The Minimal Effect of Linker Length for Fatty Acid Conjugation to a Small Protein on the Serum Half-Life Extension
Abstract
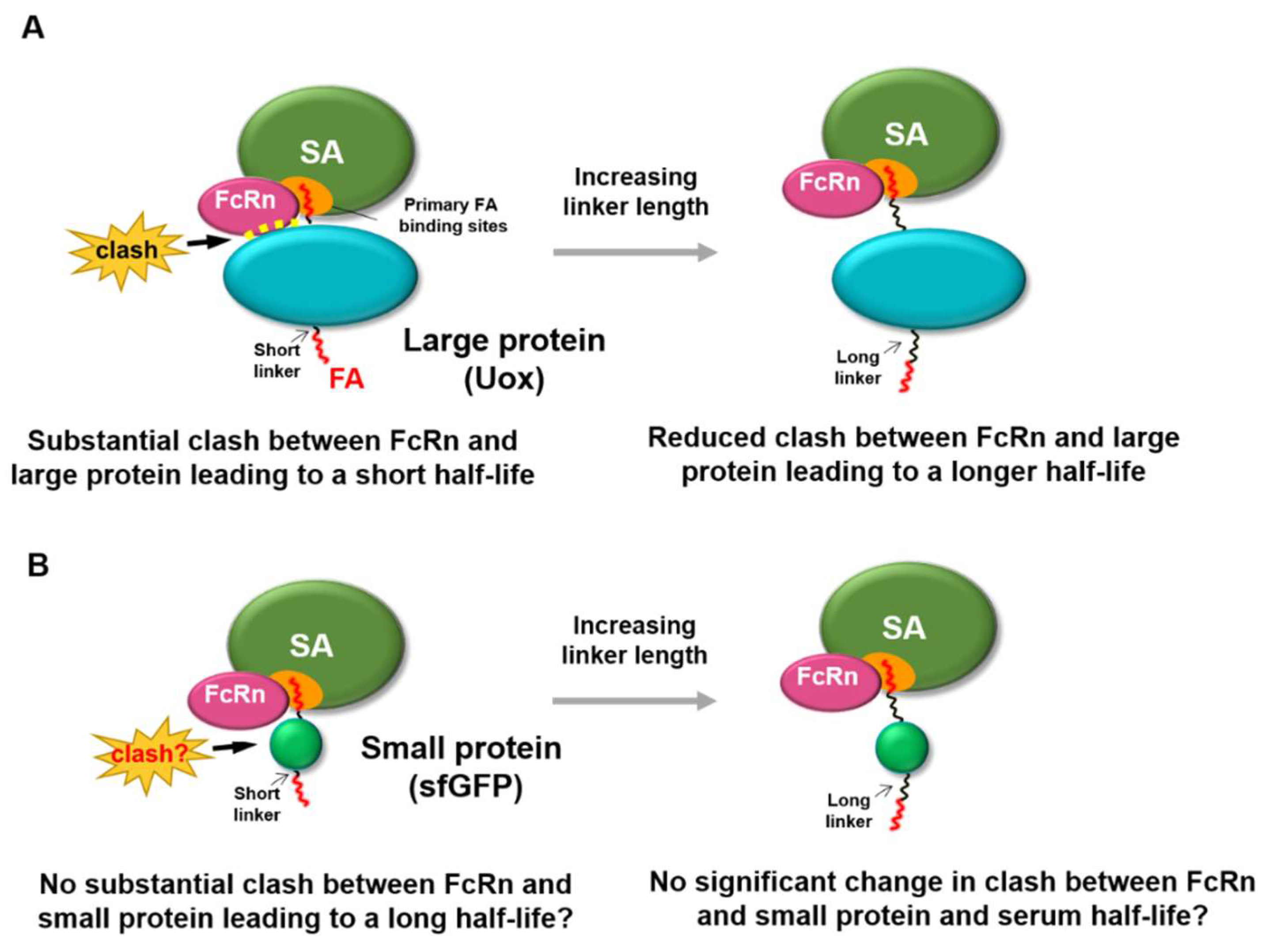
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Preparation of Purified Superfolder Green Fluorescent Protein (sfGFP) from E. coli
2.3. Preparation of sfGFP-PA Conjugates with Various Linkers
2.4. Matrix-Assisted Laser Desorption Ionization/Time-of-Flight (MALDI-TOF) Analysis of the sfGFP and sfGFP-PA Conjugates
2.5. In Vitro Serum Albumin Binding Assay
2.6. sfGFP Fluorescence Assay
2.7. Serum Half-Life Determination in Mice
2.8. FcRn/Serum Albumin/sfGFP-PA Tertiary Complex Formation Assay
3. Results and Discussion
3.1. PA-Conjugated sfGFP Conjugates with Various Linker Lengths
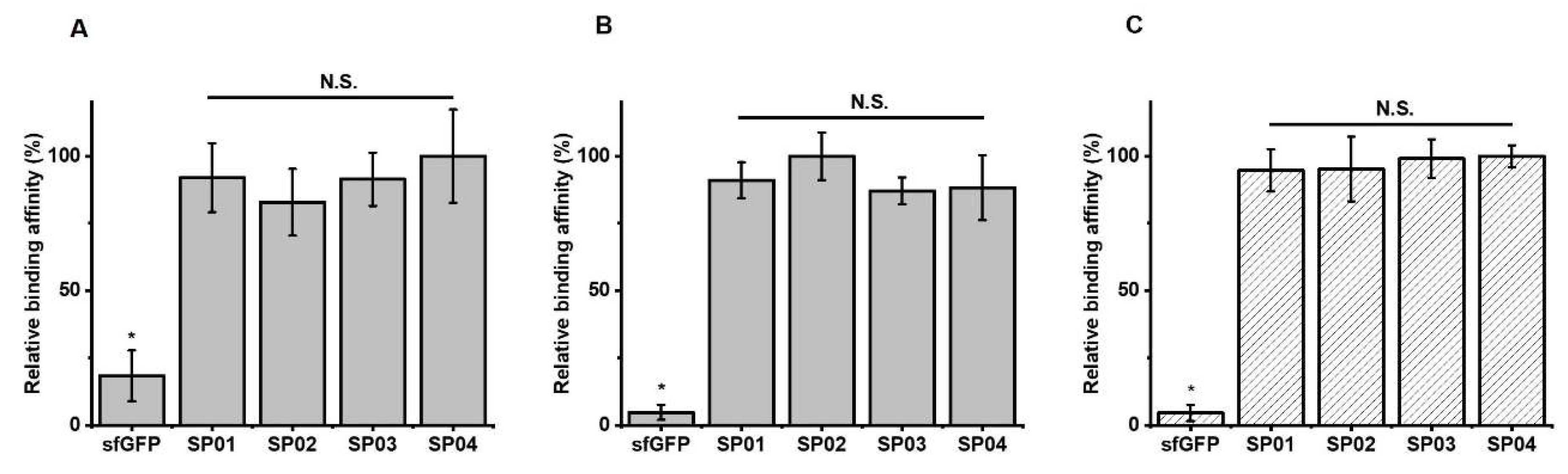
3.2. Serum Albumin Binding Affinity of sfGFP-PA Conjugates
3.3. Fluorescence of sfGFP-PA Conjugates
3.4. Serum Half-Lives of sfGFP-PA Conjugates
3.5. FcRn Binding Assays of sfGFP-PA Conjugates
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Agyei, D.; Ahmed, I.; Akram, Z.; MN Iqbal, H.; K Danquah, M. Protein and peptide biopharmaceuticals: An overview. Protein Pept. Lett. 2017, 24, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Kontermann, R.E. Strategies for extended serum half-life of protein therapeutics. Curr. Opin. Biotechnol. 2011, 22, 868–876. [Google Scholar] [CrossRef] [PubMed]
- Kontermann, R.E. Strategies to Extend Plasma Half-Lives of Recombinant Antibodies. BioDrugs 2009, 23, 93–109. [Google Scholar] [CrossRef] [PubMed]
- Jevševa, S.; Kunstelj, M.; Porekar, V.G. PEGylation of therapeutic proteins. Biotechnol. J. 2010, 5, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Lim, H.-K.; Hong, S.H.; Bae, S.M.; Choi, I.Y.; Kim, H.H. A Liquid Formulation of a Long-acting Erythropoietin Conjugate. Biotechnol. Bioprocess Eng. 2020, 25, 117–125. [Google Scholar] [CrossRef]
- Pelegri-O’Day, E.M.; Lin, E.-W.; Maynard, H.D. Therapeutic Protein–Polymer Conjugates: Advancing Beyond PEGylation. J. Am. Chem. Soc. 2014, 136, 14323–14332. [Google Scholar] [CrossRef]
- Verhoef, J.J.F.; Anchordoquy, T.J. Questioning the use of PEGylation for drug delivery. Drug Deliv. Transl. Res. 2013, 3, 499–503. [Google Scholar] [CrossRef]
- Yang, B.; Lim, S.I.; Kim, J.C.; Tae, G.; Kwon, I. Site-Specific Albumination as an Alternative to PEGylation for the Enhanced Serum Half-Life in Vivo. Biomacromolecules 2016, 17, 1811–1817. [Google Scholar] [CrossRef]
