A Novel Method to Identify Autoantibodies against Putative Target Proteins in Serum from beta-Thalassemia Major: A Pilot Study
Abstract
1. Introduction
2. Results
2.1. Patients
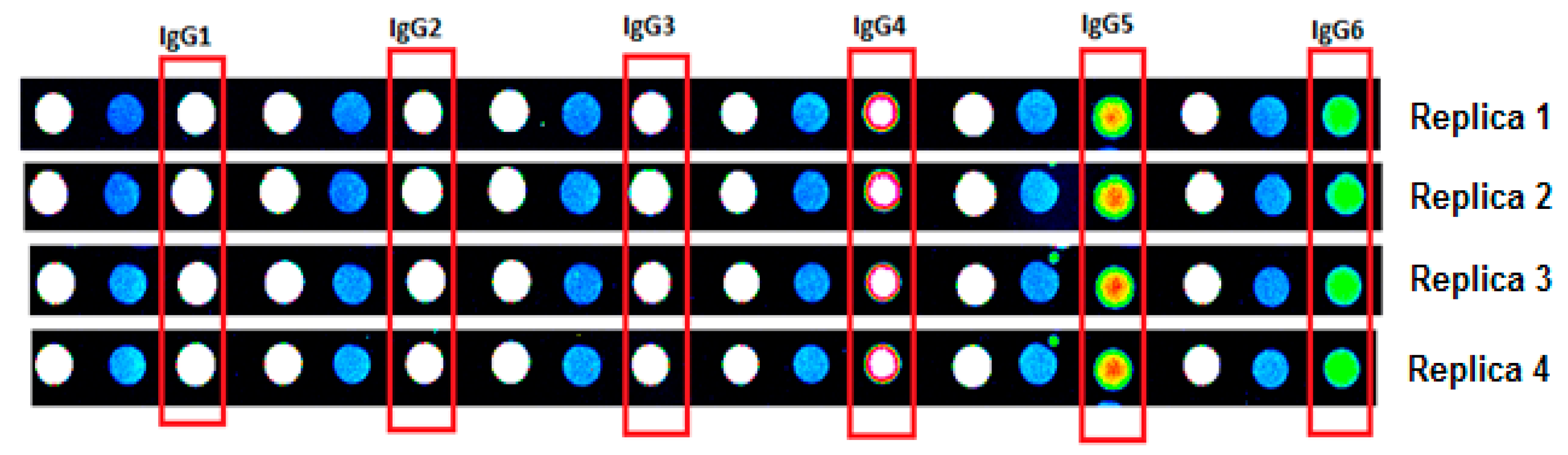
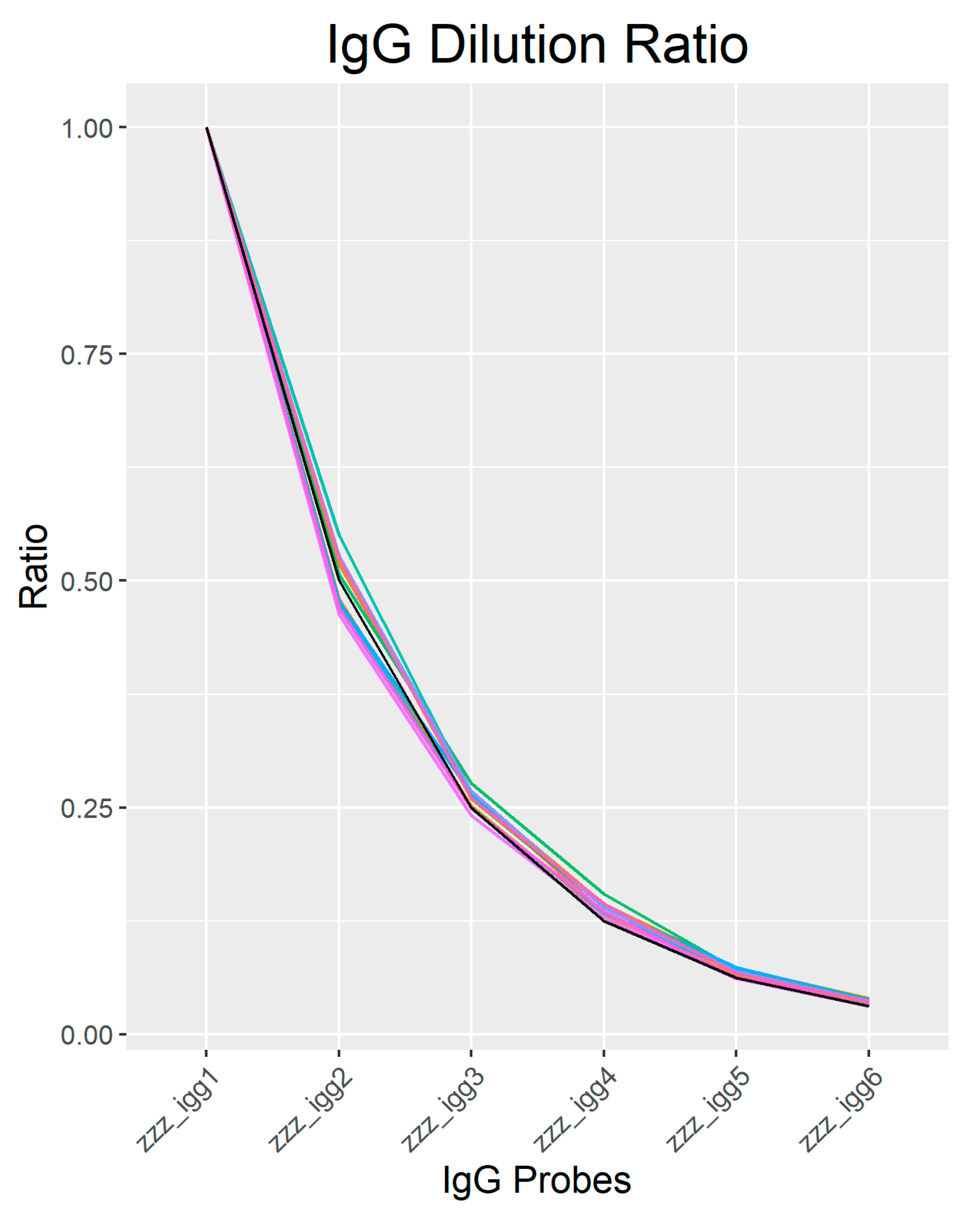
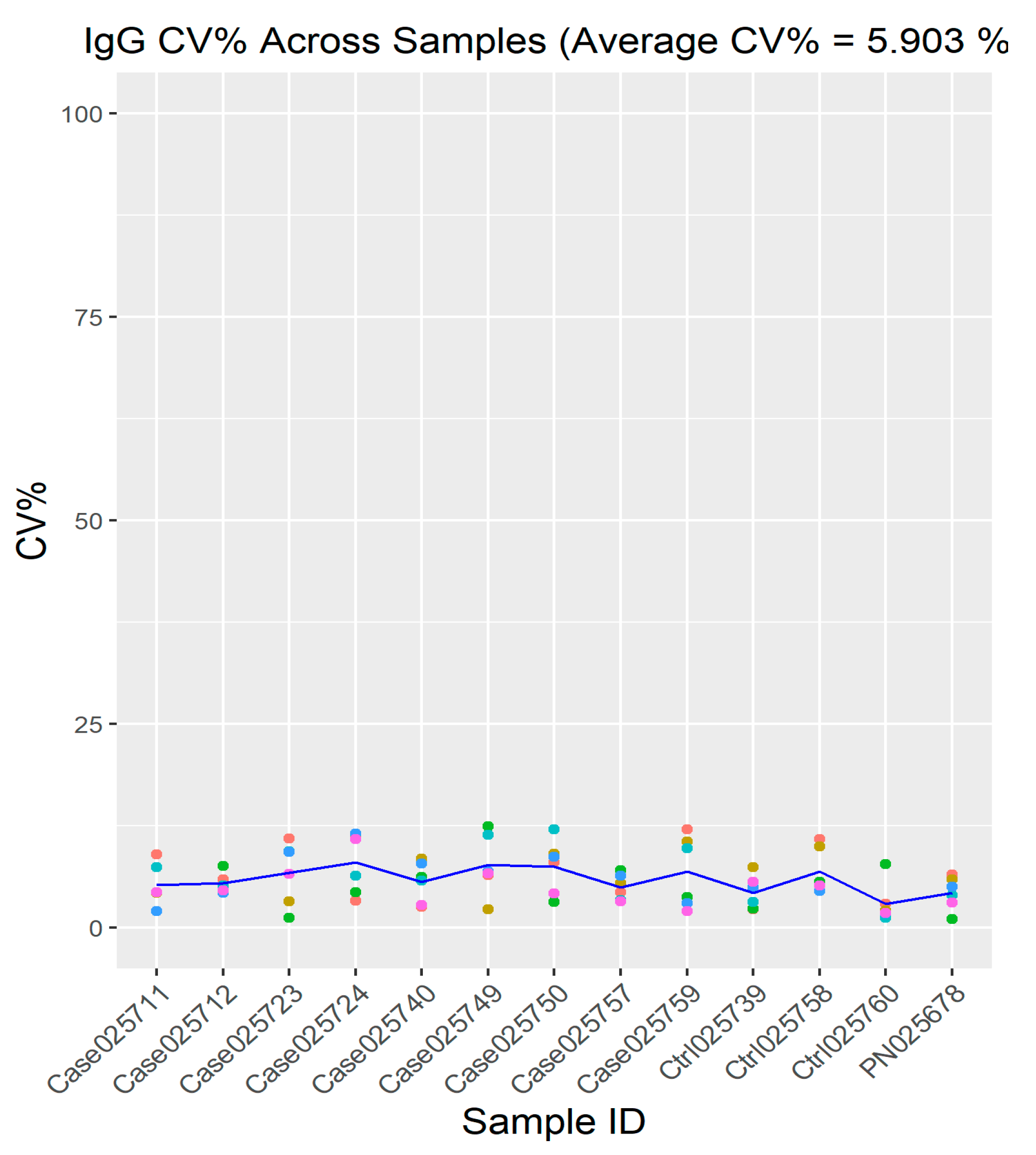
