Factors Influencing the Prevalence of Resistance-Associated Substitutions in NS5A Protein in Treatment-Naive Patients with Chronic Hepatitis C
Abstract
:1. Introduction
2. Results
2.1. Prevalence of RASs
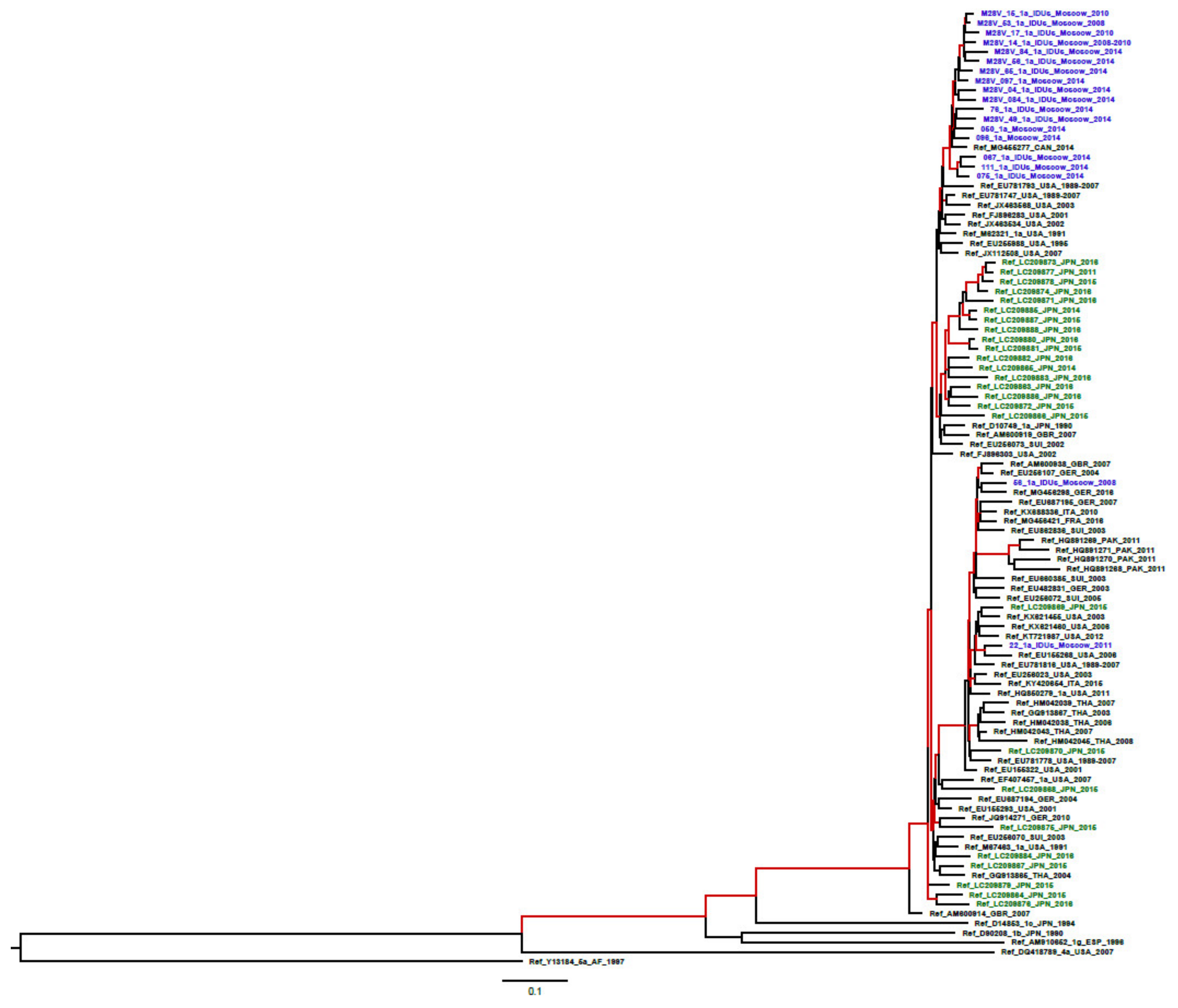
2.2. Probability of RAS Generation/Occurrence
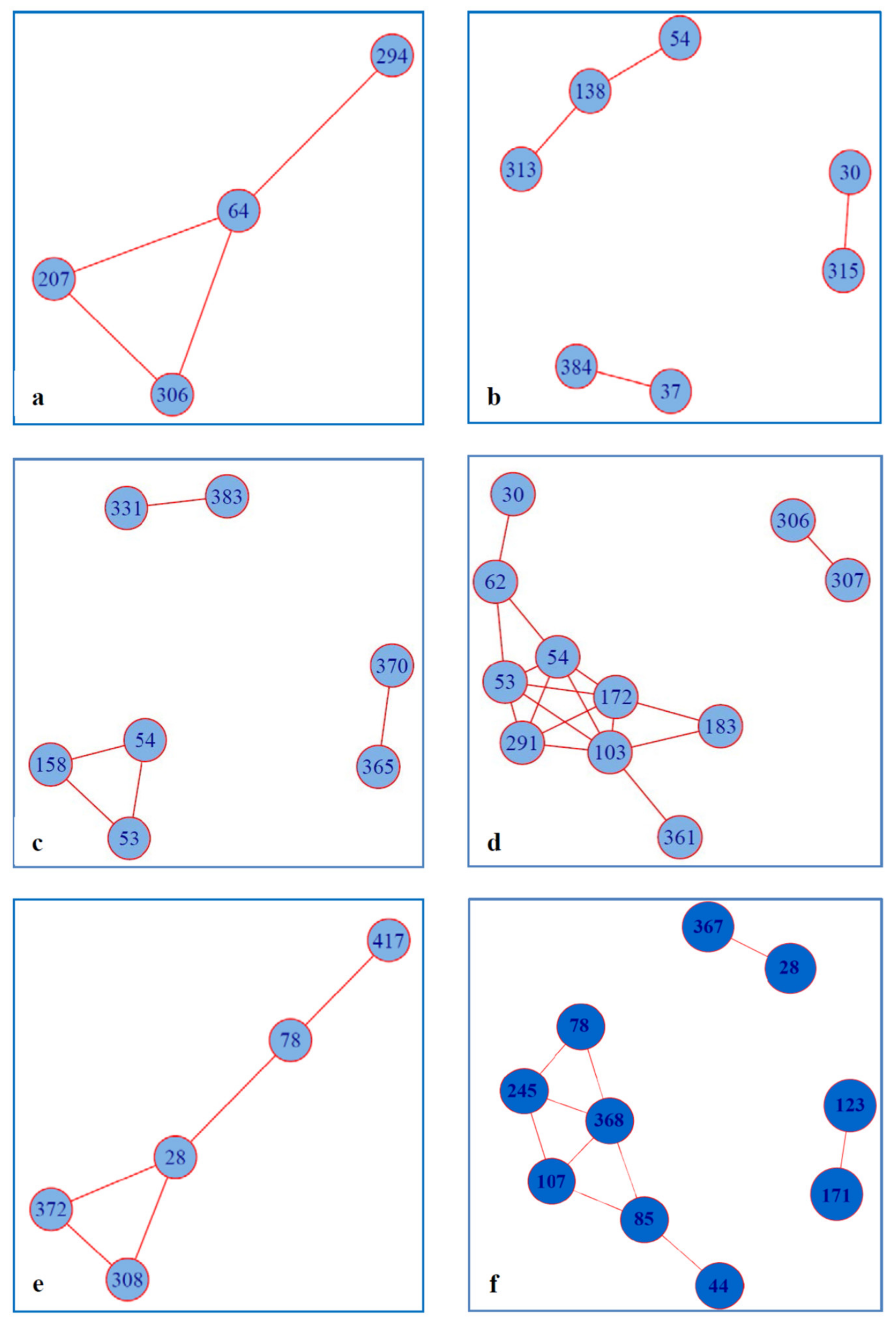
2.3. Covariance of RAS with Other Amino Acid Residues within NS5A
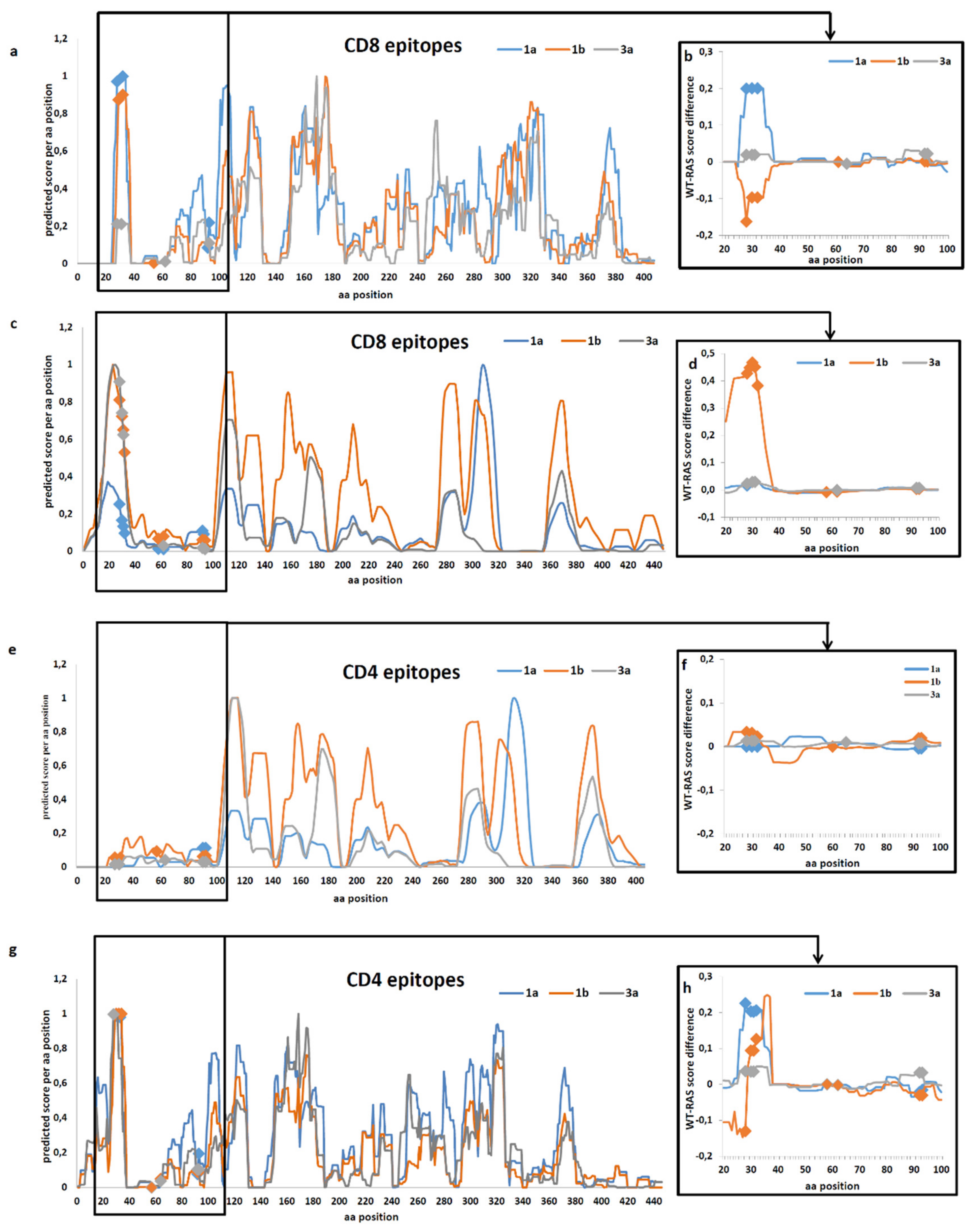
2.4. Prediction of the Effect of RAS on Immune Recognition of Epitopes
3. Discussion
4. Materials and Methods
4.1. Human Serum Samples
4.2. RNA Isolation and RT-PCR
4.3. HCV Genotyping
4.4. Amplification and Sequencing of HCV NS5A Coding Region
4.5. Retrieval of the Wild-Type and RAS Carrying HCV NS5A Sequences from Gene Bank
4.6. Phylogenetic Analysis
4.7. Analysis of Amino Acid Covariance
4.8. In Silico T-Cell Epitopic Analysis
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| HCV | Hepatitis C virus |
| CHC | Chronic hepatitis C |
| HCC | Hepatocellular carcinoma |
| DAAs | Direct-acting antivirals |
| RAS | Resistance-associated substitution |
References
- Global Health Sector Strategy on Viral Hepatitis 2016–2021. WHO, Geneva, 2016. Available online: http://apps.who.int/iris/bitstream/10665/246177/1/WHO-HIV-2016.06-eng.pdf?ua=1 (accessed on 5 February 2020).
- Wyles, D.L.; Gutierrez, J.A. Importance of HCV genotype 1 subtypes for drug resistance and response to therapy. J. Viral Hepat. 2014, 21, 229–240. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, I.M.; Asante-Appiah, E.; Wong, P.; Black, T.; Howe, A.Y.M. Prevalence and impact of baseline NS5A resistance associated variants (RAVs) on the efficacy of elbasvir/grazoprevir (EBR/GZR) against GT1a infection. Hepatol. Baltim. 2015, 62, 1393A–1394A. [Google Scholar]
- Zeuzem, S.; Mizokami, M.; Pianko, S.; Mangia, A.; Han, K.-H.; Martin, R.; Svarovskaia, E.; Dvory-Sobol, H.; Doehle, B.; Hedskog, C.; et al. NS5A resistance-associated substitutions in patients with genotype 1 hepatitis C virus: Prevalence and effect on treatment outcome. J. Hepatol. 2017, 66, 910–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lontok, E.; Harrington, P.; Howe, A.; Kieffer, T.; Lennerstrand, J.; Lenz, O.; McPhee, F.; Mo, H.; Parkin, N.; Pilot-Matias, T.; et al. Hepatitis C virus drug resistance-associated substitutions: State of the art summary. Hepatology 2015, 62, 1623–1632. [Google Scholar] [CrossRef]
