Development of Phosphorothioate DNA and DNA Thioaptamers
Abstract
:1. Introduction
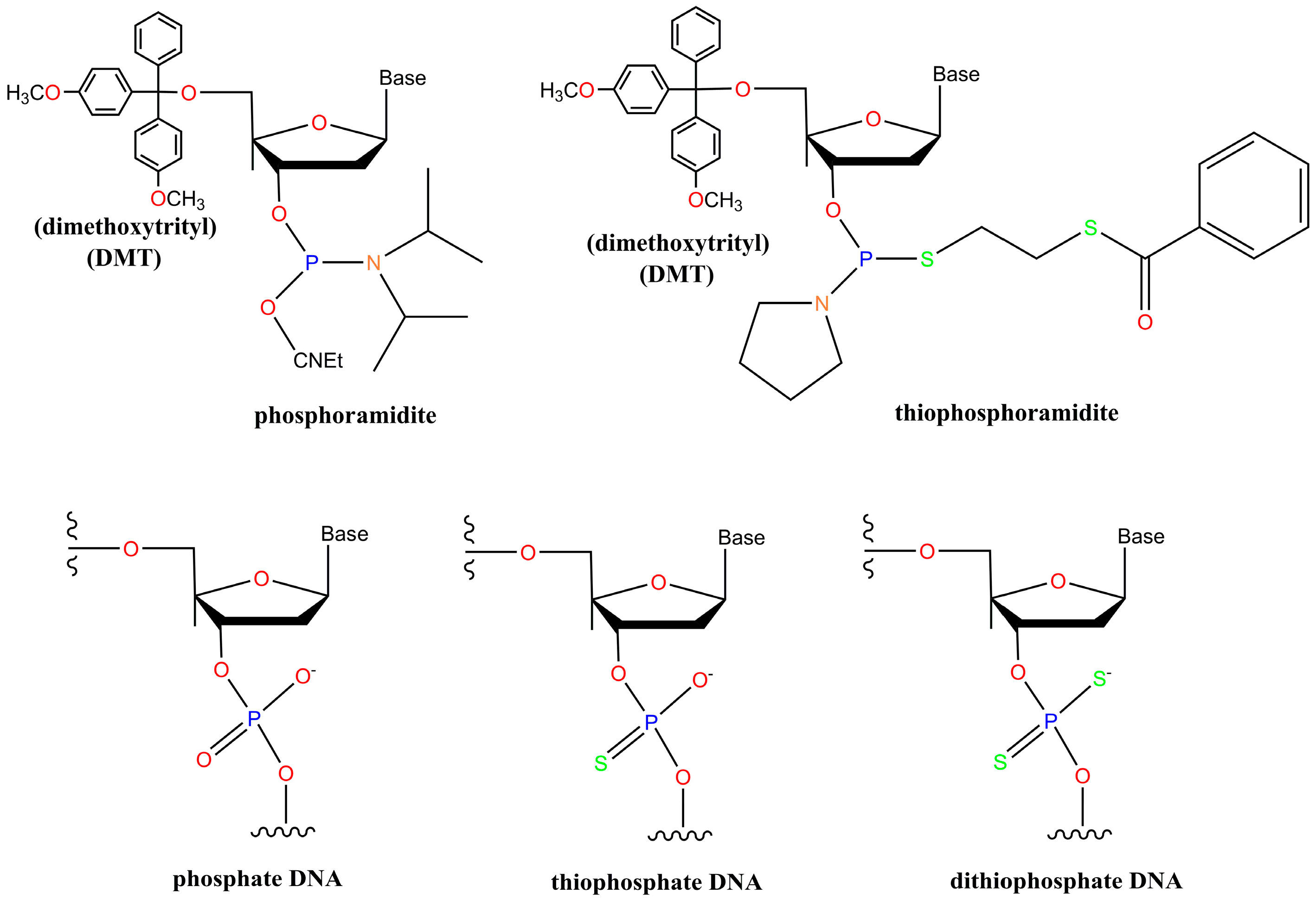
2. Chemistry of Phosphorothioate and Phosphorodithioate DNA Backbones
3. Thioaptamer Development
3.1. The Beginning
3.2. Thioaptamers Targeting Infectious Diseases
3.3. Cancer-Related Thioaptamers
3.4. Other Thioaptamers
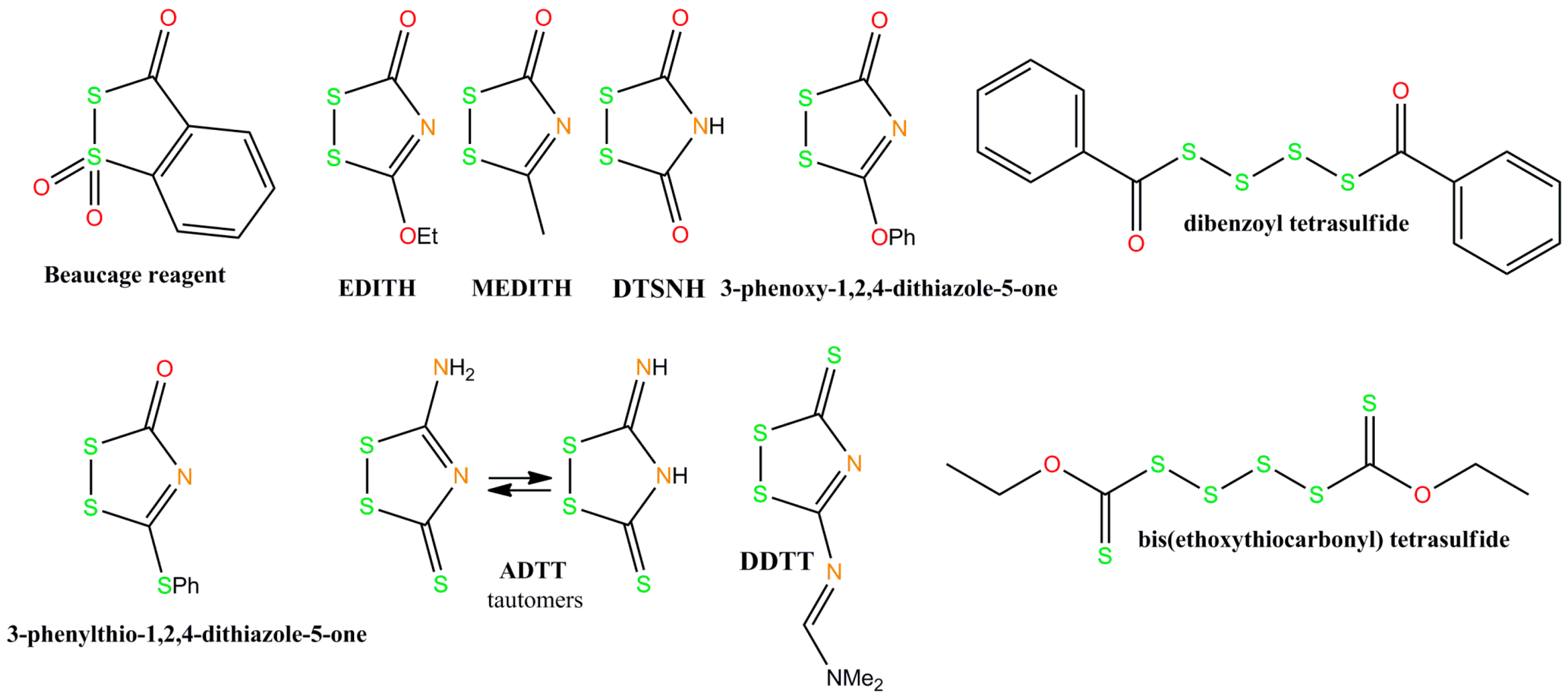
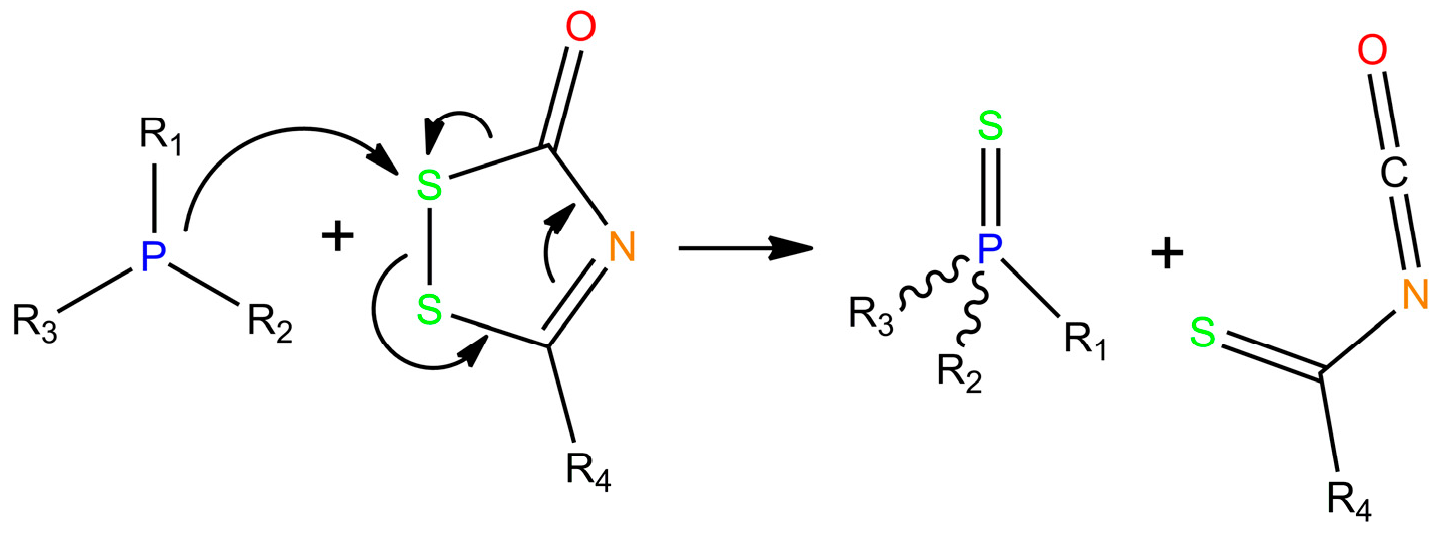
4. Recent Developments in Thioaptamer Methods
4.1. Conjugate-SELEX
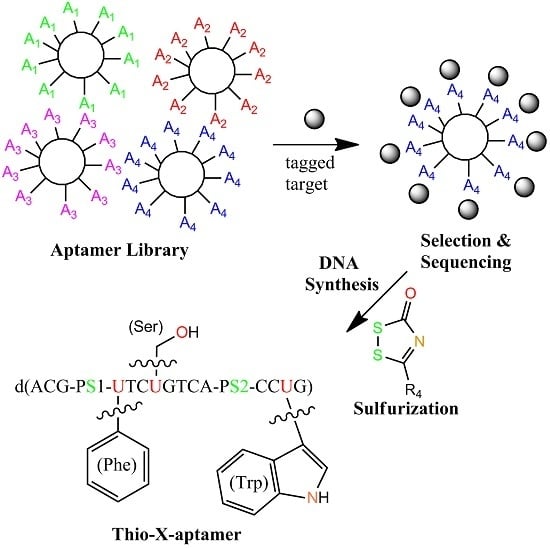
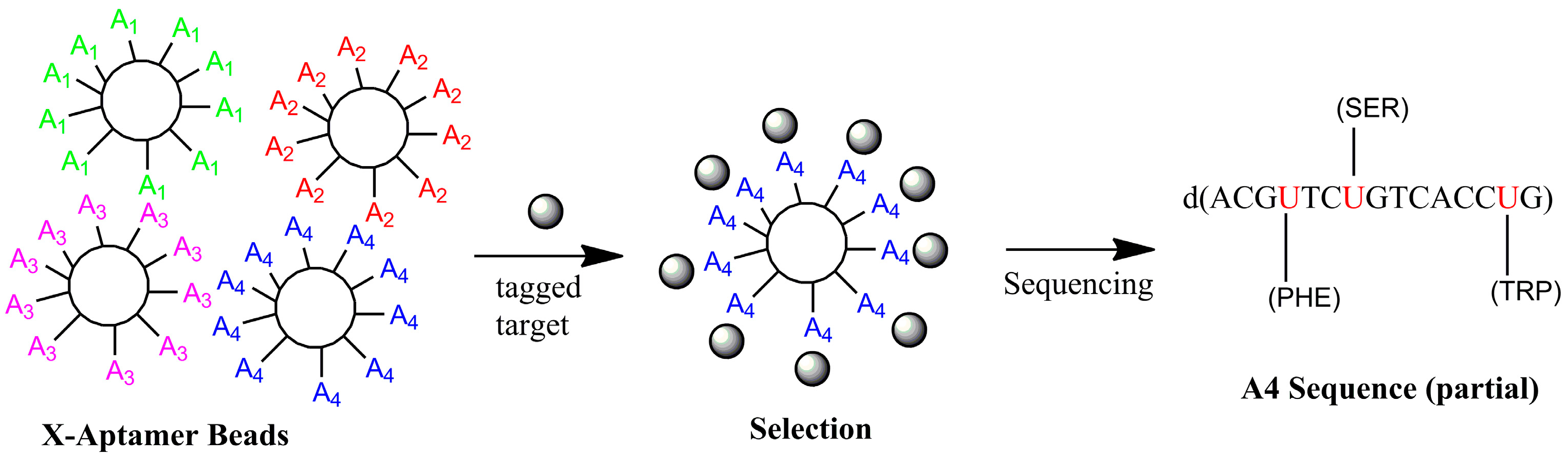
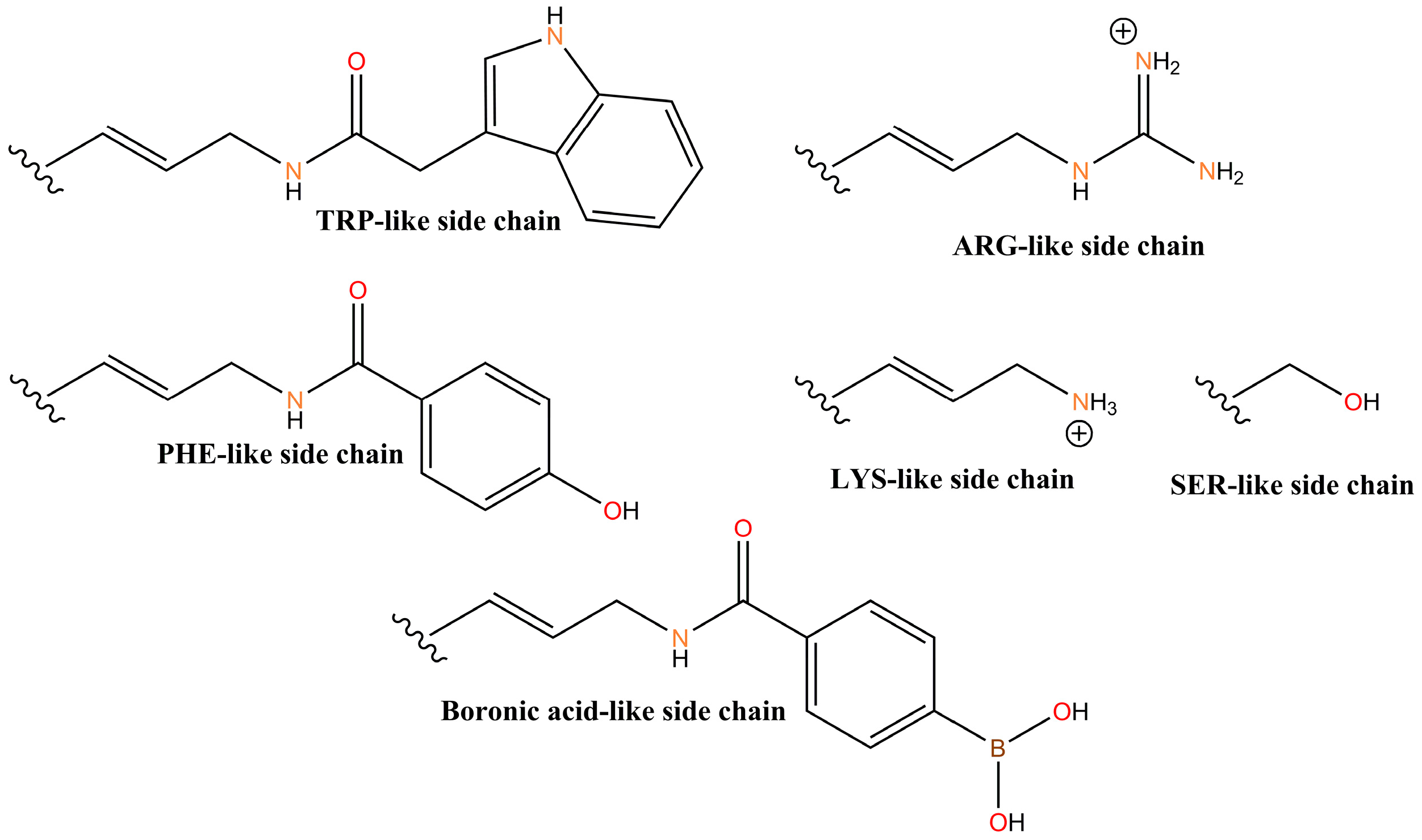
4.2. X-Aptamers
5. Conclusions
Author Contributions
Conflicts of Interest
References
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.L.; Joyce, G.F. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 1990, 344, 467–468. [Google Scholar] [CrossRef] [PubMed]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef] [PubMed]
- McKeague, M.; DeRosa, M.C. Challenges and opportunities for small molecule aptamer development. J. Nucleic Acids 2012, 2012, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.Y.; Byun, J. Nucleic acid aptamers: New methods for selection, stabilization, and application in biomedical science. Biomol. Ther. 2013, 21, 423–434. [Google Scholar] [CrossRef] [PubMed]
- Szeitner, Z.; András, J.; Gyurcsányi, R.E.; Mészáros, T. Is less more? Lessons from aptamer selection strategies. J. Pharm. Biomed. Anal. 2014, 101, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Yu, Y.; Jiang, F.; Zhou, J.; Li, Y.; Liang, C.; Dang, L.; Lu, A.; Zhang, G. Development of Cell-SELEX technology and its application in cancer diagnosis and therapy. Int. J. Mol. Sci. 2016, 17, 2079. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.-T.; Ziemer, G.; Paul, A.; Wendel, H.P. Cell-SELEX: Novel perspectives of aptamer-based therapeutics. Int. J. Mol. Sci. 2008, 9, 668–678. [Google Scholar] [CrossRef] [PubMed]
- Cerchia, L.; Giangrande, P.H.; McNamara, J.O.; Franciscis, V. Cell-Specific Aptamers for Targeted Therapies. In Nucleic Acid and Peptide Aptamers; Mayer, G., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 535, pp. 59–78. ISBN 978-1-934115-89-3. [Google Scholar]
- Ohuchi, S. Cell-SELEX Technology. BioRes. Open Access 2012, 1, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Tan, W.; Donovan, M.J.; Jiang, J. Aptamers from cell-based selection for bioanalytical applications. Chem. Rev. 2013, 113, 2842–2862. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, H.; Ding, H.; Huang, Y.; Cao, X.; Yang, G.; Li, J.; Xie, Z.; Meng, Y.; Li, X.; et al. Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX. J. Pathol. 2009, 218, 327–336. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, X.; Volk, D.E.; Lokesh, G.L.-R.; Elizondo-Riojas, M.-A.; Li, L.; Nick, A.M.; Sood, A.K.; Rosenblatt, K.P.; Gorenstein, D.G. Morph-X-Select: Morphology-based tissue aptamer selection for ovarian cancer biomarker discovery. BioTechniques 2016, 61. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Chen, Y.H.; Lennox, K.A.; Behlke, M.A.; Davidson, B.L. In vivo SELEX for identification of brain-penetrating aptamers. Mol. Ther. Nucleic Acids 2013, 2, e67. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef] [PubMed]
- Healy, J.M.; Lewis, S.D.; Kurz, M.; Boomer, R.M.; Thompson, K.M.; Wilson, C.; McCauley, T.G. Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm. Res. 2004, 21, 2234–2246. [Google Scholar] [CrossRef] [PubMed]
- Homann, M.; Göringer, H.U. Combinatorial selection of high affinity RNA ligands to live African trypanosomes. Nucleic Acids Res. 1999, 27, 2006–2014. [Google Scholar] [CrossRef] [PubMed]
- Homann, M.; Göringer, H.U. Uptake and intracellular transport of RNA aptamers in African trypanosomes suggest therapeutic “piggy-back” approach. Bioorg. Med. Chem. 2001, 9, 2571–2580. [Google Scholar] [CrossRef]
- Lorger, M.; Engstler, M.; Homann, M.; Göringer, H.U. Targeting the variable surface of African trypanosomes with variant surface glycoprotein-specific, serum-stable RNA aptamers. Eukaryot. Cell 2003, 2, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, H.; Magdesian, M.H.; Alves, M.J.M.; Colli, W. In vitro selection of RNA aptamers that bind to cell adhesion receptors of Trypanosoma cruzi and inhibit cell invasion. J. Biol. Chem. 2002, 277, 20756–20762. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.N.; Jensen, K.B.; Julin, C.M.; Weil, M.; Gold, L. High affinity ligands from in vitro selection: Complex targets. Proc. Natl. Acad. Sci. USA 1998, 95, 2902–2907. [Google Scholar] [CrossRef] [PubMed]