- Cho, J.; Lim, S.I.; Yang, B.S.; Hahn, Y.S.; Kwon, I. Generation of therapeutic protein variants with the human serum albumin binding capacity via site-specific fatty acid conjugation. Sci. Rep. 2017, 7, 18041. [Google Scholar] [CrossRef]
- Bern, M.; Sand, K.M.K.; Nilsen, J.; Sandlie, I.; Andersen, J.T. The role of albumin receptors in regulation of albumin homeostasis: Implications for drug delivery. J. Control. Release 2015, 211, 144–162. [Google Scholar] [CrossRef]
- Sleep, D.; Cameron, J.; Evans, L.R. Albumin as a versatile platform for drug half-life extension. Biochim. Biophys. Acta-Gen. Subj. 2013, 1830, 5526–5534. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.I.; Hahn, Y.S.; Kwon, I. Site-specific albumination of a therapeutic protein with multi-subunit to prolong activity in vivo. J. Control. Release 2015, 207, 93–100. [Google Scholar] [CrossRef] [PubMed]
- Zaman, R.; Islam, R.A.; Ibnat, N.; Othman, I.; Zaini, A.; Lee, C.Y.; Chowdhury, E.H. Current strategies in extending half-lives of therapeutic proteins. J. Control. Release 2019, 301, 176–189. [Google Scholar] [CrossRef]
- Kratz, F. Albumin as a drug carrier: Design of prodrugs, drug conjugates and nanoparticles. J. Control. Release 2008, 132, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Madsbad, S.; Kielgast, U.; Asmar, M.; Deacon, C.F.; Torekov, S.S.; Holst, J.J. An overview of once-weekly glucagon-like peptide-1 receptor agonists-available efficacy and safety data and perspectives for the future. Diabetes Obes. Metab. 2011, 13, 394–407. [Google Scholar] [CrossRef] [PubMed]
- Rosenstock, J.; Reusch, J.; Bush, M.; Yang, F.; Stewart, M.; Albiglutide Study Group, for the A.S. Potential of albiglutide, a long-acting GLP-1 receptor agonist, in type 2 diabetes: A randomized controlled trial exploring weekly, biweekly, and monthly dosing. Diabetes Care 2009, 32, 1880–1886. [Google Scholar] [CrossRef]
- Elsadek, B.; Kratz, F. Impact of albumin on drug delivery—New applications on the horizon. J. Control. Release 2012, 157, 4–28. [Google Scholar] [CrossRef]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic Acid: Physiological Role, Metabolism and Nutritional Implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef]
- Knudsen, L.B.; Nielsen, P.F.; Huusfeldt, P.O.; Johansen, N.L.; Madsen, K.; Pedersen, F.Z.; Thøgersen, H.; Wilken, M.; Agersø, A. Potent Derivatives of Glucagon-like Peptide-1 with Pharmacokinetic Properties Suitable for Once Daily Administration. J. Med. Chem. 2000, 43, 1664–1669. [Google Scholar] [CrossRef]