- Chen, Z.-W.; Li, H.; Ren, H.; Hu, P. Global prevalence of pre-existing HCV variants resistant to direct-acting antiviral agents (DAAs): Mining the GenBank HCV genome data. Sci. Rep. 2016, 6, 20310. [Google Scholar] [CrossRef] [Green Version]
- Wyles, D.L. Resistance to DAAs: When to Look and When It Matters. Curr. HIV/AIDS Rep. 2017, 14, 229–237. [Google Scholar] [CrossRef]
- Sarrazin, C.; Dvory-Sobol, H.; Svarovskaia, E.S.; Doehle, B.P.; Pang, P.S.; Chuang, S.-M.; Ma, J.; Ding, X.; Afdhal, N.; Kowdley, K.V.; et al. Prevalence of Resistance-Associated Substitutions in HCV NS5A, NS5B, or NS3 and Outcomes of Treatment with Ledipasvir and Sofosbuvir. Gastroenterology 2016, 151, 501–512. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, P.; Schnell, G.; Tripathi, R.; Beyer, J.; Reisch, T.; Zhang, X.; Setze, C.; Rodrigues, L.; Burroughs, M.; Redman, R.; et al. Analysis of Hepatitis C Virus Genotype 1b Resistance Variants in Japanese Patients Treated with Paritaprevir-Ritonavir and Ombitasvir. Antimicrob. Agents Chemother. 2015, 60, 1106–1113. [Google Scholar] [CrossRef] [Green Version]
- Hope, V.D.; Eramova, I.; Capurro, D.; Donoghoe, M.C. Prevalence and estimation of hepatitis B and C infections in the WHO European Region: A review of data focusing on the countries outside the European Union and the European Free Trade Association. Epidemiol. Infect. 2014, 142, 270–286. [Google Scholar] [CrossRef]
- Bailey, H.; Turkova, A.; Thorne, C. Syphilis, hepatitis C and HIV in Eastern Europe. Curr. Opin. Infect. Dis. 2017, 30, 93–100. [Google Scholar] [CrossRef] [Green Version]
- Kartashev, V.; Döring, M.; Nieto, L.; Coletta, E.; Kaiser, R.; Sierra, S. HCV EuResist Study group.New findings in HCV genotype distribution in selected West European, Russian and Israeli regions. J. Clin. Virol. 2016, 81, 82–89. [Google Scholar] [CrossRef] [PubMed]
- European Association for the Study of the Liver. EASL Recommendations on Treatment of Hepatitis C 2018. J. Hepatol. 2018, 69, 461–511. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ross-Thriepland, D.; Harris, M. Hepatitis C virus NS5A: Enigmatic but still promiscuous 10 years on! J. Gen. Virol. 2015, 96, 727–738. [Google Scholar] [CrossRef] [PubMed]