- Guo, K.-T.; Schäfer, R.; Paul, A.; Ziemer, G.; Wendel, H.P. Aptamer-based strategies for stem cell research. Mini Rev. Med. Chem. 2007, 7, 701–705. [Google Scholar] [PubMed]
- Iwagawa, T.; Ohuchi, S.P.; Watanabe, S.; Nakamura, Y. Selection of RNA aptamers against mouse embryonic stem cells. Biochimie 2012, 94, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Parekh, P.; Turner, P.; Moyer, R.W.; Tan, W. Generating aptamers for recognition of virus-infected cells. Clin. Chem. 2009, 55, 813–822. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Shangguan, D.; Wang, K.; Shi, H.; Sefah, K.; Mallikratchy, P.; Chen, H.W.; Li, Y.; Tan, W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007, 79, 4900–4907. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhu, X.; Lu, P.Y.; Rosato, R.R.; Tan, W.; Zu, Y. Oligonucleotide aptamers: New tools for targeted cancer therapy. Mol. Ther. Nucleic Acids 2014, 3, e182. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zu, Y. A highlight of recent advances in aptamer technology and its application. Molecules 2015, 20, 11959–11980. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.; Ray, P.; Liu, J.; Kuan, C.-T.; Xu, J.; Hsu, D.; Sullenger, B.A.; White, R.R.; Clary, B.M. In vivo selection against human colorectal cancer xenografts identifies an aptamer that targets RNA helicase protein DHX9. Mol. Ther. Nucleic Acids 2016, 5, e315. [Google Scholar] [CrossRef] [PubMed]
- Mai, J.; Li, X.; Zhang, G.; Huang, Y.; Xu, R.; Shen, Q.; Lokesh, G.L.; Thiviyanathan, V.; Chen, L.; Liu, H.; et al. A DNA thioaptamer with homing specificity to lymphoma bone marrow involvement. Nucleic Acids Res. 2017. under review. [Google Scholar]
- Mosing, R.K.; Mendonsa, S.D.; Bowser, M.T. Capillary electrophoresis-SELEX selection of aptamers with affinity for HIV-1 reverse transcriptase. Anal. Chem. 2005, 77, 6107–6112. [Google Scholar] [CrossRef] [PubMed]
- Drabovich, A.P.; Berezovski, M.; Okhonin, V.; Krylov, S.N. Selection of smart aptamers by methods of kinetic capillary electrophoresis. Anal. Chem. 2006, 78, 3171–3178. [Google Scholar] [CrossRef] [PubMed]
- Yufa, R.; Krylova, S.M.; Bruce, C.; Bagg, E.A.; Schofield, C.J.; Krylov, S.N. Emulsion PCR significantly improves nonequilibrium capillary electrophoresis of equilibrium mixtures-based aptamer selection: Allowing for efficient and rapid selection of aptamer to unmodified ABH2 protein. Anal. Chem. 2015, 87, 1411–1419. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.V.; Musheev, M.U.; Drabovich, A.P.; Jitkova, J.V.; Krylov, S.N. Non-SELEX: Selection of aptamers without intermediate amplification of candidate oligonucleotides. Nat. Protoc. 2006, 1, 1359–1369. [Google Scholar] [CrossRef] [PubMed]
- Berezovski, M.; Drabovich, A.; Krylova, S.M.; Musheev, M.; Okhonin, V.; Petrov, A.; Krylov, S.N. Nonequilibrium capillary electrophoresis of equilibrium mixtures: A universal tool for development of aptamers. J. Am. Chem. Soc. 2005, 127, 3165–3171. [Google Scholar] [CrossRef] [PubMed]
- Nitsche, A.; Kurth, A.; Dunkhorst, A.; Pänke, O.; Sielaff, H.; Junge, W.; Muth, D.; Scheller, F.; Stöcklein, W.; Dahmen, C.; et al. One-step selection of vaccinia virus-binding DNA aptamers by MonoLEX. BMC Biotechnol. 2007, 7, 48. [Google Scholar] [CrossRef] [PubMed]
- Dickman, M.; Hornby, D.P. Isolation of single-stranded DNA using denaturing DNA chromatography. Anal. Biochem. 2000, 284, 164–167. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Li, X.; Prow, T.W.; Reece, L.M.; Bassett, S.E.; Luxon, B.A.; Herzog, N.K.; Aronson, J.; Shope, R.E.; Leary, J.F.; et al. Immunofluorescence assay and flow-cytometry selection of bead-bound aptamers. Nucleic Acids Res. 2003, 31, e54. [Google Scholar] [CrossRef]
