. Capsazepine inhibits JAK/STAT3 signaling, tumor growth, and cell survival in prostate cancer. Oncotarget 2017, 8, 17700–17711. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Rangappa, S.; Preetham, H.D.; Chandra Nayak, S.; Gupta, V.K.; Basappa, S.; Sethi, G.; Rangappa, K.S. Targeting STAT3 signaling pathway in cancer by agents derived from Mother Nature. Semin. Cancer Biol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Baburajeev, C.; Mohan, C.D.; Patil, G.S.; Rangappa, S.; Pandey, V.; Sebastian, A.; Fuchs, J.E.; Bender, A.; Lobie, P.E.; Rangappa, K.S. Nano-cuprous oxide catalyzed one-pot synthesis of a carbazole-based STAT3 inhibitor: A facile approach via intramolecular C–N bond formation reactions. RSC Adv. 2016, 6, 36775–36785. [Google Scholar] [CrossRef]
- Baek, S.H.; Ko, J.H.; Lee, H.; Jung, J.; Kong, M.; Lee, J.W.; Lee, J.; Chinnathambi, A.; Zayed, M.E.; Alharbi, S.A.; et al. Resveratrol inhibits STAT3 signaling pathway through the induction of SOCS-1: Role in apoptosis induction and radiosensitization in head and neck tumor cells. Phytomedicine Int. J. Phytother. Phytopharm. 2016, 23, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Loh, C.Y.; Arya, A.; Naema, A.F.; Wong, W.F.; Sethi, G.; Looi, C.Y. Signal Transducer and Activator of Transcription (STATs) Proteins in Cancer and Inflammation: Functions and Therapeutic Implication. Front. Oncol. 2019, 9, 48. [Google Scholar] [CrossRef]
- Sulaiman, N.B.S.; Mohan, C.D.; Basappa, S.; Pandey, V.; Rangappa, S.; Bharathkumar, H.; Kumar, A.P.; Lobie, P.E.; Rangappa, K.S. An azaspirane derivative suppresses growth and induces apoptosis of ER-positive and ER-negative breast cancer cells through the modulation of JAK2/STAT3 signaling pathway. Int. J. Oncol. 2016, 49, 1221–1229. [Google Scholar] [CrossRef]
- Arora, L.; Kumar, A.P.; Arfuso, F.; Chng, W.J.; Sethi, G. The Role of Signal Transducer and Activator of Transcription 3 (STAT3) and Its Targeted Inhibition in Hematological Malignancies. Cancers 2018, 10, 327. [Google Scholar] [CrossRef]
- Jung, Y.Y.; Lee, J.H.; Nam, D.; Narula, A.S.; Namjoshi, O.A.; Blough, B.E.; Um, J.Y.; Sethi, G.; Ahn, K.S. Anti-myeloma Effects of Icariin Are Mediated Through the Attenuation of JAK/STAT3-Dependent Signaling Cascade. Front. Pharmacol. 2018, 9, 531. [Google Scholar] [CrossRef]
- Tan, S.M.; Li, F.; Rajendran, P.; Kumar, A.P.; Hui, K.M.; Sethi, G. Identification of beta-escin as a novel inhibitor of signal transducer and activator of transcription 3/Janus-activated kinase 2 signaling pathway that suppresses proliferation and induces apoptosis in human hepatocellular carcinoma cells. J. Pharmacol. Exp. Ther. 2010, 334, 285–293. [Google Scholar] [CrossRef]
- Lee, J.H.; Rangappa, S.; Mohan, C.D.; Sethi, G.; Lin, Z.-X.; Rangappa, K.S.; Ahn, K.S. Brusatol, a Nrf2 Inhibitor Targets STAT3 Signaling Cascade in Head and Neck Squamous Cell Carcinoma. Biomolecules 2019, 9, 550. [Google Scholar] [CrossRef]
