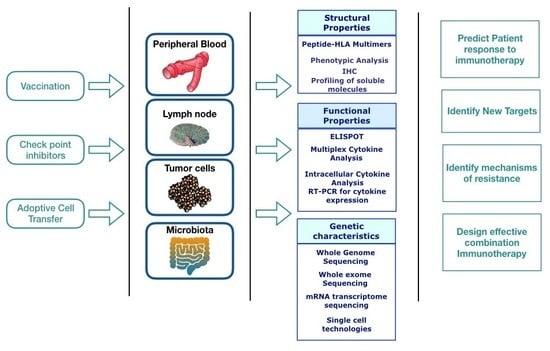
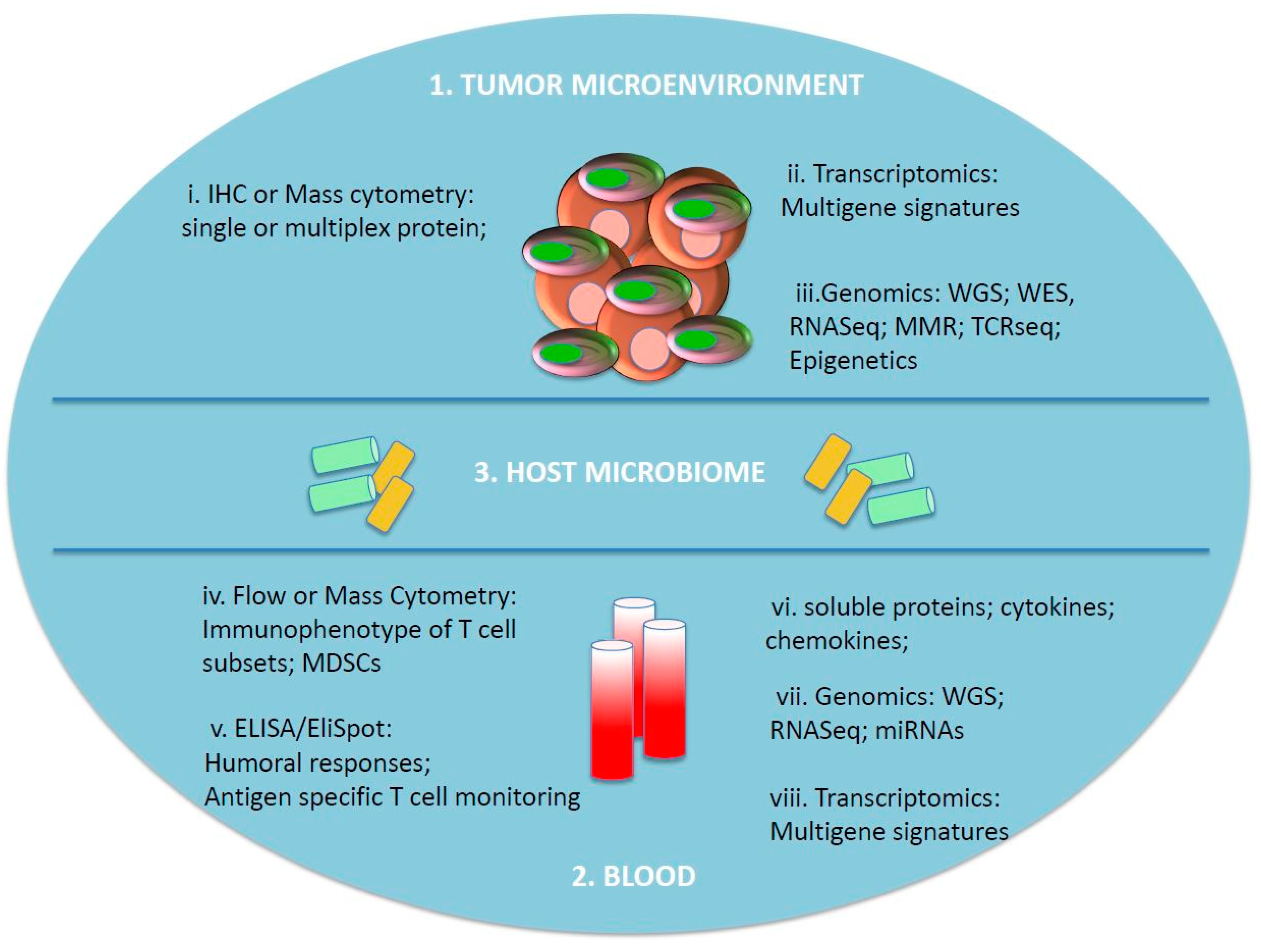
Immune Profiling of Cancer Patients Treated with Immunotherapy: Advances and Challenges
Abstract
1. Introduction
2. Immunomonitoring of Circulating Immune Cells
3. Serum Biomarkers
4. Tumor- and TME-Associated Biomarkers
5. Host Microbiome
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Schreiber, R.D.; Old, L.J.; Smyth, M.J. Cancer immunoediting: Integrating immunity’s roles in cancer suppression and promotion. Science 2011, 331, 1565–1570. [Google Scholar] [CrossRef] [PubMed]
- Koebel, C.M.; Vermi, W.; Swann, J.B.; Zerafa, N.; Rodig, S.J.; Old, L.J.; Smyth, M.J.; Schreiber, R.D. Adaptive immunity maintains occult cancer in an equilibrium state. Nature 2007, 450, 903–907. [Google Scholar] [CrossRef] [PubMed]
- Blankenstein, T. The role of tumor stroma in the interaction between tumor and immune system. Curr. Opin. Immunol. 2005, 17, 180–186. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Fridman, W.H.; Galon, J. The prognostic impact of anti-cancer immune response: A novel classification of cancer patients. Semin. Immunopathol. 2011, 33, 335–340. [Google Scholar] [CrossRef] [PubMed]
- Rotte, A.; Jin, J.Y.; Lemaire, V. Mechanistic overview of immune checkpoints to support the rational design of their combinations in cancer immunotherapy. Ann. Oncol. 2018, 29, 71–83. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.R. PD-1-targeted immunotherapy: Recent clinical findings. Clin. Adv. Hematol. Oncol. 2012, 10, 674–675. [Google Scholar] [PubMed]
- Topalian, S.L.; Hodi, F.S.; Brahmer, J.R.; Gettinger, S.N.; Smith, D.C.; McDermott, D.F.; Powderly, J.D.; Carvajal, R.D.; Sosman, J.A.; Atkins, M.B.; et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N. Engl. J. Med. 2012, 366, 2443–2454. [Google Scholar] [CrossRef] [PubMed]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.L.; Smothers, J.; Srinivasan, R.; Hoos, A. Big opportunities for small molecules in immuno-oncology. Nat. Rev. Drug Discov. 2015, 14, 603–622. [Google Scholar] [CrossRef] [PubMed]
- Wargo, J.A.; Reuben, A.; Cooper, Z.A.; Oh, K.S.; Sullivan, R.J. Immune Effects of Chemotherapy, Radiation, and Targeted Therapy and Opportunities for Combination With Immunotherapy. Semin. Oncol. 2015, 42, 601–616. [Google Scholar] [CrossRef] [PubMed]
- Iafolla, M.A.J.; Selby, H.; Warner, K.; Ohashi, P.S.; Haibe-Kains, B.; Siu, L.L. Rational design and identification of immuno-oncology drug combinations. Eur. J. Cancer 2018, 95, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Attia, P.; Phan, G.Q.; Maker, A.V.; Robinson, M.R.; Quezado, M.M.; Yang, J.C.; Sherry, R.M.; Topalian, S.L.; Kammula, U.S.; Royal, R.E.; et al. Autoimmunity correlates with tumor regression in patients with metastatic melanoma treated with anti-cytotoxic T-lymphocyte antigen-4. J. Clin. Oncol. 2005, 23, 6043–6053. [Google Scholar] [CrossRef] [PubMed]