- Lee, J.; Lee, C.; Kim, I.; Moon, H.R.; Kim, T.H.; Oh, K.T.; Lee, E.S.; Lee, K.C.; Youn, Y.S. Preparation and evaluation of palmitic acid-conjugated exendin-4 with delayed absorption and prolonged circulation for longer hypoglycemia. Int. J. Pharm. 2012, 424, 50–57. [Google Scholar] [CrossRef]
- Ramírez-Andersen, H.S.; Behrens, C.; Buchardt, J.; Fels, J.J.; Folkesson, C.G.; Jianhe, C.; Nørskov-Lauritsen, L.; Nielsen, P.F.; Reslow, M.; Rischel, C. Long-acting human growth hormone analogue by noncovalent albumin binding. Bioconjug. Chem. 2018, 29, 3129–3143. [Google Scholar] [CrossRef] [PubMed]
- Shechter, Y.; Sasson, K.; Lev-Goldman, V.; Rubinraut, S.; Rubinstein, M.; Fridkin, M. Newly Designed Modifier Prolongs the Action of Short-Lived Peptides and Proteins by Allowing Their Binding to Serum Albumin. Bioconjug. Chem. 2012, 23, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.I.; Mizuta, Y.; Takasu, A.; Hahn, Y.S.; Kim, Y.H.; Kwon, I. Site-specific fatty acid-conjugation to prolong protein half-life in vivo. J. Control. Release 2013, 170, 219–225. [Google Scholar] [CrossRef]
- Gil, M.S.; Cho, J.; Thambi, T.; Giang Phan, V.H.; Kwon, I.; Lee, D.S. Bioengineered robust hybrid hydrogels enrich the stability and efficacy of biological drugs. J. Control. Release 2017, 267, 119–132. [Google Scholar] [CrossRef]
- Milo, R.; Phillips, R. Cell Biology by the Numbers; Garland Science: New York, NY, USA, 2015; ISBN 1317230698. [Google Scholar]
- Cho, J.; Park, J.; Kim, S.; Kim, J.C.; Tae, G.; Jin, M.S.; Kwon, I. Intramolecular distance in the conjugate of urate oxidase and fatty acid governs FcRn binding and serum half-life in vivo. J. Control. Release 2020, 321, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Vogt, B. Urate oxidase (rasburicase) for treatment of severe tophaceous gout. Nephrol. Dial. Transplant. 2005, 20, 431–433. [Google Scholar] [CrossRef]
- Wang, L.-Y.; Shih, L.-Y.; Chang, H.; Jou, S.-T.; Lin, K.-H.; Yeh, T.-C.; Lin, S.-F.; Liang, D.-C. Recombinant Urate Oxidase (Rasburicase) for the Prevention and Treatment of Tumor Lysis Syndrome in Patients with Hematologic Malignancies. Acta Haematol. 2006, 115, 35–38. [Google Scholar] [CrossRef] [PubMed]
- Richette, P.; Bardin, T. Successful treatment with rasburicase of a tophaceous gout in a patient allergic to allopurinol. Nat. Clin. Pract. Rheumatol. 2006, 2, 338–342. [Google Scholar] [CrossRef]
- Edwards, N.L. Treatment-failure gout: A moving target. Arthritis Rheum. 2008, 58, 2587–2590. [Google Scholar] [CrossRef]