- Kim, C.; Lee, S.G.; Yang, W.M.; Arfuso, F.; Um, J.Y.; Kumar, A.P.; Bian, J.; Sethi, G.; Ahn, K.S. Formononetin-induced oxidative stress abrogates the activation of STAT3/5 signaling axis and suppresses the tumor growth in multiple myeloma preclinical model. Cancer Lett. 2018, 431, 123–141. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sikka, S.; Siveen, K.S.; Lee, J.H.; Um, J.Y.; Kumar, A.P.; Chinnathambi, A.; Alharbi, S.A.; Basappa; Rangappa, K.S.; et al. Cardamonin represses proliferation, invasion, and causes apoptosis through the modulation of signal transducer and activator of transcription 3 pathway in prostate cancer. Apoptosis Int. J. Program. Cell Death 2017, 22, 158–168. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.L.A.; Hirpara, J.L.; Pervaiz, S.; Eu, J.Q.; Sethi, G.; Goh, B.C. Do STAT3 inhibitors have potential in the future for cancer therapy? Expert Opin. Investig. Drugs 2017, 26, 883–887. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ahn, K.S.; Kim, C.; Shanmugam, M.K.; Siveen, K.S.; Arfuso, F.; Samym, R.P.; Deivasigamanim, A.; Lim, L.H.; Wang, L.; et al. Nimbolide-Induced Oxidative Stress Abrogates STAT3 Signaling Cascade and Inhibits Tumor Growth in Transgenic Adenocarcinoma of Mouse Prostate Model. Antioxid. Redox Signal. 2016, 24, 575–589. [Google Scholar] [CrossRef]
- Johnston, P.A.; Grandis, J.R. STAT3 signaling: Anticancer strategies and challenges. Mol. Interv. 2011, 11, 18. [Google Scholar] [CrossRef]
- Dai, X.; Ahn, K.S.; Kim, C.; Siveen, K.S.; Ong, T.H.; Shanmugam, M.K.; Li, F.; Shi, J.; Kumar, A.P.; Wang, L.Z.; et al. Ascochlorin, an isoprenoid antibiotic inhibits growth and invasion of hepatocellular carcinoma by targeting STAT3 signaling cascade through the induction of PIAS3. Mol. Oncol. 2015, 9, 818–833. [Google Scholar] [CrossRef]
- Zhang, H.-F.; Lai, R. STAT3 in cancer—Friend or foe? Cancers 2014, 6, 1408–1440. [Google Scholar] [CrossRef]
- Shanmugam, M.K.; Rajendran, P.; Li, F.; Kim, C.; Sikka, S.; Siveen, K.S.; Kumar, A.P.; Ahn, K.S.; Sethi, G. Abrogation of STAT3 signaling cascade by zerumbone inhibits proliferation and induces apoptosis in renal cell carcinoma xenograft mouse model. Mol. Carcinog. 2015, 54, 971–985. [Google Scholar] [CrossRef]
- Sebastian, A.; Pandey, V.; Mohan, C.D.; Chia, Y.T.; Rangappa, S.; Mathai, J.; Baburajeev, C.; Paricharak, S.; Mervin, L.H.; Bulusu, K.C. Novel adamantanyl-based thiadiazolyl pyrazoles targeting EGFR in triple-negative breast cancer. ACS Omega 2016, 1, 1412–1424. [Google Scholar] [CrossRef]
- Siveen, K.S.; Sikka, S.; Surana, R.; Dai, X.; Zhang, J.; Kumar, A.P.; Tan, B.K.; Sethi, G.; Bishayee, A. Targeting the STAT3 signaling pathway in cancer: Role of synthetic and natural inhibitors. Biochim. Biophys. Acta 2014, 1845, 136–154. [Google Scholar] [CrossRef]