- Hybarger, G.; Bynum, J.; Williams, R.F.; Valdes, J.J.; Chambers, J.P. A microfluidic SELEX prototype. Anal. Bioanal. Chem. 2006, 384, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Lou, X.; Qian, J.; Xiao, Y.; Viel, L.; Gerdon, A.E.; Lagally, E.T.; Atzberger, P.; Tarasow, T.M.; Heeger, A.J.; Soh, H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA 2009, 106, 2989–2994. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Ahn, J.-Y.; Jo, M.; Lee, D.; Lis, J.T.; Craighead, H.G.; Kim, S. Selection and elution of aptamers using nanoporous sol-gel arrays with integrated microheaters. Lab Chip 2009, 9, 1206. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.; Stephens, B.J.; Bonin, K.; Cubicciotti, R.; Guthold, M. A combined atomic force/fluorescence microscopy technique to select aptamers in a single cycle from a small pool of random oligonucleotides. Microsc. Res. Tech. 2007, 70, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Miyachi, Y.; Shimizu, N.; Ogino, C.; Kondo, A. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 2010, 38, e21. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-W.; Tatavarty, R.; Kim, D.W.; Jung, H.-T.; Gu, M.B. Immobilization-free screening of aptamers assisted by graphene oxide. Chem. Commun. 2012, 48, 2071–2073. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, V.-T.; Kwon, Y.S.; Kim, J.H.; Gu, M.B. Multiple GO-SELEX for efficient screening of flexible aptamers. Chem. Commun. 2014, 50, 10513–10516. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Duan, N.; Wu, S.; Hao, L.; Xia, Y.; Ma, X.; Wang, Z. Graphene oxide-assisted non-immobilized SELEX of okdaic acid aptamer and the analytical application of aptasensor. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Sayer, N.; Ibrahim, J.; Turner, K.; Tahiri-Alaoui, A.; James, W. Structural characterization of a 2′F-RNA aptamer that binds a HIV-1 SU glycoprotein, gp120. Biochem. Biophys. Res. Commun. 2002, 293, 924–931. [Google Scholar] [CrossRef]
- Spiga, F.M.; Maietta, P.; Guiducci, C. More DNA-aptamers for small drugs: A capture-SELEX coupled with surface plasmon resonance and high-throughput sequencing. ACS Comb. Sci. 2015, 17, 326–333. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Guo, L.; He, J.; Xu, H.; Xie, J. Stepping library-based post-SELEX strategy approaching to the minimized aptamer in SPR. Anal. Chem. 2017, 89, 6559–6566. [Google Scholar] [CrossRef] [PubMed]
- Raddatz, M.L.; Dolf, A.; Endl, E.; Knolle, P.; Famulok, M.; Mayer, G. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew. Chem. Int. Ed. 2008, 47, 5190–5193. [Google Scholar] [CrossRef] [PubMed]
- Mayer, G.; Ahmed, M.-S.L.; Dolf, A.; Endl, E.; Knolle, P.A.; Famulok, M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010, 5, 1993–2004. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.A.A.; Keating, G.M. Pegaptanib: In exudative age-related macular degeneration. Drugs 2005, 65, 1571–1577. [Google Scholar] [CrossRef] [PubMed]
- Ng, E.W.M.; Shima, D.T.; Calias, P.; Cunningham, E.T.; Guyer, D.R.; Adamis, A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006, 5, 123–132. [Google Scholar] [CrossRef] [PubMed]
- VEGF Inhibition Study in Ocular Neovascularization (V.I.S.I.O.N.) Clinical Trial Group. Year 2 efficacy results of 2 randomized controlled clinical trials of pegaptanib for neovascular age-related macular degeneration. Ophthalmology 2006, 113, 1508.e1–1508.e25. [Google Scholar] [CrossRef]
- Ruckman, J.; Green, L.S.; Beeson, J.; Waugh, S.; Gillette, W.L.; Henninger, D.D.; Claesson-Welsh, L.; Janjic, N. 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165) inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J. Biol. Chem. 1998, 273, 20556–20567. [Google Scholar] [CrossRef] [PubMed]
- Khati, M.; Schüman, M.; Ibrahim, J.; Sattentau, Q.; Gordon, S.; James, W. Neutralization of infectivity of diverse R5 clinical isolates of human immunodeficiency virus type 1 by gp120-binding 2’F-RNA aptamers. J. Virol. 2003, 77, 12692–12698. [Google Scholar] [CrossRef] [PubMed]
- Layzer, M.; McCaffrey, A.P.; Tanner, A.K.; Huang, Z.; Kay, M.A.; Sullenger, B.A. In vivo activity of nuclease-resistant siRNAs. RNA 2004, 10, 766–771. [Google Scholar] [CrossRef] [PubMed]
- Jellinek, D.; Green, L.S.; Bell, C.; Lynott, C.K.; Gill, N.; Vargeese, C.; Kirschenheuter, G.; McGee, D.P.C.; Abesinghe, P. Potent 2’-amino-2’-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 1995, 34, 11363–11372. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Gao, X.; Zhang, Z. Isolation and characterization of 2′-amino-modified RNA aptamers for human TNFα. Genom. Proteom. Bioinform. 2004, 2, 32–42. [Google Scholar] [CrossRef]
- Padilla, R.; Sousa, R. A Y639F/H784A T7 RNA polymerase double mutant displays superior properties for synthesizing RNAs with non-canonical NTPs. Nucleic Acids Res. 2002, 30, e138. [Google Scholar] [CrossRef] [PubMed]
- Dellafiore, M.A.; Montserrat, J.M.; Iribarren, A.M. Modified nucleoside triphosphates for in vitro selection techniques. Front. Chem. 2016, 4. [Google Scholar] [CrossRef] [PubMed]