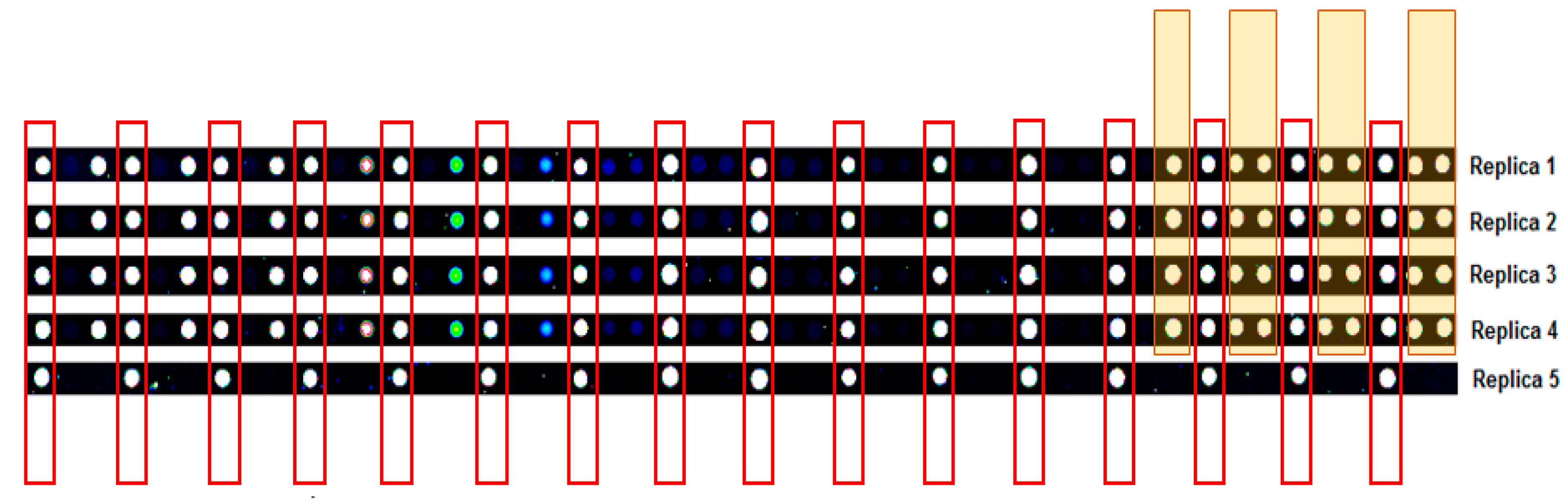
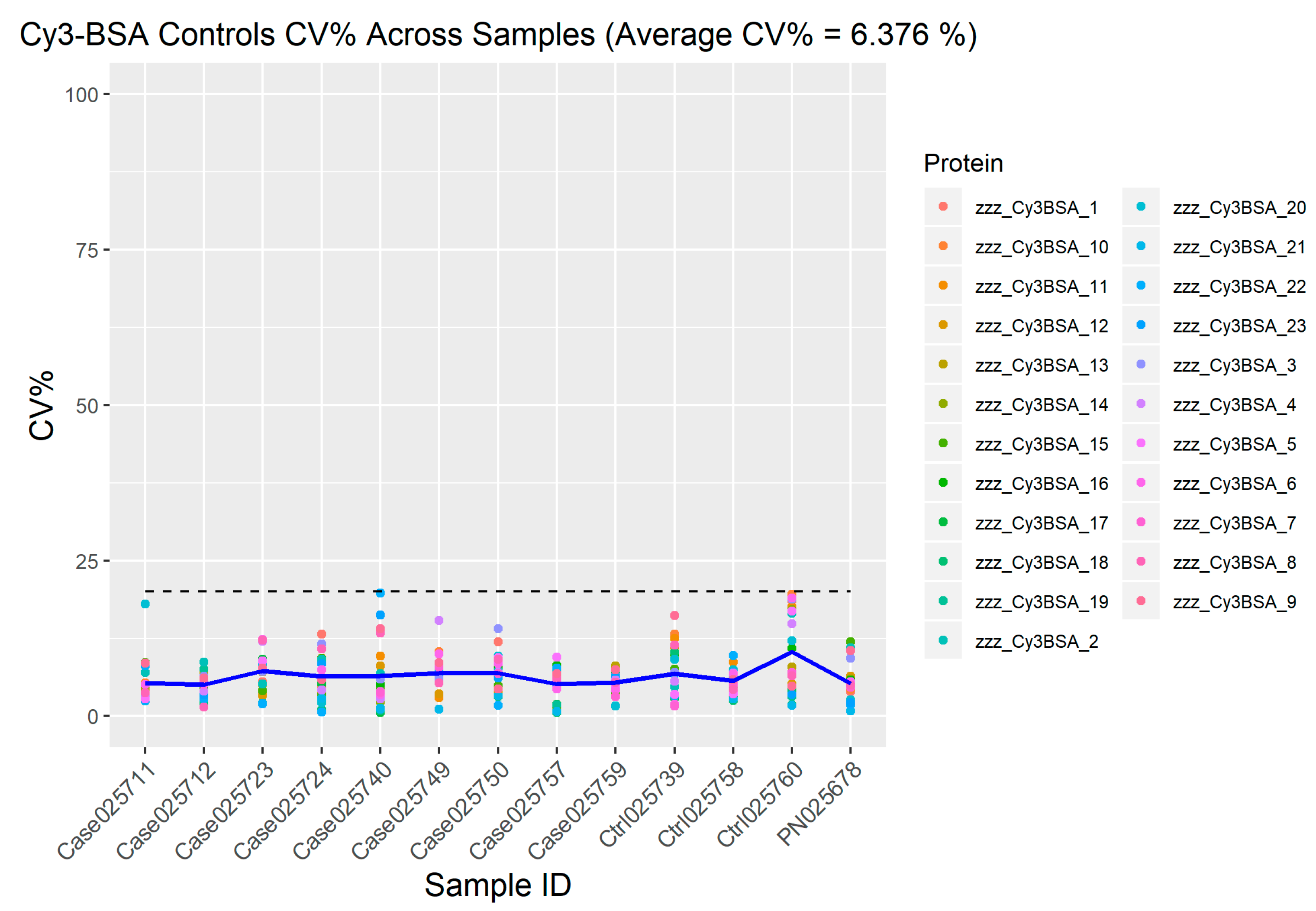
2.2. Quality Control
2.3. Cy3-BSA as Internal Control Showed Consistent Intensity across Samples
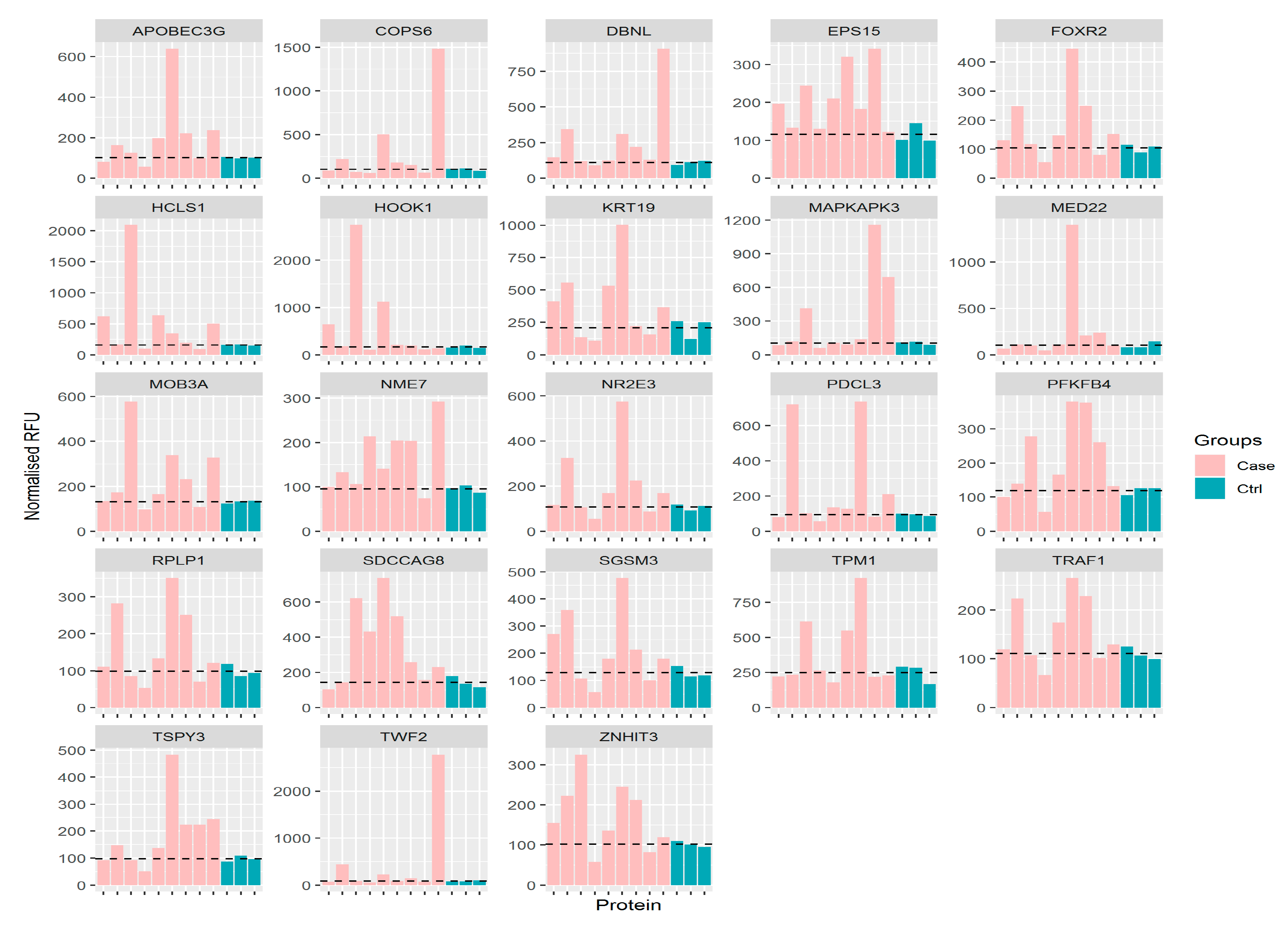
2.4. Putative Biomarkers
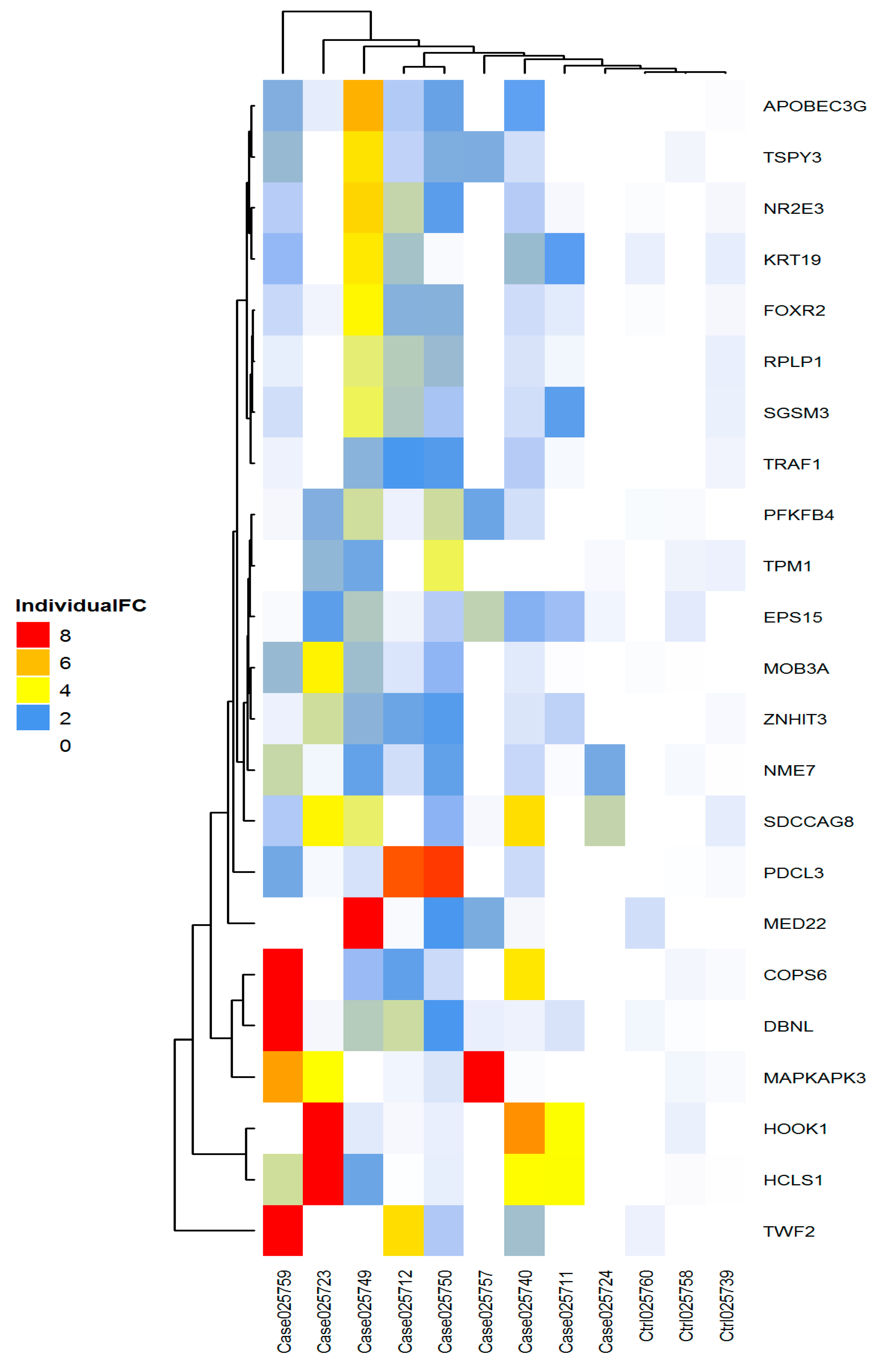
2.5. Heatmap