- Kotian, S.Y.; Mohan, C.D.; Merlo, A.A.; Rangappa, S.; Nayak, S.C.; Rai, K.M.L.; Rangappa, K.S. Small molecule based five-membered heterocycles: A view of liquid crystalline properties beyond the biological applications. J. Mol. Liq. 2020, 297, 111686. [Google Scholar] [CrossRef]
- Mohan, C.D.; Anilkumar, N.C.; Rangappa, S.; Shanmugam, M.K.; Mishra, S.; Chinnathambi, A.; Alharbi, S.A.; Bhattacharjee, A.; Sethi, G.; Kumar, A.P.; et al. Novel 1,3,4-Oxadiazole Induces Anticancer Activity by Targeting NF-κB in Hepatocellular Carcinoma Cells. Front. Oncol. 2018, 8, 42. [Google Scholar] [CrossRef]
- Nandeesh, K.N.; Swarup, H.A.; Sandhya, N.C.; Mohan, C.D.; Pavan Kumar, C.S.; Kumara, M.N.; Mantelingu, K.; Ananda, S.; Rangappa, K.S. Synthesis and antiproliferative efficiency of novel bis(imidazol-1-yl)vinyl-1,2,4-oxadiazoles. New J. Chem. 2016, 40, 2823–2828. [Google Scholar] [CrossRef]
- Kim, B.-H.; Lee, H.; Song, Y.; Park, J.-S.; Gadhe, C.G.; Choi, J.; Lee, C.-G.; Pae, A.N.; Kim, S.; Ye, S.-K. Development of Oxadiazole-Based ODZ10117 as a Small-Molecule Inhibitor of STAT3 for Targeted Cancer Therapy. J. Clin. Med. 2019, 8, 1847. [Google Scholar] [CrossRef]
- Masciocchi, D.; Villa, S.; Meneghetti, F.; Pedretti, A.; Barlocco, D.; Legnani, L.; Toma, L.; Kwon, B.-M.; Nakano, S.; Asai, A.; et al. Biological and computational evaluation of an oxadiazole derivative (MD77) as a new lead for direct STAT3 inhibitors. MedChemComm 2012, 3, 592–599. [Google Scholar] [CrossRef]
- Porta, F.; Facchetti, G.; Ferri, N.; Gelain, A.; Meneghetti, F.; Villa, S.; Barlocco, D.; Masciocchi, D.; Asai, A.; Miyoshi, N.; et al. An in vivo active 1,2,5-oxadiazole Pt(II) complex: A promising anticancer agent endowed with STAT3 inhibitory properties. Eur. J. Med. Chem. 2017, 131, 196–206. [Google Scholar] [CrossRef] [PubMed]
- Ritzén, A.; Sørensen, M.D.; Dack, K.N.; Greve, D.R.; Jerre, A.; Carnerup, M.A.; Rytved, K.A.; Bagger-Bahnsen, J. Fragment-Based Discovery of 6-Arylindazole JAK Inhibitors. ACS Med. Chem. Lett. 2016, 7, 641–646. [Google Scholar] [CrossRef] [PubMed]
- Bajusz, D.; Ferenczy, G.G.; Keserű, G.M. Discovery of Subtype Selective Janus Kinase (JAK) Inhibitors by Structure-Based Virtual Screening. J. Chem. Inf. Model. 2016, 56, 234–247. [Google Scholar] [CrossRef]
- Somu, C.; Mohan, C.D.; Ambekar, S.; Dukanya; Rangappa, S.; Baburajeev, C.P.; Sukhorukov, A.; Mishra, S.; Shanmugam, M.K.; Chinnathambi, A.; et al. Identification of a novel 1,2 oxazine that can induce apoptosis by targeting NF-κB in hepatocellular carcinoma cells. Biotechnol. Rep. 2020, 25, e00438. [Google Scholar] [CrossRef]
- Lee, J.H.; Mohan, C.D.; Shanmugam, M.K.; Rangappa, S.; Sethi, G.; Siveen, K.S.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Basappa, S.; et al. Vitexin abrogates invasion and survival of hepatocellular carcinoma cells through targeting STAT3 signaling pathway. Biochimie 2020, 175, 58–68. [Google Scholar] [CrossRef]