- Kuwahara, M.; Obika, S.; Nagashima, J.; Ohta, Y.; Suto, Y.; Ozaki, H.; Sawai, H.; Imanishi, T. Systematic analysis of enzymatic DNA polymerization using oligo-DNA templates and triphosphate analogs involving 2′,4′-bridged nucleosides. Nucleic Acids Res. 2008, 36, 4257–4265. [Google Scholar] [CrossRef] [PubMed]
- Barciszewski, J.; Medgaard, M.; Koch, T.; Kurreck, J.; Erdmann, V.A. Locked Nucleic Acid Aptamers. In Nucleic Acid and Peptide Aptamers; Mayer, G., Ed.; Humana Press: Totowa, NJ, USA, 2009; Volume 535, pp. 165–186. ISBN 978-1-934115-89-3. [Google Scholar]
- Kasahara, Y.; Irisawa, Y.; Ozaki, H.; Obika, S.; Kuwahara, M. 2′,4′-BNA/LNA aptamers: CE-SELEX using a DNA-based library of full-length 2′-O,4′-C-methylene-bridged/linked bicyclic ribonucleotides. Bioorg. Med. Chem. Lett. 2013, 23, 1288–1292. [Google Scholar] [CrossRef] [PubMed]
- Horhota, A.; Zou, K.; Ichida, J.K.; Yu, B.; McLaughlin, L.W.; Szostak, J.W.; Chaput, J.C. Kinetic analysis of an efficient DNA-dependent TNA polymerase. J. Am. Chem. Soc. 2005, 127, 7427–7434. [Google Scholar] [CrossRef] [PubMed]
- Ichida, J.K.; Zou, K.; Horhota, A.; Yu, B.; McLaughlin, L.W.; Szostak, J.W. An in vitro selection system for TNA. J. Am. Chem. Soc. 2005, 127, 2802–2803. [Google Scholar] [CrossRef] [PubMed]
- Kempeneers, V. Investigation of the DNA-dependent cyclohexenyl nucleic acid polymerization and the cyclohexenyl nucleic acid-dependent DNA polymerization. Nucleic Acids Res. 2005, 33, 3828–3836. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-H.; Chen, J.; Szostak, J.W. Enzymatic synthesis of DNA on glycerol nucleic acid templates without stable duplex formation between product and template. Proc. Natl. Acad. Sci. USA 2007, 104, 14598–14603. [Google Scholar] [CrossRef] [PubMed]
- Wittung, P.; Nielsen, P.E.; Buchardt, O.; Egholm, M.; Norde, B. DNA-like double helix formed by peptide nucleic acid. Nature 1994, 368, 561–563. [Google Scholar] [CrossRef] [PubMed]
- Lato, S.M.; Ozerova, N.D.S.; He, K.; Sergueeva, Z.; Shaw, B.R.; Burke, D.H. Boron-containing aptamers to ATP. Nucleic Acids Res. 2002, 30, 1401–1407. [Google Scholar] [CrossRef] [PubMed]
- Andreola, M.-L.; Calmels, C.; Michel, J.; Toulmé, J.-J.; Litvak, S. Towards the selection of phosphorothioate aptamers. FEBS J. 2000, 267, 5032–5040. [Google Scholar] [CrossRef]
- Yang, X.; Gorenstein, D. Progress in thioaptamer development. Curr. Drug Targets 2004, 5, 705–715. [Google Scholar] [CrossRef] [PubMed]
- Thiviyanathan, V.; Gorenstein, D.G. Aptamers and the next generation of diagnostic reagents. Proteom. Clin. Appl. 2012, 6, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.Y.; Yang, X.; Gharpure, K.M.; Hatakeyama, H.; Egli, M.; McGuire, M.H.; Nagaraja, A.S.; Miyake, T.M.; Rupaimoole, R.; Pecot, C.V.; et al. 2′-OMe-phosphorodithioate-modified siRNAs show increased loading into the RISC complex and enhanced anti-tumour activity. Nat. Commun. 2014, 5, 3459. [Google Scholar] [CrossRef] [PubMed]
- Eckstein, F. A dinucleoside phosphorothioate. Tetrahedron Lett. 1967, 8, 1157–1160. [Google Scholar] [CrossRef]
- De Clerq, E.; Eckstein, E.; Merigan, T.C. Interferon induction increased through chemical modification of a synthetic polyribonucleotide. Science 1969, 165, 1137–1139. [Google Scholar] [CrossRef]
- Burgers, P.M.; Eckstein, F. Synthesis of dinucleoside monophosphorothioates via addition of sulphur to phosphite triesters. Tetrahedron Lett. 1978, 19, 3835–3838. [Google Scholar] [CrossRef]
- Vu, H.; Hirschbein, B.L. Internucleotide phosphite sulfurization with tetraethylthiuram disulfide. Phosphorothioate oligonucleotide synthesis via phosphoramidite chemistry. Tetrahedron Lett. 1991, 32, 3005–3008. [Google Scholar] [CrossRef]
- Iyer, R.P.; Egan, W.; Regan, J.B.; Beaucage, S.L. 3H-1,2-Benzodithiole-3-one 1,1-dioxide as an improved sulfurizing reagent in the solid-phase synthesis of oligodeoxyribonucleoside phosphorothioates. J. Am. Chem. Soc. 1990, 112, 1253–1254. [Google Scholar] [CrossRef]
- Rao, M.V.; Reese, C.B.; Zhao, Z.Y. Dibenzoyl tetrasulfide: A rapid sulfur-transfer agent in the synthesis of phosphorothioate analogues of oligonucleotides. Tetrahderon Lett. 1992, 33, 4839–4842. [Google Scholar] [CrossRef]
- Stec, W.J.; Uznanski, B.; Wilk, A. Bis(O,O-diisopropoxy phosphinothioyl)disulfide: A highly efficient sulfurizing reagent for cost-effective synthesis of oligo(nucleoside phosphorothioate)s. Tetrahedron Lett. 1993, 34, 5317–5320. [Google Scholar] [CrossRef]