- Ascher, D.B.; Wielens, J.; Nero, T.L.; Doughty, L.; Morton, C.J.; Parker, M.W. Potent hepatitis C inhibitors bind directly to NS5A and reduce its affinity for RNA. Sci. Rep. 2014, 4, 4765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Boyle, D.R.; Sun, J.-H.; Nower, P.T.; Lemm, J.A.; Fridell, R.A.; Wang, C.; Romine, J.L.; Belema, M.; Nguyen, V.N.; Laurent, D.R.S.; et al. Characterizations of HCV NS5A replication complex inhibitors. Virology 2013, 444, 343–354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schinazi, R.F.; Kohler, J.J.; Nettles, J.; Amblard, F.; Hurwitz, S.J.; Bassit, L.; A Stanton, R.; Ehteshami, M. Approaches to hepatitis C treatment and cure using NS5A inhibitors. Infect. Drug Resist. 2014, 7, 41–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarrazin, C. The importance of resistance to direct antiviral drugs in HCV infection in clinical practice. J. Hepatol. 2016, 64, 486–504. [Google Scholar] [CrossRef]
- Sorbo, M.C.; Cento, V.; Di Maio, V.C.; Howe, A.Y.; García, F.; Perno, C.F.; Ceccherini-Silberstein, F. Hepatitis C virus drug resistance associated substitutions and their clinical relevance: Update 2018. Drug Resist. Updat. 2018, 37, 17–39. [Google Scholar] [CrossRef]
- Hoshino, K.; Sugiyama, M.; Date, T.; Maruwaka, S.; Arakaki, S.; Shibata, D.; Maeshiro, T.; Hokama, A.; Sakugawa, H.; Kanto, T.; et al. Phylogenetic and phylodynamic analyses of hepatitis C virus subtype 1a in Okinawa, Japan. J. Viral Hepat. 2018, 25, 976–985. [Google Scholar] [CrossRef]
- Kliemann, D.A.; Tovo, C.V.; Gorini da Veiga, A.B.; Machado, A.L.; West, J. Genetic Barrier to Direct Acting Antivirals in HCV Sequences Deposited in the European Databank. PLoS ONE 2016, 11, e0159924. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, P.; Tripathi, R.; Schnell, G.; Reisch, T.; Beyer, J.; Irvin, M.; Xie, W.; Larsen, L.; Cohen, D.; Podsadecki, T.; et al. Resistance analysis of baseline and treatment emergent variants in hepatitis C virus genotype 1 in the AVIATOR study with paritaprevir ritonavir, ombitasvir, and dasabuvir. Antimicrob. Agents Chemother. 2015, 59, 5445–5454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lahser, F.C.; Galloway, A.; Hwang, P.; Palcza, J.; Wahl, J. Interim analysis of a 3-year follow-up study of NS5A and NS3 resistance associated variants (RAVs) after treatment with grazoprevir containing regimens in patients with chronic hepatitis C virus (HCV) infection. Hepatol. Baltim. 2016, 64, 32A. [Google Scholar]