- Delyon, J.; Mateus, C.; Lefeuvre, D.; Lanoy, E.; Zitvogel, L.; Chaput, N.; Roy, S.; Eggermont, A.M.; Routier, E.; Robert, C. Experience in daily practice with ipilimumab for the treatment of patients with metastatic melanoma: An early increase in lymphocyte and eosinophil counts is associated with improved survival. Ann. Oncol. 2013, 24, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, A.M.; Ascierto, P.A.; Queirolo, P.; Pilla, L.; Ridolfi, R.; Santinami, M.; Testori, A.; Simeone, E.; Guidoboni, M.; Maurichi, A.; et al. Three-year follow-up of advanced melanoma patients who received ipilimumab plus fotemustine in the Italian Network for Tumor Biotherapy (NIBIT)-M1 phase II study. Ann. Oncol. 2015, 26, 798–803. [Google Scholar] [CrossRef] [PubMed]
- Ferrucci, P.F.; Gandini, S.; Battaglia, A.; Alfieri, S.; Di Giacomo, A.M.; Giannarelli, D.; Cappellini, G.C.; De Galitiis, F.; Marchetti, P.; Amato, G.; et al. Baseline neutrophil-to-lymphocyte ratio is associated with outcome of ipilimumab-treated metastatic melanoma patients. Br. J. Cancer 2015, 112, 1904–1910. [Google Scholar] [CrossRef] [PubMed]
- Phan, G.Q.; Yang, J.C.; Sherry, R.M.; Hwu, P.; Topalian, S.L.; Schwartzentruber, D.J.; Restifo, N.P.; Haworth, L.R.; Seipp, C.A.; Freezer, L.J.; et al. Cancer regression and autoimmunity induced by cytotoxic T lymphocyte-associated antigen 4 blockade in patients with metastatic melanoma. Proc. Natl. Acad. Sci. USA 2003, 100, 8372–8377. [Google Scholar] [CrossRef] [PubMed]
- Maker, A.V.; Attia, P.; Rosenberg, S.A. Analysis of the cellular mechanism of antitumor responses and autoimmunity in patients treated with CTLA-4 blockade. J. Immunol. 2005, 175, 7746–7754. [Google Scholar] [CrossRef] [PubMed]
- Maker, A.V.; Yang, J.C.; Sherry, R.M.; Topalian, S.L.; Kammula, U.S.; Royal, R.E.; Hughes, M.; Yellin, M.J.; Haworth, L.R.; Levy, C.; et al. Intrapatient dose escalation of anti-CTLA-4 antibody in patients with metastatic melanoma. J. Immunother. 2006, 29, 455–463. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.S.; Kahler, K.C.; Hauschild, A. Management of immune-related adverse events and kinetics of response with ipilimumab. J. Clin. Oncol. 2012, 30, 2691–2697. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, A.M.; Ascierto, P.A.; Pilla, L.; Santinami, M.; Ferrucci, P.F.; Giannarelli, D.; Marasco, A.; Rivoltini, L.; Simeone, E.; Nicoletti, S.V.; et al. Ipilimumab and fotemustine in patients with advanced melanoma (NIBIT-M1): An open-label, single-arm phase 2 trial. Lancet Oncol. 2012, 13, 879–886. [Google Scholar] [CrossRef]
- Maccalli, C.; Giannarelli, D.; Capocefalo, F.; Pilla, L.; Fonsatti, E.; Di Giacomo, A.M.; Parmiani, G.; Maio, M. Immunological markers and clinical outcome of advanced melanoma patients receiving ipilimumab plus fotemustine in the NIBIT-M1 study. Oncoimmunology 2016, 5, e1071007. [Google Scholar] [CrossRef] [PubMed]
- Calabro, L.; Morra, A.; Fonsatti, E.; Cutaia, O.; Fazio, C.; Annesi, D.; Lenoci, M.; Amato, G.; Danielli, R.; Altomonte, M.; et al. Efficacy and safety of an intensified schedule of tremelimumab for chemotherapy-resistant malignant mesothelioma: An open-label, single-arm, phase 2 study. Lancet Respir. Med. 2015, 3, 301–309. [Google Scholar] [CrossRef]