- Xu, Q.H.; Musierforsyth, K.; Hammer, R.P.; Barany, G. Use of 1,2,4-dithiazolidine-3,5-dione (DTSNH) and 3-ethoxy-1,2,4-dithiazoline-5-one (EDITH) for synthesis of phosphorothioate-containing oligodeoxyribonucleotides. Nucleic Acids Res. 1996, 24, 1602–1607. [Google Scholar]
- Zhang, Z.; Nichols, A.; Tang, J.X.; Han, Y.; Tang, J.Y. Solid phase synthesis of oligonucleotide phosphorothioate analogues using 3-methyl-1,2,4-dithiazolin-5-one (MEDITH) as a new sulfur-transfer reagent. Tetrahedron Lett. 1999, 40, 2095–2098. [Google Scholar] [CrossRef]
- Tang, J.-Y.; Han, Y.; Tang, J.X.; Zhang, Z. Large-scale synthesis of oligonucleotide phosphorothioates using 3-amino-1,2,4-dithiazole-5-thione as an efficient sulfur-transfer reagent. Org. Process Res. Dev. 2000, 4, 194–198. [Google Scholar] [CrossRef]
- Lemaître, M.M.; Murphy, A.S.; Somers, R.L. Sulfurizing reagent II and its use in synthesizing oligonucleotide phosphoramidites. Glen Rep. 2006, 18, 4–5. [Google Scholar]
- Ponomarov, O.; Laws, A.P.; Hanusek, J. 1,2,4-Dithiazole-5-ones and 5-thiones as efficient sulfurizing agents of phosphorus (III) compounds—A kinetic comparative study. Org. Biomol. Chem. 2012, 10, 8868. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.Y.-X.; Dignam, J.C.; Fong, G.W.; Li, L.; Gray, S.H.; Jacob-Samuel, B.; George, S.T. Evaluation of 3-ethoxy-1, 2, 4-dithiazoline-5-one (EDITH) as a new sulfurizing reagent in combination with labile exocyclic amino protecting groups for solid-phase oligonucleotide synthesis. Nucleic Acids Res. 1997, 25, 3590–3593. [Google Scholar] [CrossRef] [PubMed]
- Bergot, B.J.; Egan, W. Separation of synthetic phosphorothioate oligodeoxynucleotides from their oxygenated (phosphodiester) defect species by strong-anion-exchange high-performance liquid chromatography. J. Chromatogr. A 1992, 599, 35–42. [Google Scholar] [CrossRef]
- Eckstein, F. Phosphorothioates, essential components of therapeutic oligonucleotides. Nucleic Acid Ther. 2014, 24, 374–387. [Google Scholar] [CrossRef] [PubMed]
- Beaucage, S.L.; Caruthers, M.H. Deoxynucleoside phosphoramidites—A new class of key intermediates for deoxypolynucleotide synthesis. Tetrahedron Lett. 1981, 22, 1859–1862. [Google Scholar] [CrossRef]
- Gorenstein, D.; Farschtschi, N. Process for Preparing Dithiophosphate Oligonucleotide Analogs via Nucleoside Thiophosphoramidite Intermediates. Patent US5,218,088 A, 8 June 1993. [Google Scholar]
- Caruthers, M.H.; Ma, Y.-X.; Yau, E.K.; Nielsen, J.; Brill, W. Nucleoside Thiophosphoramidites. Patent US5,218,103 A, 8 June 1993. [Google Scholar]
- Caruthers, M.H.; Marshall, W.S. Polynucleotide Phosphorodithioates. Patent US5,278,302 A, 11 January 1994. [Google Scholar]
- Caruthers, M.H.; Marshall, W.S.; Brill, W.; Nielsen, J. Polynucleotide Phosphorodithioate. Patent US5,453,496 A, 26 September 1995. [Google Scholar]
- Caruthers, M.H.; Brill, W.K.D.; Yau, E.; Ma, M.; Nielsen, J. Nucleoside Thiophosphoramidites. Patent US5,684,148 A, 4 November 1997. [Google Scholar]
- Caruthers, M.H.; Brill, W.K.-D.; Yau, E.; Ma, M.; Nielsen, J. Polynucleotide Phosphorodithioate Compounds. Patent US5,602,244 A, 11 February 1997. [Google Scholar]
- Caruthers, M.H.; Brill, W.K.-D.; Yau, E.; Ma, M.; Nielsen, J. Polynucleotide Phosphorodithioate Compounds. Patent US5,750,666 A, 12 May 1998. [Google Scholar]
- Fischer, R.W.; Caruthers, M.H. Synthesis of a dinucleotide phosphoramidimidate. Tetrahedron Lett. 1995, 36, 6807–6810. [Google Scholar] [CrossRef]
- Seeberger, P.H.; Caruthers, M.H. Oxidative formation of phosphorodithiaotes via H-phosphonodithioates. Tetrahedron Lett. 1995, 36, 695–698. [Google Scholar] [CrossRef]
- Bjergrde, K.; Dahl, O.; Caruthers, M.H. Synthesis of dinucleoside phosphoramidimidates. Tetrahedron Lett. 1994, 35, 2941–2944. [Google Scholar] [CrossRef]
- Greef, C.H.; Seeberger, P.H.; Caruthers, M.H.; Beaton, G.; Bankaitis-Davis, D. Synthesis of phosphorodithioate RNA by the H-phosphonothioate method. Tetrahedron Lett. 1996, 37, 4451–4454. [Google Scholar] [CrossRef]
- Yang, X. Solid-phase synthesis of Oligodeoxynucleotide Analogs Containing Phosphorodithioate Linkages: Synthesis of Oligodeoxynucleotide Analogs Containing PS2 Linkages. In Current Protocols in Nucleic Acid Chemistry; Egli, M., Herdewijn, P., Matusda, A., Sanghvi, Y.S., Eds.; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2016; pp. 4.71.1–4.71.14. ISBN 978-0-471-14270-6. [Google Scholar]