- Fels, E.; Sundy, J.S. Refractory gout: What is it and what to do about it? Curr. Opin. Rheumatol. 2008, 20, 198–202. [Google Scholar] [CrossRef]
- Bhattacharya, A.A.; Grüne, T.; Curry, S. Crystallographic analysis reveals common modes of binding of medium and long-chain fatty acids to human serum albumin. J. Mol. Biol. 2000, 303, 721–732. [Google Scholar] [CrossRef] [PubMed]
- Curry, S.; Brick, P.; Franks, N.P. Fatty acid binding to human serum albumin: New insights from crystallographic studies. Biochim. Biophys. Acta-Mol. Cell Biol. Lipids 1999, 1441, 131–140. [Google Scholar] [CrossRef]
- Simard, J.R.; Zunszain, P.A.; Hamilton, J.A.; Curry, S. Location of High and Low Affinity Fatty Acid Binding Sites on Human Serum Albumin Revealed by NMR Drug-competition Analysis. J. Mol. Biol. 2006, 361, 336–351. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S.; Amisaki, T. Identification of High Affinity Fatty Acid Binding Sites on Human Serum Albumin by MM-PBSA Method. Biophys. J. 2008, 94, 95–103. [Google Scholar] [CrossRef]
- Fujiwara, S.; Amisaki, T. Fatty acid binding to serum albumin: Molecular simulation approaches. Biochim. Biophys. Acta-Gen. Subj. 2013, 1830, 5427–5434. [Google Scholar] [CrossRef]
- Rizzuti, B.; Bartucci, R.; Sportelli, L.; Guzzi, R. Fatty acid binding into the highest affinity site of human serum albumin observed in molecular dynamics simulation. Arch. Biochem. Biophys. 2015, 579, 18–25. [Google Scholar] [CrossRef]
- Hamilton, J.A. NMR reveals molecular interactions and dynamics of fatty acid binding to albumin. Biochim. Biophys. Acta-Gen. Subj. 2013, 1830, 5418–5426. [Google Scholar] [CrossRef]
- Pace, C.N.; Vajdos, F.; Fee, L.; Grimsley, G.; Gray, T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995, 4, 2411–2423. [Google Scholar] [CrossRef]
- Pédelacq, J.-D.; Cabantous, S.; Tran, T.; Terwilliger, T.C.; Waldo, G.S. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 2006, 24, 79–88. [Google Scholar] [CrossRef]
- Feng, G.; Mellor, R.H.; Bernstein, M.; Keller-Peck, C.; Nguyen, Q.T.; Wallace, M.; Nerbonne, J.M.; Lichtman, J.W.; Sanes, J.R. Imaging Neuronal Subsets in Transgenic Mice Expressing Multiple Spectral Variants of GFP. Neuron 2000, 28, 41–51. [Google Scholar] [CrossRef]
- Okabe, M.; Ikawa, M.; Kominami, K.; Nakanishi, T.; Nishimune, Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997, 407, 313–319. [Google Scholar] [CrossRef]
- Jung, S.-K. A Review of Image Analysis in Biochemical Engineering. Biotechnol. Bioprocess Eng. 2019, 24, 65–75. [Google Scholar] [CrossRef]