- Liakou, C.I.; Kamat, A.; Tang, D.N.; Chen, H.; Sun, J.; Troncoso, P.; Logothetis, C.; Sharma, P. CTLA-4 blockade increases IFNgamma-producing CD4+ICOShi cells to shift the ratio of effector to regulatory T cells in cancer patients. Proc. Natl. Acad. Sci. USA 2008, 105, 14987–14992. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Wagner, K.; Wolchok, J.D.; Allison, J.P. Novel cancer immunotherapy agents with survival benefit: Recent successes and next steps. Nat. Rev. Cancer 2011, 11, 805–812. [Google Scholar] [CrossRef] [PubMed]
- Vonderheide, R.H.; LoRusso, P.M.; Khalil, M.; Gartner, E.M.; Khaira, D.; Soulieres, D.; Dorazio, P.; Trosko, J.A.; Ruter, J.; Mariani, G.L.; et al. Tremelimumab in combination with exemestane in patients with advanced breast cancer and treatment-associated modulation of inducible costimulator expression on patient T cells. Clin. Cancer Res. 2010, 16, 3485–3494. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Fu, T.; Suh, W.K.; Tsavachidou, D.; Wen, S.; Gao, J.; Ng Tang, D.; He, Q.; Sun, J.; Sharma, P. CD4 T cells require ICOS-mediated PI3K signaling to increase T-Bet expression in the setting of anti-CTLA-4 therapy. Cancer Immunol. Res. 2014, 2, 167–176. [Google Scholar] [CrossRef] [PubMed]
- Menard, C.; Ghiringhelli, F.; Roux, S.; Chaput, N.; Mateus, C.; Grohmann, U.; Caillat-Zucman, S.; Zitvogel, L.; Robert, C. Ctla-4 blockade confers lymphocyte resistance to regulatory T-cells in advanced melanoma: Surrogate marker of efficacy of tremelimumab? Clin. Cancer Res. 2008, 14, 5242–5249. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Cagnon, L.; Costa-Nunes, C.M.; Baumgaertner, P.; Montandon, N.; Leyvraz, L.; Michielin, O.; Romano, E.; Speiser, D.E. Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol. Immunother. 2014, 63, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Pico de Coana, Y.; Masucci, G.; Hansson, J.; Kiessling, R. Myeloid-derived suppressor cells and their role in CTLA-4 blockade therapy. Cancer Immunol. Immunother. 2014, 63, 977–983. [Google Scholar] [CrossRef] [PubMed]
- Simeone, E.; Gentilcore, G.; Giannarelli, D.; Grimaldi, A.M.; Caraco, C.; Curvietto, M.; Esposito, A.; Paone, M.; Palla, M.; Cavalcanti, E.; et al. Immunological and biological changes during ipilimumab treatment and their potential correlation with clinical response and survival in patients with advanced melanoma. Cancer Immunol. Immunother. 2014, 63, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Tarhini, A.A.; Edington, H.; Butterfield, L.H.; Lin, Y.; Shuai, Y.; Tawbi, H.; Sander, C.; Yin, Y.; Holtzman, M.; Johnson, J.; et al. Immune monitoring of the circulation and the tumor microenvironment in patients with regionally advanced melanoma receiving neoadjuvant ipilimumab. PLoS ONE 2014, 9, e87705. [Google Scholar] [CrossRef] [PubMed]
- Weide, B.; Martens, A.; Zelba, H.; Stutz, C.; Derhovanessian, E.; Di Giacomo, A.M.; Maio, M.; Sucker, A.; Schilling, B.; Schadendorf, D.; et al. Myeloid-derived suppressor cells predict survival of patients with advanced melanoma: Comparison with regulatory T cells and NY-ESO-1- or melan-A-specific T cells. Clin. Cancer Res. 2014, 20, 1601–1609. [Google Scholar] [CrossRef] [PubMed]