- King, D.J.; Ventura, D.A.; Brasier, A.R.; Gorenstein, D.G. Novel combinatorial selection of phosphorothioate oligonucleotide aptamers. Biochemistry 1998, 37, 16489–16493. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Fennewald, S.; Luxon, B.A.; Aronson, J.; Herzog, N.K.; Gorenstein, D.G. Aptamers containing thymidine 3′-O-phosphorodithioates: Synthesis and binding to nuclear factor-κB. Bioorg. Med. Chem. Lett. 1999, 9, 3357–3362. [Google Scholar] [CrossRef]
- Volk, D.E.; Yang, X.; Fennewald, S.M.; King, D.J.; Bassett, S.; Venkitachalam, S.; Herzog, N.; Luxon, B.A.; Gorenstein, D.G. Solution structure and design of dithiophosphate backbone aptamers targeting transcription factor NF-κB. Bioorg. Chem. 2002, 30, 396–419. [Google Scholar] [CrossRef]
- Volk, D.E.; Power, T.D.; Gorenstein, D.G.; Luxon, B.A. An ab initio study of phosphorothioate and phosphorodithioate interactions with sodium cation. Tetrahedron Lett. 2002, 43, 4443–4447. [Google Scholar] [CrossRef]
- Marshall, W.S.; Caruthers, M.H. Phosphorodithioate DNA as a potential therapeutic drug. Science 1993, 259, 1564–1570. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Hodge, R.P.; Luxon, B.A.; Shope, R.; Gorenstein, D.G. Separation of synthetic oligonucleotide dithioates from monothiophosphate impurities by anion-exchange chromatography on a mono-Q column. Anal. Biochem. 2002, 306, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Abeydeera, N.D.; Egli, M.; Cox, N.; Mercier, K.; Conde, J.N.; Pallan, P.S.; Mizurini, D.M.; Sierant, M.; Hibti, F.-E.; Hassell, T.; et al. Evoking picomolar binding in RNA by a single phosphorodithioate linkage. Nucleic Acids Res. 2016, 44, 8052–8064. [Google Scholar] [CrossRef] [PubMed]
- King, D.J.; Bassett, S.E.; Li, X.; Fennewald, S.A.; Herzog, N.K.; Luxon, B.A.; Shope, R.; Gorenstein, D.G. Combinatorial selection and binding of phosphorothioate aptamers targeting human NF-κB RelA(p65) and p50. Biochemistry 2002, 41, 9696–9706. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Bassett, S.E.; Li, X.; Luxon, B.A.; Herzog, N.K.; Shope, R.E.; Aronson, J.; Prow, T.W.; Leary, J.F.; Kirby, R.; et al. Construction and selection of bead-bound combinatorial oligonucleotide phosphorothioate and phosphorodithioate aptamer libraries designed for rapid PCR-based sequencing. Nucleic Acids Res. 2002, 30, e132. [Google Scholar] [CrossRef] [PubMed]
- Somasunderam, A.; Ferguson, M.R.; Rojo, D.R.; Thiviyanathan, V.; Li, X.; O’Brien, W.A.; Gorenstein, D.G. Combinatorial selection, inhibition and antiviral activity of DNA thioaptamers targeting the RNase H domain of HIV-1 reverse transcriptase. Biochemistry 2005, 44, 10388–10395. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, M.R.; Rojo, D.R.; Somasunderam, A.; Thiviyanathan, V.; Ridley, B.; Yang, X.; Gorenstein, D.G. Delivery of double-stranded DNA thioaptamers into HIV-1 infected cells for antiviral activity. Biochem. Biophys. Res. Commun. 2006, 344, 792–797. [Google Scholar] [CrossRef] [PubMed]
- Vaillant, A.; Juteau, J.-M.; Lu, H.; Liu, S.; Lackman-Smith, C.; Ptak, R.; Jiang, S. Phosphorothioate oligonucleotides inhibit human immunodeficiency virus type 1 fusion by blocking gp41 core formation. Antimicrob. Agents Chemother. 2006, 50, 1393–1401. [Google Scholar] [CrossRef] [PubMed]
- Karpuj, M.V.; Giles, K.; Gelibter, S.; Scott, M.R.; Lingappa, V.R.; Szoka, F.C.; Peretz, D.; Denetclaw, W.; Prusiner, S.B. Phosphorothioate oligonucleotides reduce PrPsc levels and prion infectivity in cultured cells. Mol. Med. 2007, 13, 190. [Google Scholar] [CrossRef] [PubMed]
- King, D.J.; Safar, J.G.; Legname, G.; Prusiner, S.B. Thioaptamer interactions with prion proteins: Sequence-specific and non-specific binding sites. J. Mol. Biol. 2007, 369, 1001–1014. [Google Scholar] [CrossRef] [PubMed]
- Fennewald, S.M.; Scott, E.P.; Zhang, L.H.; Aronson, J.F.; Gorenstein, D.G.; Luxon, B.A.; Shope, R.E.; Beasley, D.W.C.; Herzog, N.K. Thioaptamer decoy targeting AP-1 proteins influences cytokine expression and the outcome of arenavirus infections. J. Gen. Virol. 2007, 88, 981–990. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.H.; Ren, J.; Liu, C.-M.; Liu, P.K. Intracellular gene transcription factor protein-guided MRI by DNA aptamers in vivo. FASEB J. 2014, 28, 464–473. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lee, M.S.; Watowich, S.J.; Gorenstein, D.G. Combinatorial selection of a RNA thioaptamer that binds to Venezuelan equine encephalitis virus capsid protein. FEBS Lett. 2007, 581, 2497–2502. [Google Scholar] [CrossRef] [PubMed]