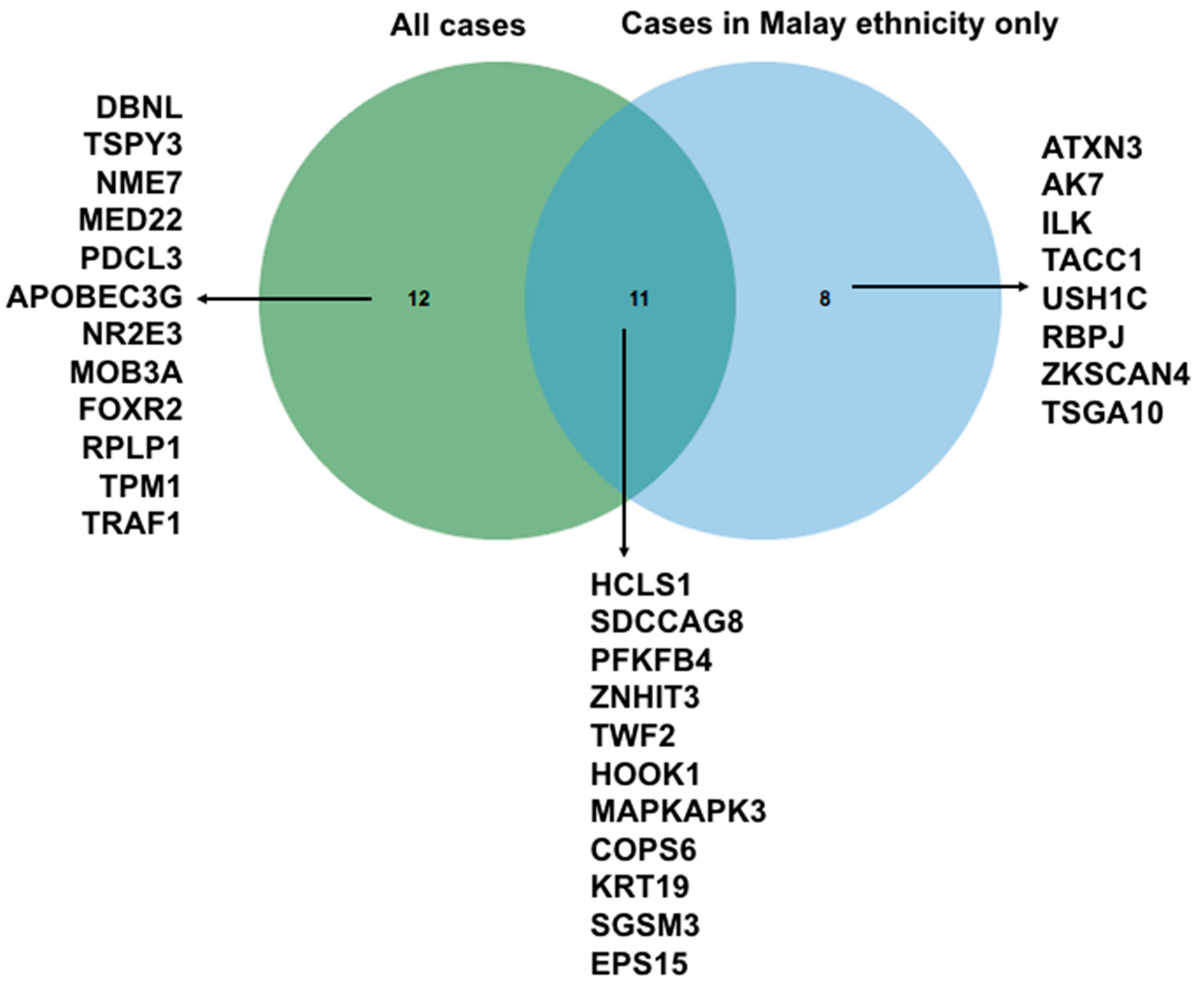
2.6. Autoantibody Signature Based on Malay Ethnicity Cases
2.7. Autoantibody Biomarkers Showed Relevance to Thalassaemia Major Diseases
2.8. Clustering of Biomarkers
3. Discussion
4. Materials and Methods
4.1. Study Design
4.2. Serum/Plasma Dilution
4.3. Biomarker Assay
4.4. Array Washing after Serum Binding
4.5. Incubation with Cy3-anti IgG
4.6. Bioinformatics Analysis
Image Analysis: Raw Data Extraction
4.7. Data Handling and Pre-Processing
- (i = spot number and j = sample number)
- Load all Cy3-BSA across all samples, j, into an i X j matrix X.