- Wyles, D.; Mangia, A.; Cheng, W.; Shafran, S.; Schwabe, C.; Ouyang, W.; Hedskog, C.; McNally, J.; Brainard, D.M.; Doehle, B.; et al. Long-term persistence of HCV NS5A resistance associated substitutions after treatment with the HCV NS5A inhibitor, ledipasvir, without sofosbuvir. Antivir. Ther. 2018, 23, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Akuta, N.; Suzuki, F.; Sezaki, H.; Kobayashi, M.; Fujiyama, S.; Kawamura, Y.; Hosaka, T.; Kobayashi, M.; Saitoh, S.; Suzuki, Y.; et al. Complex Association of Virus- and Host-Related Factors with Hepatocellular Carcinoma Rate following Hepatitis C Virus Clearance. J. Clin. Microbiol. 2019, 57, e01463-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sullivan, J.C.; De Meyer, S.; Bartels, D.J.; Dierynck, I.; Zhang, E.Z.; Spanks, J.; Tigges, A.M.; Ghys, A.; Dorrian, J.; Adda, N.; et al. Evolution of treatment-emergent resistant variants in telaprevir phase 3 clinical trials. Clin. Infect. Dis. 2013, 57, 221–229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akuta, N.; Suzuki, F.; Hirakawa, M.; Kawamura, Y.; Sezaki, H.; Suzuki, Y.; Hosaka, T.; Kobayashi, M.; Kobayashi, M.; Saitoh, S.; et al. Amino acid substitutions in hepatitis C virus core region predict hepatocarcinogenesis following eradication of HCV RNA by antiviral therapy. J. Med. Virol. 2011, 83, 1016–1022. [Google Scholar] [CrossRef] [PubMed]
- Ogata, F.; Akuta, N.; Kobayashi, M.; Fujiyama, S.; Kawamura, Y.; Sezaki, H.; Hosaka, T.; Kobayashi, M.; Saitoh, S.; Suzuki, Y.; et al. Amino acid substitutions in the hepatitis C virus core region predict hepatocarcinogenesis following eradication of HCV RNA by all-oral direct-acting antiviral regimens. J. Med. Virol. 2018, 90, 1087–1093. [Google Scholar] [CrossRef]
- Fourati, S.; Pawlotsky, J.M. Virologic tools for HCV drug resistance testing. Viruses 2015, 7, 6346–6359. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Chen, Q.; Hao, F.; Wan, Z.; Guo, H.; Lu, R.; Mao, L.; Du, H.; Lu, J.; Zhang, C. Subtype distribution of Hepatitis C virus in Jiangsu, China. J. Med. Virol. 2016, 88, 498–505. [Google Scholar] [CrossRef]