- Yang, M.H.; Jung, S.H.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Sethi, G.; Ahn, K.S. Attenuation of STAT3 Signaling Cascade by Daidzin Can Enhance the Apoptotic Potential of Bortezomib against Multiple Myeloma. Biomolecules 2020, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Hirpara, J.L.; Eu, J.-Q.; Sethi, G.; Wang, L.; Goh, B.-C.; Wong, A.L. Targeting STAT3 and oxidative phosphorylation in oncogene-addicted tumors. Redox Biol. 2019, 25, 101073. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, C.; Ko, J.-H.; Jung, Y.Y.; Jung, S.H.; Kim, E.; Kong, M.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; et al. Casticin inhibits growth and enhances ionizing radiation–induced apoptosis through the suppression of STAT3 signaling cascade. J. Cell. Biochem. 2019, 120, 9787–9798. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, C.; Lee, J.; Um, J.-Y.; Sethi, G.; Ahn, K.S. Arctiin is a pharmacological inhibitor of STAT3 phosphorylation at tyrosine 705 residue and potentiates bortezomib-induced apoptotic and anti-angiogenic effects in human multiple myeloma cells. Phytomedicine Int. J. Phytother. Phytopharm. 2019, 55, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Chai, E.Z.P.; Shanmugam, M.K.; Arfuso, F.; Dharmarajan, A.; Wang, C.; Kumar, A.P.; Samy, R.P.; Lim, L.H.K.; Wang, L.; Goh, B.C.; et al. Targeting transcription factor STAT3 for cancer prevention and therapy. Pharmacol. Ther. 2016, 162, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Mohan, C.D.; Deivasigamani, A.; Jung, Y.Y.; Rangappa, S.; Basappa, S.; Chinnathambi, A.; Alahmadi, T.A.; Alharbi, S.A.; Garg, M.; et al. Brusatol suppresses STAT3-driven metastasis by downregulating epithelial-mesenchymal transition in hepatocellular carcinoma. J. Adv. Res. 2020. [Google Scholar] [CrossRef]
- Fathi, M.A.A.; Abd El-Hafeez, A.A.; Abdelhamid, D.; Abbas, S.H.; Montano, M.M.; Abdel-Aziz, M. 1,3,4-oxadiazole/chalcone hybrids: Design, synthesis, and inhibition of leukemia cell growth and EGFR, Src, IL-6 and STAT3 activities. Bioorganic Chem. 2019, 84, 150–163. [Google Scholar] [CrossRef]
- Khanam, R.; Hejazi, I.I.; Shahabuddin, S.; Bhat, A.R.; Athar, F. Pharmacokinetic evaluation, molecular docking and in vitro biological evaluation of 1, 3, 4-oxadiazole derivatives as potent antioxidants and STAT3 inhibitors. J. Pharm. Anal. 2019, 9, 133–141. [Google Scholar] [CrossRef]
- Nirvanappa, A.C.; Mohan, C.D.; Rangappa, S.; Ananda, H.; Sukhorukov, A.Y.; Shanmugam, M.K.; Sundaram, M.S.; Nayaka, S.C.; Girish, K.S.; Chinnathambi, A.; et al. Novel Synthetic Oxazines Target NF-κB in Colon Cancer In Vitro and Inflammatory Bowel Disease In Vivo. PLoS ONE 2016, 11, e0163209. [Google Scholar] [CrossRef]