- Sort spot intensities in each column j of X to get Xsort
- Take the mean across each row i of Xsort to get < Xi >
- The intensity-based normalization on each sample was calculated based on the Equation (3) below:
- Calculate the sum of the mean across each row i, Σ < Xi >
- For each sample, k, calculate the sum of all Cy3-BSA controls, Σ Xk
- For each sample, k,
4.8. Data Analysis
4.9. Unsupervised Clustering on Normalised RFU for 23 Biomarkers across 12 Samples
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cappellini, M.D.; Cohen, A.; Eleftheriou, A.; Piga, A.; Porter, J.; Taher, A. Guidelines for the Clinical Management of Thalassaemia (Internet), 2nd Revised ed.; Thalassaemia International Federation: Nicosia, Cyprus, 2008. Available online: https://www.ncbi.nlm.nih.gov/books/NBK173968/ (accessed on 10 September 2019).
- Thong, M.K.; Law, H.Y.; Ng, I.S. Molecular heterogeneity of beta-thalassaemia in Malaysia: A practical approach to diagnosis. Ann. Acad. Med. Singap. 1996, 25, 79–83. [Google Scholar]
- George, E. Thalassaemia Carrier Diagnosis in Malaysia; Thalassaemia Diagnostic Service: Kuala Lumpur, Malaysia, 1998. [Google Scholar]
- Rund, D.; Rachmilewitz, E. Beta-thalassemia. N. Engl. J. Med. 2005, 353, 1135–1146. [Google Scholar] [CrossRef] [PubMed]
- Chakrabarti, A.; Bhattacharya, D.; Basu, A.; Basu, S.; Saha, S.; Halder, S. Differential expression of red cell proteins in hemoglobinopathy. Proteom. Clin. Appl. 2011, 5, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Goodman, S.R.; Daescu, O.; Kakhniashvili, D.G.; Zivanic, M. The proteomics and interactomics of human erythrocytes. Exp. Biol. Med. 2013, 238, 509–518. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Righetti, P.G.; Zolla, L. The red blood cell proteome and interactome: An update. J. Proteome Res. 2010, 9, 144–163. [Google Scholar] [CrossRef]
- Bhattacharya, D.; Saha, S.; Basu, S.; Chakravarty, S.; Chakravarty, A.; Banerjee, D.; Chakrabarti, A. Differential regulation of redox proteins and chaperones in HbE beta-thalassemia erythrocyte proteome. Proteom.-Clin. Appl. 2010, 4, 480–488. [Google Scholar]
- Hatairaktham, S.; Srisawat, C.; Siritanaratkul, N.; Chiangjong, W.; Fucharoen, S.; Thongboonkerd, V.; Kalpravidh, R.W. Differential plasma proteome profiles of mild versus severe β-thalassemia/Hb E. Ann. Hematol. 2013, 92, 365–377. [Google Scholar] [CrossRef]
- Zhang, D.Y.; Ye, F.; Gao, L.; Liu, X.; Zhao, X.; Che, Y.; Wang, H.; Wang, L.; Wu, J.; Song, D.; et al. Proteomics, pathway array and signaling network-based medicine in cancer. Cell Div. 2009, 4, 20. [Google Scholar] [CrossRef]
- Altinoz, M.A.; Gedikoglu, G.; Deniz, G. β-Thalassemia trait association with autoimmune diseases: β-globin locus proximity to the immunity genes or role of hemorphins? Immunopharmacol. Immunotoxicol. 2012, 34, 181–190. [Google Scholar] [CrossRef]
- Gnjatic, S.; Ritter, E.; Buchler, M.W.; Giese, N.A.; Brors, B.; Frei, C.; Murray, A.; Halama, N.; Zornig, I.; Chen, Y.T.; et al. Seromic profling of ovarian and pancreatic cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 5088–5093. [Google Scholar] [CrossRef]
- Blackburn, J.M.; Shoko, A. Protein function microarrays for customised systems-oriented proteome analysis. Methods Mol. Biol. 2011, 785, 305–330. [Google Scholar] [PubMed]
- Stempfer, R.; Syed, P.; Vierlinger, K.; Pichler, R.; Meese, E.; Leidinger, P.; Ludwig, N.; Kriegner, A.; Nohammer, C.; Weinhausel, A. Tumour auto-antibody screening: Performance of protein microarrays using SEREX derived antigens. BMC Cancer 2010, 10, 627. [Google Scholar] [CrossRef] [PubMed]
- Liew, J.; Amir, A.; Chen, Y.; Fong, M.Y.; Razali, R.; Lau, Y.L. Autoantibody profile of patients infected with knowlesi malaria. Clin. Chim. Acta 2015, 448, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Soe, H.J.; Yong, Y.K.; Al-Obaidi, M.M.; Raju, C.S.; Gudimella, R.; Manikam, R.; Sekaran, S.D. Identifying protein biomarkers in predicting disease severity of dengue virus infection using immune-related protein microarray. Medicine 2018, 97, e9713. [Google Scholar] [CrossRef]
- Da Gama Duarte, J.; Parakh, S.; Andrews, M.C.; Woods, K.; Pasam, A.; Tutuka, C.; Ostrouska, S.; Blackburn, J.M.; Behren, A.; Cebon, J. Autoantibodies May Predict Immune-Related Toxicity: Results from a Phase I Study of Intralesional Bacillus Calmette–Guérin followed by Ipilimumab in Patients with Advanced Metastatic Melanoma. Front. Immunol. 2018, 9, 411. [Google Scholar] [CrossRef]
- Zaenker, P.; Lo, J.; Pearce, R.; Cantwell, P.; Cowell, L.; Lee, M.; Quirk, C.; Law, H.; Gray, H.; Ziman, M. A diagnostic autoantibody signature for primary cutaneous melanoma. Oncotarget 2018, 9, 30539–30551. [Google Scholar] [CrossRef][Green Version]