- Gandham, S.H.A.; Volk, D.E.; Rao, L.G.L.; Neerathilingam, N.; Gorenstein, D.G. Thioaptamers targeting Dengue virus type-2 envelope protein domain III. Biochem. Biophys. Res. Commun. 2014, 453, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Lee, M.S.; Copland, J.A.; Luxon, B.A.; Gorenstein, D.G. Combinatorial selection of a single stranded DNA thioaptamer targeting TGF-β-1 protein. Bioorg. Med. Chem. Lett. 2008, 18, 1835–1839. [Google Scholar] [CrossRef] [PubMed]
- Matharu, Z.; Patel, D.; Gao, Y.; Haque, A.; Zhou, Q.; Revzin, A. Detecting transforming growth factor-β release from liver cells using an aptasensor integrated with microfluidics. Anal. Chem. 2014, 86, 8865–8872. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Tang, Y.; Xing, D. One-step homogeneous protein detection based on aptamer probe and fluorescence cross-correlation spectroscopy. Anal. Chem. 2011, 83, 2906–2912. [Google Scholar] [CrossRef] [PubMed]
- Bock, L.C.; Griffin, L.C.; Latham, J.A.; Vermaas, E.H.; Toole, J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 1992, 355, 564–566. [Google Scholar] [CrossRef] [PubMed]
- Tasset, D.M.; Kubik, M.F.; Steiner, W. Oligonucleotide inhibitors of human thrombin that bind distinct epitopes. J. Mol. Biol. 1997, 272, 688–698. [Google Scholar] [CrossRef] [PubMed]
- Somasunderam, A.; Thiviyanathan, V.; Tanaka, T.; Li, X.; Neerathilingam, M.; Lokesh, G.L.; Mann, A.; Peng, Y.; Ferrari, M.; Klostergaard, J.; et al. Combinatorial selection of DNA thioaptamers targeted to the HA binding domain of CD44. Biochemistry 2010, 49, 9106–9112. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Wang, X.; Ding, B.; Cai, H.; Wang, X.; Fan, Y.; Li, Y.; Liu, S.; Nie, S.; Lu, Q. Thioaptamer-conjugated CD44-targeted delivery system for the treatment of breast cancer in vitro and in vivo. J. Drug Target. 2016, 24, 359–371. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.P.; Somasunderam, A.; Nieves-Alicea, R.; Li, X.; Hu, A.; Sood, A.K.; Ferrari, M.; Gorenstein, D.G.; Tanaka, T. Identification of thioaptamer ligand against E-selectin: Potential application for inflamed vasculature imaging. PLoS ONE 2010, 5, e13050. [Google Scholar] [CrossRef] [PubMed]
- Bendas, G.; Borsig, L. Cancer cell adhesion and metastasis: Selectins, integrins, and the inhibitory potential of heparins. Int. J. Cell Biol. 2012, 2012, 676731. [Google Scholar] [CrossRef] [PubMed]
- Berg, E.L.; Robinson, M.K.; Mansson, O.; Butcher, E.C.; Magnani, J.L. A carbohydrate domain common to both sialyl Le (a) and sialyl Le (X) is recognized by the endothelial cell leukocyte adhesion molecule ELAM-1. J. Biol. Chem. 1991, 266, 14869–14872. [Google Scholar] [PubMed]
- Ehrhardt, C.; Kneuer, C.; Bakowsky, U. Selectins-an emerging target for drug delivery. Adv. Drug Deliv. Rev. 2004, 56, 527–549. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.P.; Somasunderam, A.; Liu, X.; Gorenstein, D.G.; Ferrari, M. E-selectin-targeted porous silicon particle for nanoparticle delivery to the bone marrow. Adv. Mater. 2011, 23, H278–H282. [Google Scholar] [CrossRef] [PubMed]
- Mann, A.P.; Bhavane, R.C.; Somasunderam, A.; Montalvo-Ortiz, B.L.; Ghaghada, K.B.; Volk, D.; Nieves-Alicea, R.; Suh, K.S.; Ferrari, M.; Annapragada, A.; et al. Thioaptamer conjugated liposomes for tumor vasculature targeting. Oncotarget 2011, 2, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Zong, H.; Sen, S.; Zhang, G.; Gorenstein, D.G.; Liu, X.; Ferrari, M.; Roboz, G.J.; Shen, H.; Guzman, M.L. Novel multistage nanoparticle drug delivery to ablate leukemia stem cells in their niche. Blood 2012, 120, 2631. [Google Scholar]
- Zong, H.; Sen, S.; Zhang, G.; Mu, C.; Albayati, Z.F.; Gorenstein, D.G.; Liu, X.; Ferrari, M.; Crooks, P.A.; Roboz, G.J.; et al. In vivo targeting of leukemia stem cells by directing parthenolide-loaded nanoparticles to the bone marrow niche. Leukemia 2016, 30, 1582–1586. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.-A.; Hasan, N.; Mann, A.P.; Zheng, W.; Zhao, L.; Morris, L.; Zhu, W.; Zhao, Y.D.; Suh, K.S.; Dooley, W.C.; et al. Blocking the adhesion cascade at the premetastatic niche for prevention of breast cancer metastasis. Mol. Ther. 2015, 23, 1044–1054. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Tsolmon, B.; Zheng, W.; Zhao, L.; Volk, D.E.; Lokesh, G.L.-R.; Hagensick, C.; Gupta, V.; Morris, L.; Zhao, Y.D.; et al. Safety evaluation of intravenously administered mono-thioated aptamer against E-selectin in mice. Toxicol. Appl. Pharmacol. 2015, 287, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Kamal, M.; Kang, S.-A.; Zhang, R.; Lokesh, G.L.R.; Thiviyanathan, V.; Hasan, N.; Woo, S.; Zhao, Y.; Leslie, M.; et al. E-selectin targeting PEGylated-thioaptamer prevents breast cancer metastases. Mol. Ther. Nucleic Acids 2016, 5, e399. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, D.; Koenig, A.; Jennings, S.; Hicke, B.; Han, H.-L.; Fitzwater, T.; Chang, Y.-F.; Varki, N.; Parma, D.; Varki, A. Calcium-dependent oligonucleotide antagonists specific for L-selectin. Proc. Natl. Acad. Sci. USA 1996, 93, 5883–5887. [Google Scholar] [CrossRef] [PubMed]