- Lu, J.; Feng, Y.; Chen, L.; Zeng, Z.; Liu, X.; Cai, W.; Wang, H.; Guo, X.; Zhou, H.; Tao, W.; et al. Subtype-Specific Prevalence of Hepatitis C Virus NS5A Resistance Associated Substitutions in Mainland China. Front. Microbiol. 2019, 10, 535. [Google Scholar] [CrossRef]
- Hernandez, D.; Zhou, N.; Ueland, J.; Monikowski, A.; McPhee, F. Natural prevalence of NS5A polymorphisms in subjects infected with hepatitis C virus genotype 3 and their effects on the antiviral activity of NS5A inhibitors. J. Clin. Virol. 2013, 57, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Pawlotsky, J.M. Retreatment of Hepatitis C Virus-Infected Patients with Direct-Acting Antiviral Failures. Semin. Liver Dis. 2019. [Google Scholar] [CrossRef] [PubMed]
- Bagaglio, S.; Andolina, A.; Merli, M.; Uberti-Foppa, C.; Morsica, G. Frequency of Natural Resistance within NS5a Replication Complex Domain in Hepatitis C Genotypes 1a, 1b: Possible Implication of Subtype-Specific Resistance Selection in Multiple Direct Acting Antivirals Drugs Combination Treatment. Hagedorn C, ed. Viruses 2016, 8, 91. [Google Scholar] [CrossRef] [PubMed]
- Messina, J.P.; Humphreys, I.; Flaxman, A.; Brown, A.; Cooke, G.; Pybus, O.G.; Barnes, E. Global distribution and prevalence of hepatitis C virus genotypes. Hepatology 2015, 61, 77–87. [Google Scholar] [CrossRef] [Green Version]
- Isaeva, O.V.; Kichatova, V.S.; Karlsen, A.A.; Solonin, S.A.; Dmitriev, P.N.; Kyuregyan, K.K.; Mikhailov, M.I. Multi-year dynamics of spread of hepatitis C virus genotypes in Moscow region. J. Microbiol. Epidemiol. Immunobiol. 2016, 4, 35–42. [Google Scholar]
- Carrasco, I.; Arias, A.; Benítez-Gutiérrez, L. Baseline NS5A resistance associated substitutions may impair DAA response in real-world hepatitis C patients. J. Med. Virol. 2018, 90, 532–536. [Google Scholar] [CrossRef]
- Sun, J.-H.; O’Boyle, D.R.; Zhang, Y.; Wang, C.; Nower, P.; Valera, L.; Roberts, S.; Nettles, R.E.; Fridell, R.A.; Gao, M. Impact of a baseline polymorphism on the emergence of resistance to the hepatitis C virus nonstructural protein 5a replication complex inhibitor, BMS-790052. Hepatology 2012, 55, 1692–1699. [Google Scholar] [CrossRef] [Green Version]
- Recommendations for Testing, Managing, and Treating Hepatitis C|HCV Guidance. Available online: http://hcvguidelines.org/ (accessed on 5 February 2020).