- Suwarnalata, G.; Tan, A.H.; Isa, H.; Gudimella, R.; Anwar, A.; Loke, M.F.; Mahadeva, S.; Lim, S.-Y.; Vadivelu, J. Augmentation of Autoantibodies by Helicobacter pylori in Parkinson’s Disease Patients May Be Linked to Greater Severity. PLoS ONE 2016, 11, e0153725. [Google Scholar] [CrossRef][Green Version]
- Koscielny, G.; An, P.; Carvalho-Silva, D.; Cham, J.A.; Fumis, L.; Gasparyan, R.; Hasan, S.; Karamanis, N.; Maguire, M.; Papa, E.; et al. Open Targets: A platform for therapeutic target identification and validation. Nucleic Acids Res. 2017, 45, D985–D994. [Google Scholar] [CrossRef]
- Gharagozloo, M.; Karimi, M.; Amirghofran, Z. Double-faced cell-mediated immunity in β-thalassemia major: Stimulated phenotype versus suppressed activity. Ann. Hematol. 2009, 88, 21. [Google Scholar] [CrossRef]
- Nyberg, F.; Sanderson, K.; Glämsta, E.L. The hemorphins: A new class of opioid peptides derived from the blood protein hemoglobin. Pept. Sci. 1997, 43, 147–156. [Google Scholar] [CrossRef]
- Noulsri, E.; Lerdwana, S.; Fucharoen, S.; Pattanapanyasat, K. Pheno- typic characterization of circulating CD4/CD8 T-lymphocytes in beta- thalassemia patients. Asian Pac. J. Allergy Immunol. 2014, 32, 261–269. [Google Scholar] [CrossRef] [PubMed]
- Bozdogan, G.; Erdem, E.; Demirel, G.Y.; Yildirmak, Y. The role of Treg cells and FoxP3 expression in immunity of beta-thalassemia major AND beta-thalassemia trait patients. Pediatr. Hematol. Oncol. 2010, 27, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Taniuchi, I.; Kitamura, D.; Maekawa, Y.; Fukuda, T.; Kishi, H.; Watanabe, T. Antigen-receptor induced clonal expansion and deletion of lymphocytes are impaired in mice lacking HS1 protein, a substrate of the antigen-receptor-coupled tyrosine kinases. EMBO J. 1995, 14, 3664–3678. [Google Scholar] [CrossRef]
- Corthay, A. How do regulatory T cells work? Scand. J. Immunol. 2009, 70, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Cunningham-Rundles, S.; Giardina, P.J.; Grady, R.W.; Califano, C.; McKenzie, P.; De Sousa, M. Effect of transfusional iron overload on immune response. J. Infect. Dis. 2000, 182 (Suppl. 1), S115–S121. [Google Scholar] [CrossRef]
- Poole, J.; Daniels, G. Blood group antibodies and their significance in transfusion medicine. Transfus. Med. Rev. 2007, 21, 58–71. [Google Scholar] [CrossRef]
- Shamsian, B.; Arzanian, M.T.; Shamshiri, A.R.; Alavi, S.; Khojasteh, O. Frequency of red cell alloimmunization in patients with beta-major thalassemia in an Iranian referral hospital. Iran J. Pediatr. 2008, 18, 149–153. [Google Scholar]
- Van Regenmortel, M. The concept and operational definitionof protein epitopes. Philos. Trans. R. Soc. Lond. Ser. B 1989, 323, 451–466. [Google Scholar]
- Yaniv, G.; Twig, G.; Shor, D.B.-A.; Furer, A.; Sherer, Y.; Mozes, O.; Komisar, O.; Slonimsky, E.; Klang, E.; Lotan, E.; et al. A volcanic explosion of autoantibodies in systemic lupus erythematosus: A diversity of 180 different antibodies found in SLE patients. Autoimmun. Rev. 2015, 14, 75–79. [Google Scholar] [CrossRef]
- Burska, A.N.; Hunt, L.; Boissinot, M.; Strollo, R.; Ryan, B.J.; Vital, E.; Nissim, A.; Winyard, P.G.; Emery, P.; Ponchel, F. Autoantibodies to posttranslational modifications in rheumatoid arthritis. Mediat. Inflamm. 2014. [Google Scholar] [CrossRef]
- Chazenbalk, G.D.; Pichurin, P.; Chen, C.-R.; Latrofa, F.; Johnstone, A.P.; McLachlan, S.M.; Rapoport, B. Thyroid-stimulating autoantibodies in Graves disease preferentially recognize the free A subunit, not the thyrotropin holoreceptor. J. Clin. Investig. 2002, 110, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Fayyaz, A.; Kurien, B.T.; Scofield, R.H. Autoantibodies in Sjögren’s Syndrome. Rheum. Dis. Clin. N. Am. 2016. [Google Scholar] [CrossRef] [PubMed]
- Zachou, K.; Rigopoulou, E.; Dalekos, G.N. Autoantibodies and autoantigens in autoimmune hepatitis: Important tools in clinical practice and to study pathogenesis of the disease. J. Autoimmune Dis. 2004. [Google Scholar] [CrossRef]
- Burbelo, P.D.; Gordon, S.M.; Waldman, M.; Edison, J.D.; Little, D.J.; Stitt, R.S.; Olson, S.W. Autoantibodies are present before the clinical diagnosis of systemic sclerosis. PLoS ONE 2019, 14, e0214202. [Google Scholar] [CrossRef]
- Zang, R.; Li, Y.; Jin, R.; Wang, X.; Lei, Y.; Che, Y.; He, J.; Liu, C.; Zheng, S.; Zhou, F.; et al. Enhancement of diagnostic performance in lung cancers by combining CEA and CA125 with autoantibodies detection. OncoImmunology 2019, 1–8. [Google Scholar] [CrossRef]
- Bassaro, L.; Russell, S.J.; Pastwa, E.; Somiari, S.A.; Somiari, R.I. Screening for Multiple Autoantibodies in Plasma of Patients with Breast Cancer. Cancer Genom. Proteom. 2017, 14, 427–435. Available online: http://www.ncbi.nlm.nih.gov/pubmed/29109092 (accessed on 5 March 2020).