- Martens, A.; Wistuba-Hamprecht, K.; Geukes Foppen, M.; Yuan, J.; Postow, M.A.; Wong, P.; Romano, E.; Khammari, A.; Dreno, B.; Capone, M.; et al. Baseline Peripheral Blood Biomarkers Associated with Clinical Outcome of Advanced Melanoma Patients Treated with Ipilimumab. Clin. Cancer Res. 2016, 22, 2908–2918. [Google Scholar] [CrossRef] [PubMed]
- Parmiani, G.; Russo, V.; Maccalli, C.; Parolini, D.; Rizzo, N.; Maio, M. Peptide-based vaccines for cancer therapy. Hum. Vaccines Immunother. 2014, 10, 3175–3178. [Google Scholar] [CrossRef] [PubMed]
- Butterfield, L.H.; Buffo, M.J. Immunologic monitoring of cancer vaccine trials using the ELISPOT assay. Methods Mol. Biol. 2014, 1102, 71–82. [Google Scholar] [CrossRef] [PubMed]
- Parmiani, G.; Pilla, L.; Corti, A.; Doglioni, C.; Cimminiello, C.; Bellone, M.; Parolini, D.; Russo, V.; Capocefalo, F.; Maccalli, C. A pilot Phase I study combining peptide-based vaccination and NGR-hTNF vessel targeting therapy in metastatic melanoma. Oncoimmunology 2014, 3, e963406. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.S.; Hamid, O.; Chasalow, S.D.; Wu, D.Y.; Parker, S.M.; Galbraith, S.; Gnjatic, S.; Berman, D. Ipilimumab increases activated T cells and enhances humoral immunity in patients with advanced melanoma. J. Immunother. 2012, 35, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Adamow, M.; Ginsberg, B.A.; Rasalan, T.S.; Ritter, E.; Gallardo, H.F.; Xu, Y.; Pogoriler, E.; Terzulli, S.L.; Kuk, D.; et al. Integrated NY-ESO-1 antibody and CD8+ T-cell responses correlate with clinical benefit in advanced melanoma patients treated with ipilimumab. Proc. Natl. Acad. Sci. USA 2011, 108, 16723–16728. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Ginsberg, B.; Page, D.; Li, Y.; Rasalan, T.; Gallardo, H.F.; Xu, Y.; Adams, S.; Bhardwaj, N.; Busam, K.; et al. CTLA-4 blockade increases antigen-specific CD8(+) T cells in prevaccinated patients with melanoma: Three cases. Cancer Immunol. Immunother. 2011, 60, 1137–1146. [Google Scholar] [CrossRef] [PubMed]
- Kirkwood, J.M.; Butterfield, L.H.; Tarhini, A.A.; Zarour, H.; Kalinski, P.; Ferrone, S. Immunotherapy of cancer in 2012. CA Cancer J. Clin. 2012, 62, 309–335. [Google Scholar] [CrossRef] [PubMed]
- Hannani, D.; Vetizou, M.; Enot, D.; Rusakiewicz, S.; Chaput, N.; Klatzmann, D.; Desbois, M.; Jacquelot, N.; Vimond, N.; Chouaib, S.; et al. Anticancer immunotherapy by CTLA-4 blockade: Obligatory contribution of IL-2 receptors and negative prognostic impact of soluble CD25. Cell Res. 2015, 25, 208–224. [Google Scholar] [CrossRef] [PubMed]
- Maccalli, C.; Scaramuzza, S.; Parmiani, G. TNK cells (NKG2D+ CD8+ or CD4+ T lymphocytes) in the control of human tumors. Cancer Immunol. Immunother. 2009, 58, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Paschen, A.; Baingo, J.; Schadendorf, D. Expression of stress ligands of the immunoreceptor NKG2D in melanoma: Regulation and clinical significance. Eur. J. Cell Biol. 2014, 93, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Baragano Raneros, A.; Suarez-Alvarez, B.; Lopez-Larrea, C. Secretory pathways generating immunosuppressive NKG2D ligands: New targets for therapeutic intervention. Oncoimmunology 2014, 3, e28497. [Google Scholar] [CrossRef] [PubMed]