- Yin, C.; Goonawardane, N.; Stewart, H.; Harris, M. A role for domain I of the hepatitis C virus NS5A protein in virus assembly. PLoS Pathog. 2018, 14, e1006834. [Google Scholar] [CrossRef] [Green Version]
- Lim, P.J. Correlation between NS5A dimerization and hepatitis C virus replication. J. Biol. Chem. 2012, 287, 30861–30873. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharya, D.; Ansari, I.H.; Mehle, A.; Striker, R. Fluorescence resonance energy transfer-based intracellular assay for the conformation of hepatitis C virus drug target NS5A. J. Virol. 2012, 86, 8277–8286. [Google Scholar] [CrossRef] [Green Version]
- Love, R.A.; Brodsky, O.; Hickey, M.J.; Wells, P.A.; Cronin, C.N. Crystal structure of a novel dimeric form of NS5A domain I protein from hepatitis C virus. J. Virol. 2009, 83, 4395–4403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, J. Hepatitis C virus nonstructural protein 5A: Biochemical characterization of a novel structural class of RNA-binding proteins. J. Virol. 2010, 84, 12480–12491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartlett, C.; Curd, A.; Peckham, M.; Harris, M. Visualisation and analysis of hepatitis C virus non-structural proteins using super-resolution microscopy. Sci. Rep. 2018, 8, 13604. [Google Scholar] [CrossRef] [PubMed]
- Bilello, J.P.; Lallos, L.B.; McCarville, J.F.; La Colla, M.; Serra, I.; Chapron, C.; Gillum, J.M.; Pierra, C.; Standring, D.N.; Seifer, M. In vitro activity and resistance profile of samatasvir, a novel NS5A replication inhibitor of hepatitis C virus. Antimicrob. Agents Chemother. 2014, 58, 4431–4442. [Google Scholar] [CrossRef] [Green Version]
- Aurora, R.; Donlin, M.J.; Cannon, N.A.; Tavis, J.E. Genome-wide hepatitis C virus amino acid covariance networks can predict response to antiviral therapy in humans. J. Clin. Investig. 2009, 119, 225–236. [Google Scholar] [CrossRef]
- Li, Z.; Chen, Z.W.; Li, H.; Ren, H.; Hu, P. Prevalence of hepatitis C virus-resistant association substitutions to direct-acting antiviral agents in treatment-naïve hepatitis C genotype 1b-infected patients in western China. Infect. Drug Resist. 2017, 10, 377–392. [Google Scholar] [CrossRef] [Green Version]
- Knops, E.; Sierra, S.; Kalaghatgi, P.; Heger, E.; Kaiser, R.; Kalinina, O.V. Epistatic Interactions in NS5A of Hepatitis C Virus Suggest Drug Resistance Mechanisms. Genes 2018, 9, 343. [Google Scholar] [CrossRef] [Green Version]
- Abravanel, F.; Métivier, S.; Chauveau, M.; Péron, J.-M.; Izopet, J. Transmission of HCV NS5A inhibitor-resistant variants among HIV-infected men who have sex with men. Clin. Infect. Dis. 2016, 63, 1271–1272. [Google Scholar] [CrossRef] [Green Version]
- Van De Vijver, D.A.; Wensing, A.M.; Angarano, G.; Asjö, B.; Balotta, C.; Boeri, E.; Camacho, R.; Chaix, M.-L.; Costagliola, D.; De Luca, A.; et al. The calculated genetic barrier for antiretroviral drug resistance substitutions is largely similar for different HIV-1 subtypes. J. Acquir. Immune Defic. Syndr. 2006, 41, 352–360. [Google Scholar] [CrossRef] [Green Version]
- Pascu, M.; Martus, P.; Höhne, M.; Wiedenmann, B.; Hopf, U.; Schreier, E.; Berg, T. Sustained virological response in hepatitis C virus type 1b infected patients is predicted by the number of mutations within the NS5A-ISDR: A meta-analysis focused on geographical differences. Gut 2004, 53, 1345–1351. [Google Scholar] [CrossRef]