- Ahn, G.; Yu, G.; Abdullah, A.; Kim, Y.; Lee, D. Controlling the Release Profile Through Phase Control of Calcium Phosphate-Alginate Core-shell Nanoparticles in Gene Delivery. Macromol. Res. 2019, 27, 579–585. [Google Scholar] [CrossRef]
- Yang, B.; Kim, J.C.; Seong, J.; Tae, G.; Kwon, I. Comparative studies of the serum half-life extension of a protein via site-specific conjugation to a species-matched or-mismatched albumin. Biomater. Sci. 2018, 6, 2092–2100. [Google Scholar] [CrossRef] [PubMed]
- Datta-Mannan, A. Mechanisms Influencing the Pharmacokinetics and Disposition of Monoclonal Antibodies and Peptides. Drug Metab. Dispos. 2019, 47, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Nilsen, J.; Bern, M.; Sand, K.M.K.; Grevys, A.; Dalhus, B.; Sandlie, I.; Andersen, J.T. Human and mouse albumin bind their respective neonatal Fc receptors differently. Sci. Rep. 2018, 8, 14648. [Google Scholar] [CrossRef]
- Seijsing, J.; Lindborg, M.; Höidén-Guthenberg, I.; Bönisch, H.; Guneriusson, E.; Frejd, F.Y.; Abrahmsén, L.; Ekblad, C.; Löfblom, J.; Uhlén, M.; et al. An engineered affibody molecule with pH-dependent binding to FcRn mediates extended circulatory half-life of a fusion protein. Proc. Natl. Acad. Sci. USA 2014, 111, 17110–17115. [Google Scholar] [CrossRef]
- Hu, Y.-B.; Dammer, E.B.; Ren, R.-J.; Wang, G. The endosomal-lysosomal system: From acidification and cargo sorting to neurodegeneration. Transl. Neurodegener. 2015, 4, 18. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cho, J.; Park, J.; Tae, G.; Jin, M.S.; Kwon, I. The Minimal Effect of Linker Length for Fatty Acid Conjugation to a Small Protein on the Serum Half-Life Extension. Biomedicines 2020, 8, 96. https://doi.org/10.3390/biomedicines8050096
Cho J, Park J, Tae G, Jin MS, Kwon I. The Minimal Effect of Linker Length for Fatty Acid Conjugation to a Small Protein on the Serum Half-Life Extension. Biomedicines. 2020; 8(5):96. https://doi.org/10.3390/biomedicines8050096
Chicago/Turabian StyleCho, Jinhwan, Junyong Park, Giyoong Tae, Mi Sun Jin, and Inchan Kwon. 2020. "The Minimal Effect of Linker Length for Fatty Acid Conjugation to a Small Protein on the Serum Half-Life Extension" Biomedicines 8, no. 5: 96. https://doi.org/10.3390/biomedicines8050096
APA StyleCho, J., Park, J., Tae, G., Jin, M. S., & Kwon, I. (2020). The Minimal Effect of Linker Length for Fatty Acid Conjugation to a Small Protein on the Serum Half-Life Extension. Biomedicines, 8(5), 96. https://doi.org/10.3390/biomedicines8050096