- Ashwini, N.; Garg, M.; Mohan, C.D.; Fuchs, J.E.; Rangappa, S.; Anusha, S.; Swaroop, T.R.; Rakesh, K.S.; Kanojia, D.; Madan, V.; et al. Synthesis of 1,2-benzisoxazole tethered 1,2,3-triazoles that exhibit anticancer activity in acute myeloid leukemia cell lines by inhibiting histone deacetylases, and inducing p21 and tubulin acetylation. Bioorganic Med. Chem. 2015, 23, 6157–6165. [Google Scholar] [CrossRef]
- Lee, J.H.; Kim, C.; Kim, S.-H.; Sethi, G.; Ahn, K.S. Farnesol inhibits tumor growth and enhances the anticancer effects of bortezomib in multiple myeloma xenograft mouse model through the modulation of STAT3 signaling pathway. Cancer Lett. 2015, 360, 280–293. [Google Scholar] [CrossRef] [PubMed]
- Sethi, G.; Chatterjee, S.; Rajendran, P.; Li, F.; Shanmugam, M.K.; Wong, K.F.; Kumar, A.P.; Senapati, P.; Behera, A.K.; Hui, K.M.; et al. Inhibition of STAT3 dimerization and acetylation by garcinol suppresses the growth of human hepatocellular carcinoma in vitro and in vivo. Mol. Cancer 2014, 13, 66. [Google Scholar] [CrossRef] [PubMed]
- Niu, G.; Wright, K.L.; Ma, Y.; Wright, G.M.; Huang, M.; Irby, R.; Briggs, J.; Karras, J.; Cress, W.D.; Pardoll, D.; et al. Role of Stat3 in Regulating p53 Expression and Function. Mol. Cell. Biol. 2005, 25, 7432. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.P.; Bajaj, S.; Maity, T.K.; Singh, J. Synthesis and Evaluation of Anticancer Activity of 1, 3, 4-Oxadiazole Derivatives against Ehrlich Ascites Carcinoma Bearing Mice and Their Correlation with Histopathology of Liver. Receptor 2017, 15, 16. [Google Scholar] [CrossRef]
- Omar, F.A.; Mahfouz, N.M.; Rahman, M.A. Design, synthesis and antiinflammatory activity of some 1,3,4-oxadiazole derivatives. Eur. J. Med. Chem. 1996, 31, 819–825. [Google Scholar] [CrossRef]
- Tiwari, A.; Gopalan Kutty, N.; Kumar, N.; Chaudhary, A.; Vasanth Raj, P.; Shenoy, R.; Mallikarjuna Rao, C. Synthesis and evaluation of selected 1,3,4-oxadiazole derivatives for in vitro cytotoxicity and in vivo anti-tumor activity. Cytotechnology 2016, 68, 2553–2565. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Malojirao, V.H.; Vigneshwaran, V.; Thirusangu, P.; Mahmood, R.; Prabhakar, B.T. The tumor antagonistic steroidal alkaloid Solanidine prompts the intrinsic suicidal signal mediated DFF-40 nuclear import and nucleosomal disruption. Life Sci. 2018, 199, 139–150. [Google Scholar] [CrossRef] [PubMed]
- Baburajeev, C.P.; Dhananjaya Mohan, C.; Ananda, H.; Rangappa, S.; Fuchs, J.E.; Jagadish, S.; Sivaraman Siveen, K.; Chinnathambi, A.; Ali Alharbi, S.; Zayed, M.E.; et al. Development of Novel Triazolo-Thiadiazoles from Heterogeneous “Green” Catalysis as Protein Tyrosine Phosphatase 1B Inhibitors. Sci. Rep. 2015, 5, 14195. [Google Scholar] [CrossRef] [PubMed]