- Kichatova, V.S.; Kyuregyan, K.K.; Soboleva, N.V.; Karlsen, A.A.; Isaeva, O.V.; Isaguliants, M.G.; Mikhailov, M.I. Frequency of Interferon-Resistance Conferring Substitutions in Amino Acid Positions 70 and 91 of Core Protein of the Russian HCV 1b Isolates Analyzed in the T-Cell Epitopic Context. J. Immunol. Res. 2018. Article ID 7685371. [Google Scholar] [CrossRef] [PubMed]
- Cuypers, L.; Li, G.; Neumann-Haefelin, C.; Piampongsant, S.; Libin, P.; Van Laethem, K.; Vandamme, A.M.; Theys, K. Mapping the genomic diversity of HCV subtypes 1a and 1b: Implications of structural and immunological constraints for vaccine and drug development. Virus Evol. 2016, 2, vew024. [Google Scholar] [CrossRef] [PubMed]
- Ikram, A.; Obaid, A.; Awan, F.M.; Hanif, R.; Naz, A.; Paracha, R.Z.; Lange, A.; Janjua, H.A. Identification of drug resistance and immune-driven variations in hepatitis C virus (HCV) NS3/4A, NS5A and NS5B regions reveals a new approach toward personalized medicine. Antiviral Res. 2017, 137, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Fytili, P.; Dalekos, G.; Schlaphoff, V.; Suneetha, P.; Sarrazin, C.; Zauner, W.; Zachou, K.; Berg, T.; Manns, M.; Klade, C.; et al. Cross-genotype-reactivity of the immunodominant HCV CD8 T-cell epitope NS3-1073. Vaccine 2008, 26, 3818–3826. [Google Scholar] [CrossRef] [PubMed]
- Merani, S.; Petrovic, D.; James, I.; Chopra, A.; Cooper, N.; Freitas, E.; Rauch, A.; Di Iulio, J.; John, M.; Lucas, M.; et al. Effect of immune pressure on hepatitis C virus evolution: Insights from a single-source outbreak. Hepatology 2011, 53, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Ohno, T.; Mizokami, M. Genotyping by type-specific primers that can type HCV types 1-6. Methods Mol. Med. 1999, 19, 159–164. [Google Scholar] [CrossRef] [PubMed]
- Anisimova, M.; Gascuel, O. Approximate Likelihood-Ratio Test for Branches: A Fast, Accurate, and Powerful Alternative. Syst. Biol. 2006, 55, 539–552. [Google Scholar] [CrossRef]
- Drummond, A.J.; Rambaut, A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol. Biol. 2007, 7, 214. [Google Scholar] [CrossRef] [Green Version]
- Shapiro, B.; Rambaut, A.; Drummond, A.J. Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol. Biol. Evol. 2006, 23, 7–9. [Google Scholar] [CrossRef] [Green Version]
- Shen, W.; Li, Y. A novel algorithm for detecting multiple covariance and clustering of biological sequences. Sci. Rep. 2016, 6, 30425. [Google Scholar] [CrossRef] [Green Version]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. Int. J. Complex. Syst. 2005, 1695, 1–9. [Google Scholar]
- González-Galarza, F.F.; Takeshita, L.Y.; Santos, E.J.; Kempson, F.; Maia, M.H.T.; Da Silva, A.L.S.; E Silva, A.L.T.; Ghattaoraya, G.S.; Alfirevic, A.; Jones, A.R.; et al. Allele frequency net 2015 update: New features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acid Res. 2015, 28, D784–D788. [Google Scholar] [CrossRef] [PubMed]
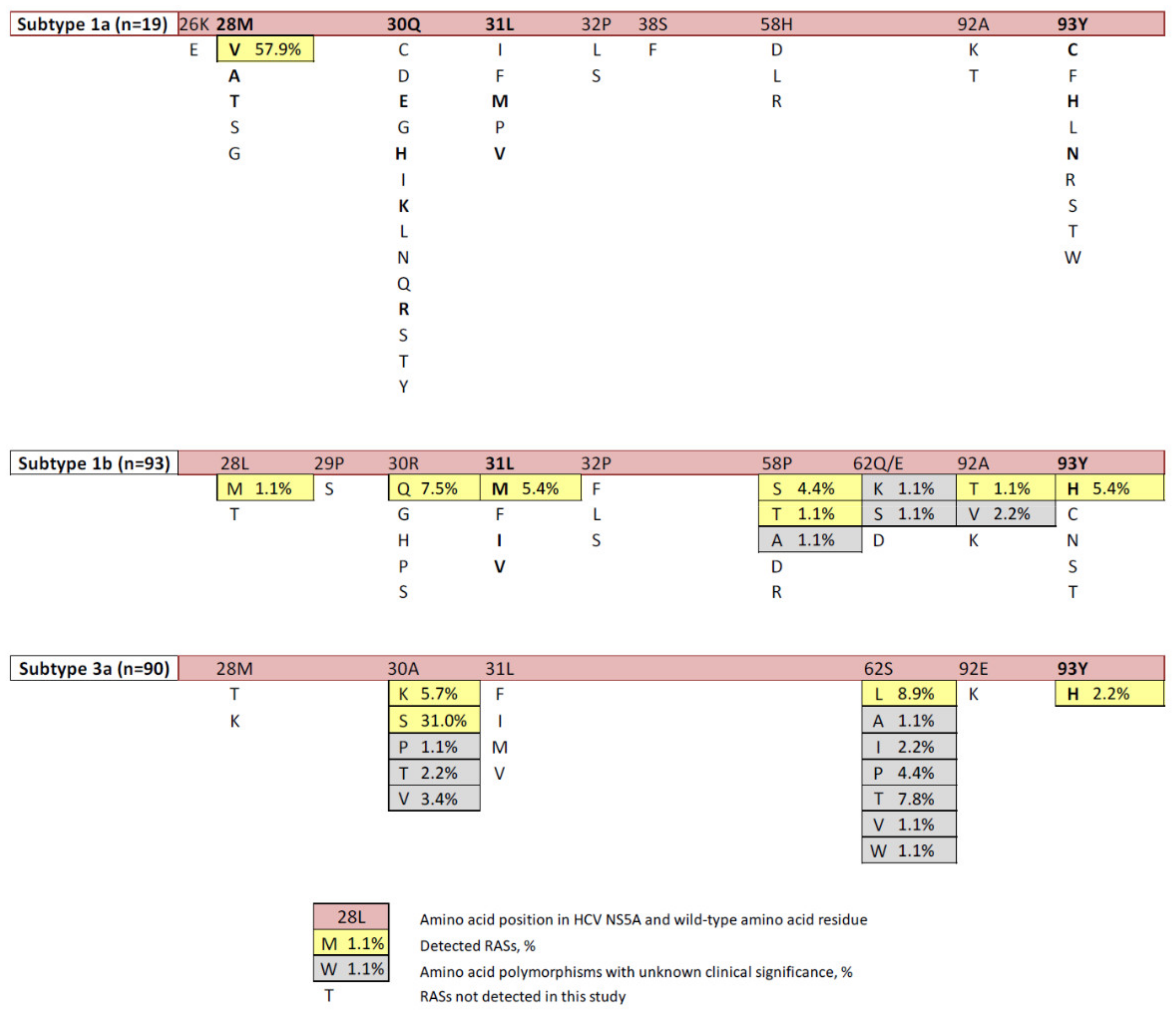
| HCV Genotype (nn of Sequences) | RAS | Nn (%) | Nt Substitution | Genetic Barrier to Resistance ** | |||||
|---|---|---|---|---|---|---|---|---|---|
| Codon, Wild Type (Prevalence, %) | Codon, RAS (Prevalence, %) | Pattern of Substitution | Nt Substitution Type * | ||||||