- Qiu, J.; Keyser, B.; Lin, Z.T.; Wu, T. Autoantibodies as potential biomarkers in breast cancer. Biosensors 2018, 8, 67. [Google Scholar] [CrossRef]
- Atkins, M.B.; Philips, G.K. Emerging monoclonal antibodies for the treatment of renal cell carcinoma (RCC). Expert Opin. Emerg. Drugs 2016, 21, 243–254. [Google Scholar] [CrossRef]
- Jones, K.L.; Van de Water, J. Maternal autoantibody related autism: Mechanisms and pathways. Mol. Psychiatry 2019. [Google Scholar] [CrossRef]
- Chimenti, M.S.; Triggianese, P.; De Martino, E.; Conigliaro, P.; Fonti, G.L.; Sunzini, F.; Caso, F.; Perricone, C.; Costa, L.; Perricone, R. An update on pathogenesis of psoriatic arthritis and potential therapeutic targets. Expert Rev. Clin. Immunol. 2019, 1–14. [Google Scholar] [CrossRef]
- Lewis, M.J.; Mcandrew, M.B.; Wheeler, C.; Workman, N.; Agashe, P.; Koopmann, J.; Vyse, T.J. Autoantibodies targeting TLR and SMAD pathways de fi ne new subgroups in systemic lupus erythematosus. J. Autoimmun. 2018, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.; Wang, J.; Wei, Q.; Du, Y.P.; Qiu, H.P.; Yang, C.; Hou, Y.C. Tropomyosin-1 promotes cancer cell apoptosis via the p53-mediated mitochondrial pathway in renal cell carcinoma. Oncol. Lett. 2018, 15, 7060–7068. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2010, 39, D561–D568. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.; Feng, Q.; Li, J.; Huang, Y.; Li, D. A Double Heterozygote for (δβ) 0-Thalassemia and Codons 41/42 (–TTCT) Behaves as a Homozygote for the Frameshift Mutation in a Chinese Family. Hemoglobin 2007, 31, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Koya, Y.; Liu, W.; Yamakita, Y.; Senga, T.; Shibata, K.; Yamashita, M.; Nawa, A.; Kikkawa, F.; Kajiyama, H. Hematopoietic lineage cell-specific protein 1 (HS1), a hidden player in migration, invasion, and tumor formation, is over-expressed in ovarian carcinoma cells. Oncotarget 2018, 9, 32609. [Google Scholar] [CrossRef] [PubMed]
- dos Remedios, C.G.; Chhabra, D.; Kekic, M.; Dedova, I.V.; Tsubakihara, M.; Berry, D.A.; Nosworthy, N.J. Actin Binding Proteins: Regulation of Cytoskeletal Microfilaments. Physiol. Rev. 2003, 83, 433–473. [Google Scholar] [CrossRef]
- Maciver, S.K. Maciver Lab Web Page. 2004. Available online: http://www.bms.ed.ac.uk/research/others/smaciver/Encyclop/encycloABP.htm (accessed on 12 December 2019).
- Bolstad, B.M.; Irizarry, R.; Astrand, M.; Speed, T.P. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003, 19, 185–193. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical Grouping to Optimize an Objective Function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]

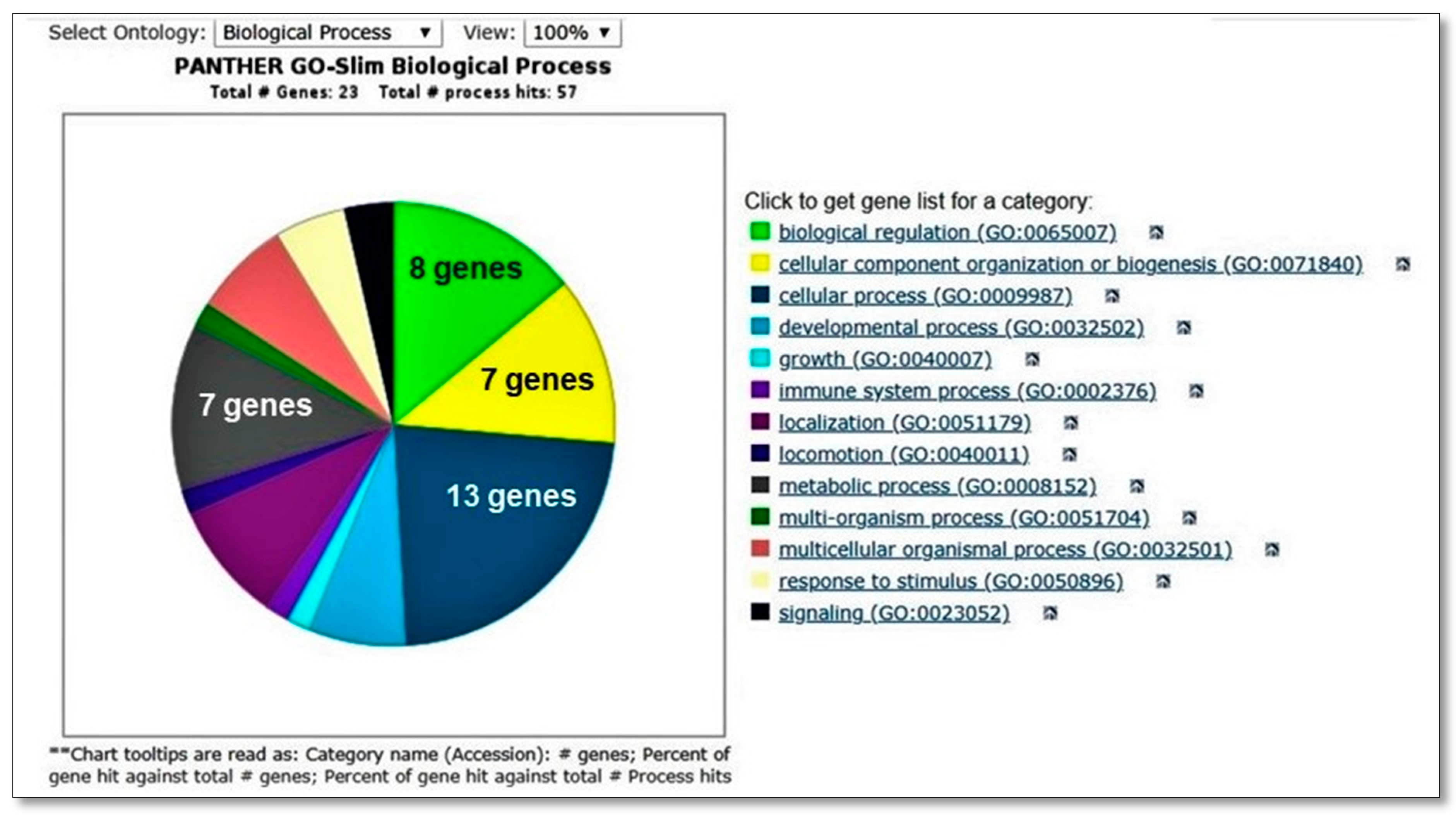
| Case ID | Hemoglobin | WBC Count | Total Bilirubin | Mutation Analysis | Biomarkers |
|---|---|---|---|---|---|
| (g/dL) | (×109 /L) | (mol/L) | |||
| 025757 | 9.1 | 6.1 | 41 | β+ IVS 1-5 [G-C] β variant codon 26 [G-A] | MAPKAPK3, TSPY3, PFKFB4 |
| 025724 | 8.6 | 6.8 | 21 | Not available | SDCCAG8, NME7 |
| 025712 | 12.4 | 8.9 | 15 | Not available | DBNL, TWF2, PDCL3, NR2E3, ZNHIT3 |
| 025711 | 11.1 | 5.3 | 34 | α SEA deletion α+ α-thal 3.7Kb deletion | HCLS1, HOOK1 |
| 025740 | 9.9 | 6.4 | 74 | β+ IVS 1-5 [G>C] β variant Codon 26 (GAG>GTG) | HCLS1, SDCCAG8, HOOK1, COPS6 |