- Jinushi, M.; Hodi, F.S.; Dranoff, G. Therapy-induced antibodies to MHC class I chain-related protein A antagonize immune suppression and stimulate antitumor cytotoxicity. Proc. Natl. Acad. Sci. USA 2006, 103, 9190–9195. [Google Scholar] [CrossRef] [PubMed]
- Maccalli, C.; Giannarelli, D.; Chiarucci, C.; Cutaia, O.; Giacobini, G.; Hendrickx, W.; Amato, G.; Annesi, D.; Bedognetti, D.; Altomonte, M.; et al. Soluble NKG2D ligands are biomarkers associated with the clinical outcome to immune checkpoint blockade therapy of metastatic melanoma patients. Oncoimmunology 2017, 6, e1323618. [Google Scholar] [CrossRef] [PubMed]
- Blay, J.Y.; Negrier, S.; Combaret, V.; Attali, S.; Goillot, E.; Merrouche, Y.; Mercatello, A.; Ravault, A.; Tourani, J.M.; Moskovtchenko, J.F.; et al. Serum level of interleukin 6 as a prognosis factor in metastatic renal cell carcinoma. Cancer Res. 1992, 52, 3317–3322. [Google Scholar] [PubMed]
- Tartour, E.; Blay, J.Y.; Dorval, T.; Escudier, B.; Mosseri, V.; Douillard, J.Y.; Deneux, L.; Gorin, I.; Negrier, S.; Mathiot, C.; et al. Predictors of clinical response to interleukin-2—Based immunotherapy in melanoma patients: A French multiinstitutional study. J. Clin. Oncol. 1996, 14, 1697–1703. [Google Scholar] [CrossRef] [PubMed]
- Sabatino, M.; Kim-Schulze, S.; Panelli, M.C.; Stroncek, D.; Wang, E.; Taback, B.; Kim, D.W.; Deraffele, G.; Pos, Z.; Marincola, F.M.; et al. Serum vascular endothelial growth factor and fibronectin predict clinical response to high-dose interleukin-2 therapy. J. Clin. Oncol. 2009, 27, 2645–2652. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Hegde, P.S.; Clynes, R.; Foukas, P.G.; Harari, A.; Kleen, T.O.; Kvistborg, P.; Maccalli, C.; Maecker, H.T.; Page, D.B.; et al. Novel technologies and emerging biomarkers for personalized cancer immunotherapy. J. Immunother. Cancer 2016, 4, 3. [Google Scholar] [CrossRef] [PubMed]
- Kelderman, S.; Heemskerk, B.; van Tinteren, H.; van den Brom, R.R.; Hospers, G.A.; van den Eertwegh, A.J.; Kapiteijn, E.W.; de Groot, J.W.; Soetekouw, P.; Jansen, R.L.; et al. Lactate dehydrogenase as a selection criterion for ipilimumab treatment in metastatic melanoma. Cancer Immunol. Immunother. 2014, 63, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Costes, A.; Sanchez-Cabo, F.; Kirilovsky, A.; Mlecnik, B.; Lagorce-Pages, C.; Tosolini, M.; Camus, M.; Berger, A.; Wind, P.; et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006, 313, 1960–1964. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Tosolini, M.; Kirilovsky, A.; Berger, A.; Bindea, G.; Meatchi, T.; Bruneval, P.; Trajanoski, Z.; Fridman, W.H.; Pages, F.; et al. Histopathologic-based prognostic factors of colorectal cancers are associated with the state of the local immune reaction. J. Clin. Oncol. 