- Rajendran, P.; Li, F.; Shanmugam, M.K.; Vali, S.; Abbasi, T.; Kapoor, S.; Ahn, K.S.; Kumar, A.P.; Sethi, G. Honokiol inhibits signal transducer and activator of transcription-3 signaling, proliferation, and survival of hepatocellular carcinoma cells via the protein tyrosine phosphatase SHP-1. J. Cell. Physiol. 2012, 227, 2184–2195. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, G.; Yao, F.; He, Y.; Liang, G.; Zhang, Y.; Hu, B.; Wu, Y.; Li, Y.; Liu, H. Growth inhibition and apoptosis induced by osthole, a natural coumarin, in hepatocellular carcinoma. PLoS ONE 2012, 7, e37865. [Google Scholar] [CrossRef]
- Yue, Q.; Zhou, X.; Leng, Q.; Zhang, L.; Cheng, B.; Zhang, X. 7-ketocholesterol-induced caspase-mediated apoptosis in Saccharomyces cerevisiae. FEMS Yeast Res. 2013, 13, 796–803. [Google Scholar] [CrossRef]
- Gurupadaswamy, H.D.; Thirusangu, P.; Vijay Avin, B.R.; Vigneshwaran, V.; Prashanth Kumar, M.V.; Abhishek, T.S.; Lakshmi Ranganatha, V.; Khanum, S.A.; Prabhakar, B.T. DAO-9 (2,5-di(4-aryloylaryloxymethyl)-1,3,4-oxadiazole) exhibits p53 induced apoptogenesis through caspase-3 mediated endonuclease activity in murine carcinoma. Biomed. Pharmacother. 2014, 68, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Ranganatha, V.L.; Vijay Avin, B.R.; Thirusangu, P.; Prashanth, T.; Prabhakar, B.T.; Khanum, S.A. Synthesis, angiopreventive activity, and in vivo tumor inhibition of novel benzophenone-benzimidazole analogs. Life Sci. 2013, 93, 904–911. [Google Scholar] [CrossRef] [PubMed]
- Shanmugam, M.K.; Manu, K.A.; Ong, T.H.; Ramachandran, L.; Surana, R.; Bist, P.; Lim, L.H.K.; Prem Kumar, A.; Hui, K.M.; Sethi, G. Inhibition of CXCR4/CXCL12 signaling axis by ursolic acid leads to suppression of metastasis in transgenic adenocarcinoma of mouse prostate model. Int. J. Cancer 2011, 129, 1552–1563. [Google Scholar] [CrossRef] [PubMed]
- Mohan, C.D.; Bharathkumar, H.; Dukanya; Rangappa, S.; Shanmugam, M.K.; Chinnathambi, A.; Alharbi, S.A.; Alahmadi, T.A.; Bhattacharjee, A.; Lobie, P.E.; et al. N-Substituted Pyrido-1,4-Oxazin-3-Ones Induce Apoptosis of Hepatocellular Carcinoma Cells by Targeting NF-κB Signaling Pathway. Front. Pharmacol. 2018, 9, 1125. [Google Scholar] [CrossRef]
- Sawhney, M.; Rohatgi, N.; Kaur, J.; Shishodia, S.; Sethi, G.; Gupta, S.D.; Deo, S.V.S.; Shukla, N.K.; Aggarwal, B.B.; Ralhan, R. Expression of NF-κB parallels COX-2 expression in oral precancer and cancer: Association with smokeless tobacco. Int. J. Cancer 2007, 120, 2545–2556. [Google Scholar] [CrossRef]
- Siveen, K.S.; Mustafa, N.; Li, F.; Kannaiyan, R.; Ahn, K.S.; Kumar, A.P.; Chng, W.J.; Sethi, G. Thymoquinone overcomes chemoresistance and enhances the anticancer effects of bortezomib through abrogation of NF-κB regulated gene products in multiple myeloma xenograft mouse model. Oncotarget 2014, 5, 634–648. [Google Scholar] [CrossRef]
- Keerthy, H.K.; Mohan, S.; Basappa; Bharathkumar, H.; Rangappa, S.; Svensson, F.; Bender, A.; Mohan, C.D.; Rangappa, K.S.; Bhatnagar, R. Triazole-Pyridine Dicarbonitrile Targets Phosphodiesterase 4 to Induce Cytotoxicity in Lung Carcinoma Cells. Chem. Biodivers. 2019, 16, e1900234. [Google Scholar] [CrossRef]