| 1а (19) | M28V | 11 (57.9%) | ATG | 100% | GTG | 100% | A_ _→G_ _ | Ts | 1 |
| 1b (93) | L28M | 1 (1.1%) | CTG | 85.9% | ATG | 100% | C_ _ →A_ _ | Tv | 2.5 |
| CTA | 8.7% | C_A→A_G | Tv+ Ts | 3.5 | |||||
| R30Q | 7 (7.5%) | CGG | 68.6% | CAA | 14.3% | _GG→ _ AA or _G_→ _ A_ | Ts+ Ts or Ts | 2 or 1 | |
| CGA | 25.6% | _G_→ _ A_ or _GA→ _ AG | Ts or Ts+ Ts | 1 or 2 | |||||
| CGC | 2.3% | CAG | 85.7% | _GC→_AA or _GC→_AG | Ts+Tv | 3.5 | |||
| CGT | 1.2% | _GT→_AA or _GT→_AG | Ts+Tv | 3.5 | |||||
| L31M | 5 (5.4%) | TTG | 37.5% | ATG | 100% | T_ _ →A_ _ | Tv | 2.5 | |
| TTA | 35.2% | T_ A →A_ G | Tv+ Ts | 3.5 | |||||
| CTA | 10.2% | C_A→A_G | Tv + Ts | 3.5 | |||||
| CTG | 6.8% | C_ _ →A_ _ | Tv | 2.5 | |||||
| P58S | 4 (4.4%) | CCA | 84.0% | TCA | 100% | C_ _→T_ _ | Ts | 1 | |
| CCG | 9.2% | C_G→T_ A | Ts+ Ts | 2 | |||||
| CCC | 1.1% | C_C→T_ A | Ts+Tv | 3.5 | |||||
| P58T | 1 (1.1%) | CCA | 84.0% | ACA | 100% | C_ _→ A_ _ | Tv | 2.5 | |
| CCG | 9.2% | C_G→A_A | Tv+ Ts | 3.5 | |||||
| CCC | 1.1% | C_C→A_A | Tv+ Tv | 5 | |||||
| A92T | 1 (1.1%) | GCG | 60.0% | ACG | 100% | G_ _→A_ _ | Ts | 1 | |
| GCA | 32.2% | G_A→A_G | Ts+ Ts | 2 | |||||
| GCC | 2.2% | G_C→A_G | Ts+Tv | 3.5 | |||||
| GCT | 1.1% | G_T→A_G | Ts+Tv | 3.5 | |||||
| Y93H | 5 (5.4%) | TAC | 93.2% | CAC | 100% | T_ _→C_ _ | Ts | 1 | |
| TAT | 4.5% | T_T →C_C | Ts+ Ts | 2 | |||||
| 3a (90) | А30К | 5 (5.7%) | GCG | 92.2% | AAG | 100% | GC_→AA_ | Ts+Tv | 3.5 |
| GCA | 5.9% | GCA→AAG | Ts+Tv+ Ts | 4.5 | |||||
| GCC | 1.9% | GCC→AAG | Ts+Tv+Tv | 6 | |||||
| A30S | 26 (31.0%) | GCG | 92.2% | TCG | 100% | G_ _→T_ _ | Tv | 2.5 | |
| GCA | 5.9% | G_A→T_G | Tv + Ts | 3.5 | |||||
| GCC | 1.9% | G_C→T_G | Tv+ Tv | 5 | |||||
| S62L | 8 (8.9%) | TCA | TTA | 75% | _C_→_T_ | Ts | 1 | ||
| 100% | TTG | 12.5% | _CA→_TG | Ts+ Ts | 2 | ||||
| CTA | 12.5% | TC_→CT_ | Ts+ Ts | 2 | |||||
| Y93H | 2 (2.2%) | TAC | 100% | CAC | 100% | T_ _→C_ _ | Ts | 1 | |
| HCV1a | HCV1b | HCV3a | |||
|---|---|---|---|---|---|
| RAS | Wild Type | RAS | Wild Type | RAS | Wild Type |
| 28V-78R | 28M-78K | 30Q-315V | 30R-315I | 30S-62S | 30A-62L |
| 28V-308L | 28M-308R | ||||
| 28V-372L | 28M-372V | ||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kyuregyan, K.K.; Kichatova, V.S.; Karlsen, A.A.; Isaeva, O.V.; Solonin, S.A.; Petkov, S.; Nielsen, M.; Isaguliants, M.G.; Mikhailov, M.I. Factors Influencing the Prevalence of Resistance-Associated Substitutions in NS5A Protein in Treatment-Naive Patients with Chronic Hepatitis C. Biomedicines 2020, 8, 80. https://doi.org/10.3390/biomedicines8040080
Kyuregyan KK, Kichatova VS, Karlsen AA, Isaeva OV, Solonin SA, Petkov S, Nielsen M, Isaguliants MG, Mikhailov MI. Factors Influencing the Prevalence of Resistance-Associated Substitutions in NS5A Protein in Treatment-Naive Patients with Chronic Hepatitis C. Biomedicines. 2020; 8(4):80. https://doi.org/10.3390/biomedicines8040080
Chicago/Turabian StyleKyuregyan, Karen K., Vera S. Kichatova, Anastasiya A. Karlsen, Olga V. Isaeva, Sergei A. Solonin, Stefan Petkov, Morten Nielsen, Maria G. Isaguliants, and Mikhail I. Mikhailov. 2020. "Factors Influencing the Prevalence of Resistance-Associated Substitutions in NS5A Protein in Treatment-Naive Patients with Chronic Hepatitis C" Biomedicines 8, no. 4: 80. https://doi.org/10.3390/biomedicines8040080