2011, 29, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Galon, J.; Angell, H.K.; Bedognetti, D.; Marincola, F.M. The continuum of cancer immunosurveillance: prognostic, predictive, and mechanistic signatures. Immunity 2013, 39, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Camus, M.; Tosolini, M.; Mlecnik, B.; Pages, F.; Kirilovsky, A.; Berger, A.; Costes, A.; Bindea, G.; Charoentong, P.; Bruneval, P.; et al. Coordination of intratumoral immune reaction and human colorectal cancer recurrence. Cancer Res. 2009, 69, 2685–2693. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Bindea, G.; Angell, H.K.; Maby, P.; Angelova, M.; Tougeron, D.; Church, S.E.; Lafontaine, L.; Fischer, M.; Fredriksen, T.; et al. Integrative Analyses of Colorectal Cancer Show Immunoscore Is a Stronger Predictor of Patient Survival Than Microsatellite Instability. Immunity 2016, 44, 698–711. [Google Scholar] [CrossRef] [PubMed]
- Mlecnik, B.; Van den Eynde, M.; Bindea, G.; Church, S.E.; Vasaturo, A.; Fredriksen, T.; Lafontaine, L.; Haicheur, N.; Marliot, F.; Debetancourt, D.; et al. Comprehensive Intrametastatic Immune Quantification and Major Impact of Immunoscore on Survival. J. Natl. Cancer Inst. 2018, 110. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Chen, X. Immunoregulatory role of B7-H1 in chronicity of inflammatory responses. Cell. Mol. Immunol. 2006, 3, 179–187. [Google Scholar] [PubMed]
- Hirsch, F.R.; McElhinny, A.; Stanforth, D.; Ranger-Moore, J.; Jansson, M.; Kulangara, K.; Richardson, W.; Towne, P.; Hanks, D.; Vennapusa, B.; et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J. Thorac. Oncol. 2017, 12, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.R.; Tykodi, S.S.; Chow, L.Q.; Hwu, W.J.; Topalian, S.L.; Hwu, P.; Drake, C.G.; Camacho, L.H.; Kauh, J.; Odunsi, K.; et al. Safety and activity of anti-PD-L1 antibody in patients with advanced cancer. N. Engl. J. Med. 2012, 366, 2455–2465. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B. Current Perspectives in Immunotherapy for Non-Small Cell Lung Cancer. Semin. Oncol. 2015, 42 (Suppl. 2), S11–S18. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodriguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csoszi, T.; Fulop, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Reik, W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 2007, 447, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.R.; Hammers, H.; Lipson, E.J. Nivolumab: Targeting PD-1 to bolster antitumor immunity. Future Oncol. 2015, 11, 1307–1326. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; von Minckwitz, G.; Brase, J.C.; Sinn, B.V.; Gade, S.; Kronenwett, R.; Pfitzner, B.M.; Salat, C.; Loi, S.; Schmitt, W.D.; et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. 2015, 33, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Issa-Nummer, Y.; Darb-Esfahani, S.; Loibl, S.; Kunz, G.; Nekljudova, V.; Schrader, I.; Sinn, B.V.; Ulmer, H.U.; Kronenwett, R.; Just, M.; et al. Prospective validation of immunological infiltrate for prediction of response to neoadjuvant chemotherapy in HER2-negative breast cancer--a substudy of the neoadjuvant GeparQuinto trial. PLoS ONE 2013, 8, e79775. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, R.; Tanaka, M.; Yano, A.; Tse, G.M.; Yamaguchi, M.; Koura, K.; Kanomata, N.; Kawaguchi, A.; Akiba, J.; Naito, Y.; et al. Tumor-infiltrating lymphocytes are important pathologic predictors for neoadjuvant chemotherapy in patients with breast cancer. Hum. Pathol. 2012, 43, 1688–1694. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.; Chen, J.Q.; Liu, C.; Malu, S.; Creasy, C.; Tetzlaff, M.T.; Xu, C.; McKenzie, J.A.; Zhang, C.; Liang, X.; et al. Loss of PTEN Promotes Resistance to T Cell-Mediated Immunotherapy. Cancer Discov. 2016, 6, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Tran, E.; Ahmadzadeh, M.; Lu, Y.C.; Gros, A.; Turcotte, S.; Robbins, P.F.; Gartner, J.J.; Zheng, Z.; Li, Y.F.; Ray, S.; et al. Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 2015, 350, 1387–1390. [Google Scholar] [CrossRef] [PubMed]
- Kelderman, S.; Schumacher, T.N.; Kvistborg, P. Mismatch Repair-Deficient Cancers Are Targets for Anti-PD-1 Therapy. Cancer Cell 2015, 28, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Kelderman, S.; Kvistborg, P. Tumor antigens in human cancer control. Biochim. Biophys. Acta 2016, 1865, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Kvistborg, P.; Philips, D.; Kelderman, S.; Hageman, L.; Ottensmeier, C.; Joseph-Pietras, D.; Welters, M.J.; van der Burg, S.; Kapiteijn, E.; Michielin, O.; et al. Anti-CTLA-4 therapy broadens the melanoma-reactive CD8+ T cell response. Sci. Transl. Med. 2014, 6, 254ra128. [Google Scholar] [CrossRef] [PubMed]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Mennonna, D.; Maccalli, C.; Romano, M.C.; Garavaglia, C.; Capocefalo, F.; Bordoni, R.; Severgnini, M.; De Bellis, G.; Sidney, J.; Sette, A.; et al. T cell neoepitope discovery in colorectal cancer by high throughput profiling of somatic mutations in expressed genes. Gut 2015, 66, 454–463. [Google Scholar] [CrossRef] [PubMed]
- Trajanoski, Z.; Maccalli, C.; Mennonna, D.; Casorati, G.; Parmiani, G.; Dellabona, P. Somatically mutated tumor antigens in the quest for a more efficacious patient-oriented immunotherapy of cancer. Cancer Immunol. Immunother. 2015, 64, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Hussein, Y.R.; Weigelt, B.; Levine, D.A.; Schoolmeester, J.K.; Dao, L.N.; Balzer, B.L.; Liles, G.; Karlan, B.; Kobel, M.; Lee, C.H.; et al. Clinicopathological analysis of endometrial carcinomas harboring somatic POLE exonuclease domain mutations. Mod. Pathol. 2015, 28, 505–514. [Google Scholar] [CrossRef] [PubMed]
- Mehnert, J.M.; Panda, A.; Zhong, H.; Hirshfield, K.; Damare, S.; Lane, K.; Sokol, L.; Stein, M.N.; Rodriguez-Rodriquez, L.; Kaufman, H.L.; et al. Immune activation and response to pembrolizumab in POLE-mutant endometrial cancer. J. Clin. Investig. 2016, 126, 2334–2340. [Google Scholar] [CrossRef] [PubMed]
- Roh, W.; Chen, P.L.; Reuben, A.; Spencer, C.N.; Prieto, P.A.; Miller, J.P.; Gopalakrishnan, V.; Wang, F.; Cooper, Z.A.; Reddy, S.M.; et al. Integrated molecular analysis of tumor biopsies on sequential CTLA-4 and PD-1 blockade reveals markers of response and resistance. Sci. Transl. Med. 2017, 9, eaah3560. [Google Scholar] [CrossRef] [PubMed]