- Becker, S.; Groner, B.; Müller, C.W. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature 1998, 394, 145–151. [Google Scholar] [CrossRef]
- Huang, T.-T.; Lan, Y.-W.; Chen, C.-M.; Ko, Y.-F.; Ojcius, D.M.; Martel, J.; Young, J.D.; Chong, K.-Y. Antrodia cinnamomea induces anti-tumor activity by inhibiting the STAT3 signaling pathway in lung cancer cells. Sci. Rep. 2019, 9, 5145. [Google Scholar] [CrossRef]




| Entry | Oxadiazole | Boronic acid | Biphenyl-1,3,4-oxadiazole |
|---|---|---|---|
| 1 | 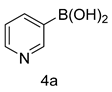 | 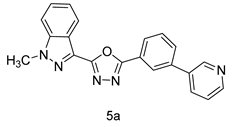 | |
| 2 | 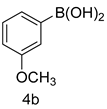 | 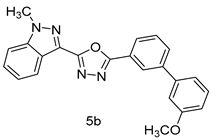 | |
| 3 |  | 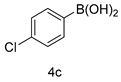 | 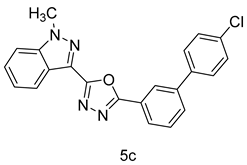 |
| 4 | 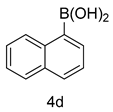 |  | |
| 5 | 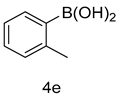 | 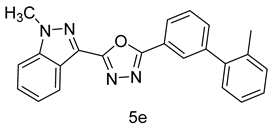 | |
| 6 |  | 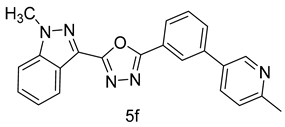 | |
| 7 |  |  | |
| 8 | 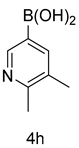 | 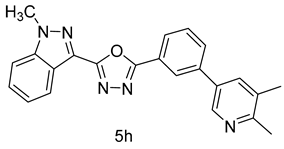 | |
| 9 |  |  | |
| 10 |  |  | |
| 11 |  | 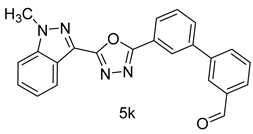 | |
| 12 |  | 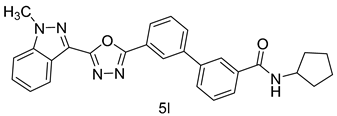 | |
| 13 |  |  |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malojirao, V.H.; Girimanchanaika, S.S.; Shanmugam, M.K.; Sherapura, A.; Dukanya; Metri, P.K.; Vigneshwaran, V.; Chinnathambi, A.; Alharbi, S.A.; Rangappa, S.; et al. Novel 1,3,4-oxadiazole Targets STAT3 Signaling to Induce Antitumor Effect in Lung Cancer. Biomedicines 2020, 8, 368. https://doi.org/10.3390/biomedicines8090368
Malojirao VH, Girimanchanaika SS, Shanmugam MK, Sherapura A, Dukanya, Metri PK, Vigneshwaran V, Chinnathambi A, Alharbi SA, Rangappa S, et al. Novel 1,3,4-oxadiazole Targets STAT3 Signaling to Induce Antitumor Effect in Lung Cancer. Biomedicines. 2020; 8(9):368. https://doi.org/10.3390/biomedicines8090368
Chicago/Turabian StyleMalojirao, Vikas H., Swamy S. Girimanchanaika, Muthu K. Shanmugam, Ankith Sherapura, Dukanya, Prashant K. Metri, Vellingiri Vigneshwaran, Arunachalam Chinnathambi, Sulaiman Ali Alharbi, Shobith Rangappa, and et al. 2020. "Novel 1,3,4-oxadiazole Targets STAT3 Signaling to Induce Antitumor Effect in Lung Cancer" Biomedicines 8, no. 9: 368. https://doi.org/10.3390/biomedicines8090368
APA StyleMalojirao, V. H., Girimanchanaika, S. S., Shanmugam, M. K., Sherapura, A., Dukanya, Metri, P. K., Vigneshwaran, V., Chinnathambi, A., Alharbi, S. A., Rangappa, S., Mohan, C. D., Basappa, Prabhakar, B. T., & Rangappa, K. S. (2020). Novel 1,3,4-oxadiazole Targets STAT3 Signaling to Induce Antitumor Effect in Lung Cancer. Biomedicines, 8(9), 368. https://doi.org/10.3390/biomedicines8090368