| 025723 | 11.7 | 7.8 | 44 | β codon 41/42 [-TTCT] β variant codon 26 [G-A] | HCLS1. SDCCAG8, ZNHIT3, HOOK1, MAPKAPK3, MOB3A, PFKFB4, ZNHIT3 |
| 025749 | 8.1 | 5.4 | 99 | β codon 41/42 [-TTCT] | SDCCAG8, TSPY3, PFKFB4, APOBEC3G, APOBEC3G NR2E3KRT19, FOXR2, RPLP1, SGSM3, HCLS1, DBNL, ZNHIT3, NME7 |
| 025759 | 9.6 | 15.4 | 81 | β codon 41/42 [-TTCT] β+ -28 [A-G] | HCLS1, DBNL, NME7, TWF2, MAPKAPK3, COPS6, TSPY3 |
| 025750 | 9.0 | 10.2 | 44 | β codon 41/42 [-TTCT] β codon 71/72 [+A] | PFKFB4, PDCL3, TPM1, DBNL, TSPY3, ZNHIT3, NME7 |
| No. | Proteins | Penetrance Frequency (* Case) | Penetrance Fold Change (* Case) | Mean (Control) |
|---|---|---|---|---|
| 1 | HCLS1 | 5 | 5.33 | 157.83 |
| 2 | DBNL | 4 | 4.07 | 109.26 |
| 3 | SDCCAG8 | 4 | 4.01 | 144.09 |
| 4 | TSPY3 | 4 | 3.02 | 97.43 |
| 5 | PFKFB4 | 4 | 2.7 | 119.35 |
| 6 | ZNHIT3 | 4 | 2.46 | 102.29 |
| 7 | NME7 | 4 | 2.39 | 95.36 |
| 8 | TWF2 | 3 | 13.38 | 85.86 |
| 9 | HOOK1 | 3 | 9.21 | 163.31 |
| 10 | MAPKAPK3 | 3 | 7.21 | 104.63 |
| 11 | COPS6 | 3 | 7.08 | 103.80 |
| 12 | MED22 | 3 | 5.95 | 103.00 |
| 13 | PDCL3 | 3 | 5.89 | 94.35 |
| 14 | APOBEC3G | 3 | 3.58 | 102.11 |
| 15 | NR2E3 | 3 | 3.49 | 107.26 |
| 16 | KRT19 | 3 | 3.32 | 209.80 |
| 17 | MOB3A | 3 | 3.17 | 130.69 |
| 18 | FOXR2 | 3 | 3.00 | 104.71 |
| 19 | RPLP1 | 3 | 2.97 | 99.18 |
| 20 | SGSM3 | 3 | 2.85 | 129.13 |
| 21 | TPM1 | 3 | 2.79 | 248.36 |
| 22 | EPS15 | 3 | 2.61 | 115.74 |
| 23 | TRAF1 | 3 | 2.16 | 110.63 |
| Disease Full Name | No. of Associated Targets | All Targets |
|---|---|---|
| Thalassemia | 4 | EPS15 HCLS1 KRT19 TPM1 |
| Beta-thal and related diseases | 3 | EPS15 HCLS1 TPM1 |
| Beta-thal | 2 | HCLS1 TPM1 |
| Beta-thal intermedia | 1 | HCLS1 |
| Hereditary persistence of fetal Hb- beta-thal | 1 | EPS15 |
| Delta-beta-thal | 1 | EPS15 |
| Beta-thal associated with another Hb anomaly | 1 | EPS15 |
| Beta-thal major | 1 | HCLS1 |
| Alpha-thal | 1 | KRT19 |
| Alpha-thal and related diseases | 1 | KRT19 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sumera, A.; Anuar, N.D.; Radhakrishnan, A.K.; Ibrahim, H.; Rutt, N.H.; Ismail, N.H.; Tan, T.-M.; Baba, A.A. A Novel Method to Identify Autoantibodies against Putative Target Proteins in Serum from beta-Thalassemia Major: A Pilot Study. Biomedicines 2020, 8, 97. https://doi.org/10.3390/biomedicines8050097
Sumera A, Anuar ND, Radhakrishnan AK, Ibrahim H, Rutt NH, Ismail NH, Tan T-M, Baba AA. A Novel Method to Identify Autoantibodies against Putative Target Proteins in Serum from beta-Thalassemia Major: A Pilot Study. Biomedicines. 2020; 8(5):97. https://doi.org/10.3390/biomedicines8050097
Chicago/Turabian StyleSumera, Afshan, Nur Diana Anuar, Ammu Kutty Radhakrishnan, Hishamshah Ibrahim, Nurul H. Rutt, Nur Hafiza Ismail, Ti-Myen Tan, and Abdul Aziz Baba. 2020. "A Novel Method to Identify Autoantibodies against Putative Target Proteins in Serum from beta-Thalassemia Major: A Pilot Study" Biomedicines 8, no. 5: 97. https://doi.org/10.3390/biomedicines8050097
APA StyleSumera, A., Anuar, N. D., Radhakrishnan, A. K., Ibrahim, H., Rutt, N. H., Ismail, N. H., Tan, T.-M., & Baba, A. A. (2020). A Novel Method to Identify Autoantibodies against Putative Target Proteins in Serum from beta-Thalassemia Major: A Pilot Study. Biomedicines, 8(5), 97. https://doi.org/10.3390/biomedicines8050097