- Simoni, Y.; Becht, E.; Fehlings, M.; Loh, C.Y.; Koo, S.L.; Teng, K.W.W.; Yeong, J.P.S.; Nahar, R.; Zhang, T.; Kared, H.; et al. Bystander CD8(+) T cells are abundant and phenotypically distinct in human tumour infiltrates. Nature 2018, 557, 575–579. [Google Scholar] [CrossRef] [PubMed]
- Behbehani, G.K.; Samusik, N.; Bjornson, Z.B.; Fantl, W.J.; Medeiros, B.C.; Nolan, G.P. Mass Cytometric Functional Profiling of Acute Myeloid Leukemia Defines Cell-Cycle and Immunophenotypic Properties That Correlate with Known Responses to Therapy. Cancer Discov. 2015, 5, 988–1003. [Google Scholar] [CrossRef] [PubMed]
- Bendall, S.C.; Nolan, G.P. From single cells to deep phenotypes in cancer. Nat. Biotechnol. 2012, 30, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Han, A.; Glanville, J.; Hansmann, L.; Davis, M.M. Linking T-cell receptor sequence to functional phenotype at the single-cell level. Nat. Biotechnol. 2014, 32, 684–692. [Google Scholar] [CrossRef] [PubMed]
- Shalek, A.K.; Satija, R.; Adiconis, X.; Gertner, R.S.; Gaublomme, J.T.; Raychowdhury, R.; Schwartz, S.; Yosef, N.; Malboeuf, C.; Lu, D.; et al. Single-cell transcriptomics reveals bimodality in expression and splicing in immune cells. Nature 2013, 498, 236–240. [Google Scholar] [CrossRef] [PubMed]
- Hamid, O.; Schmidt, H.; Nissan, A.; Ridolfi, L.; Aamdal, S.; Hansson, J.; Guida, M.; Hyams, D.M.; Gomez, H.; Bastholt, L.; et al. A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J. Transl. Med. 2011, 9, 204. [Google Scholar] [CrossRef] [PubMed]
- Flower, K.J.; Ghaem-Maghami, S.; Brown, R. Is there a Role for Epigenetic Enhancement of Immunomodulatory Approaches to Cancer Treatment? Curr. Cancer Drug Targets 2018, 18, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Paulos, C.M.; Kaiser, A.; Wrzesinski, C.; Hinrichs, C.S.; Cassard, L.; Boni, A.; Muranski, P.; Sanchez-Perez, L.; Palmer, D.C.; Yu, Z.; et al. Toll-like receptors in tumor immunotherapy. Clin. Cancer Res. 2007, 13, 5280–5289. [Google Scholar] [CrossRef] [PubMed]
- Berman, D.; Parker, S.M.; Siegel, J.; Chasalow, S.D.; Weber, J.; Galbraith, S.; Targan, S.R.; Wang, H.L. Blockade of cytotoxic T-lymphocyte antigen-4 by ipilimumab results in dysregulation of gastrointestinal immunity in patients with advanced melanoma. Cancer Immunol. 2010, 10, 11. [Google Scholar]
- Vetizou, M.; Pitt, J.M.; Daillere, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pilla, L.; Maccalli, C. Immune Profiling of Cancer Patients Treated with Immunotherapy: Advances and Challenges. Biomedicines 2018, 6, 76. https://doi.org/10.3390/biomedicines6030076
Pilla L, Maccalli C. Immune Profiling of Cancer Patients Treated with Immunotherapy: Advances and Challenges. Biomedicines. 2018; 6(3):76. https://doi.org/10.3390/biomedicines6030076
Chicago/Turabian StylePilla, Lorenzo, and Cristina Maccalli. 2018. "Immune Profiling of Cancer Patients Treated with Immunotherapy: Advances and Challenges" Biomedicines 6, no. 3: 76. https://doi.org/10.3390/biomedicines6030076
APA StylePilla, L., & Maccalli, C. (2018). Immune Profiling of Cancer Patients Treated with Immunotherapy: Advances and Challenges. Biomedicines, 6(3), 76. https://doi.org/10.3390/biomedicines6030076