- Hicke, B.J.; Watson, S.R.; Koenig, A.; Lynott, C.K.; Bargatze, R.F.; Chang, Y.F.; Ringquist, S.; Moon-McDermott, L.; Jennings, S.; Fitzwater, T.; et al. DNA aptamers block L-selectin function in vivo. Inhibition of human lymphocyte trafficking in SCID mice. J. Clin. Investig. 1996, 98, 2688–2692. [Google Scholar] [CrossRef] [PubMed]
- Gutsaeva, D.R.; Parkerson, J.B.; Yerigenahally, S.D.; Kurz, J.C.; Schaub, R.G.; Ikuta, T.; Head, C.A. Inhibition of cell adhesion by anti-P-selectin aptamer: A new potential therapeutic agent for sickle cell disease. Blood 2011, 117, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Sheriff, S.; Lewis, K.B.; Tinch, S.L.; Cho, J.; Balasubramaniam, A.; Kennedy, M.A. HMGA-targeted phosphorothioate DNA aptamers increase sensitivity to gemcitabine chemotherapy in human pancreatic cancer cell lines. Cancer Lett. 2012, 315, 18–27. [Google Scholar] [CrossRef] [PubMed]
- Reeves, R.; Nissen, M.S. The AT-DNA-binding domain of mammalian high mobility group I chromosomal proteins. A novel peptide motif for recognizing DNA structure. J. Biol. Chem. 1990, 265, 8573–8582. [Google Scholar] [PubMed]
- Liau, S.-S.; Whang, E. HMGA1 is a molecular determinant of chemoresistance to gemcitabine in pancreatic adenocarcinoma. Clin. Cancer Res. 2008, 14, 1470–1477. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Ueda, Y.; Akaboshi, S.; Hino, Y.; Sekita, Y.; Nakao, M. HMGA2 maintains oncogenic Ras-induced epithelial-mesenchymal transition in human pancreatic cancer cells. Am. J. Pathol. 2009, 174, 854–868. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Duan, J.; Cao, B.; Zhang, L.; Lu, X.; Wang, F.; Yao, F.; Zhu, Z.; Yuan, W.; Wang, C.; et al. Selection of a novel DNA thioaptamer against HER2 structure. Clin. Transl. Oncol. 2015, 17, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Puglisi, F.; Fontanella, C.; Amoroso, V.; Bianchi, G.V.; Bisagni, G.; Falci, C.; Fontana, A.; Generali, D.; Gianni, L.; Grassadonia, A.; et al. Current challenges in HER2-positive breast cancer. Crit. Rev. Oncol./Hematol. 2016, 98, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Li, B.T.; Ross, D.S.; Aisner, D.L.; Chaft, J.E.; Hsu, M.; Kako, S.L.; Kris, M.G.; Varella-Garcia, M.; Arcila, M.E. HER2 amplification and HER2 mutation are distinct molecular targets in lung cancers. J. Thorac. Oncol. 2016, 11, 414–419. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.-L.; Lee, M.-Y.; Chao, W.-R.; Han, C.-P. The status of HER2 amplification and Kras mutations in mucinous ovarian carcinoma. Hum. Genom. 2016, 10. [Google Scholar] [CrossRef] [PubMed]
- Abrahao-Machado, L.F.; Scapulatempo-Neto, C. HER2 testing in gastric cancer: An update. World J. Gastroenterol. 2016, 22, 4619. [Google Scholar] [CrossRef] [PubMed]
- Arnould, L.; Gelly, M.; Penault-Llorca, F.; Benoit, L.; Bonnetain, F.; Migeon, C.; Cabaret, V.; Fermeaux, V.; Bertheau, P.; Garnier, J.; et al. Trastuzumab-based treatment of HER2-positive breast cancer: An antibody-dependent cellular cytotoxicity mechanism? Br. J. Cancer 2006, 94, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Sartore-Bianchi, A.; Trusolino, L.; Martino, C.; Bencardino, K.; Lonardi, S.; Bergamo, F.; Zagonel, V.; Leone, F.; Depetris, I.; Martinelli, E.; et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): A proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 738–746. [Google Scholar] [CrossRef]
- Liu, Z.; Duan, J.-H.; Song, Y.-M.; Ma, J.; Wang, F.-D.; Lu, X.; Yang, X.-D. Novel HER2 aptamer selectively delivers cytotoxic drug to HER2-positive breast cancer cells in vitro. J. Transl Med. 2012, 10, 148. [Google Scholar] [CrossRef] [PubMed]
- Mangala, L.S.; Wang, H.; Jian, D.; Wu, S.Y.; Somasunderam, A.; Volk, D.E.; Lokesh, G.L.R.; Li, X.; Pradeep, S.; Yang, X.; et al. Improving vascular maturation using non-coding RNAs increases anti-tumor effect of chemotherapy. JCI Insight 2016, 1, e87754. [Google Scholar] [CrossRef] [PubMed]
- Maji, S.; Chaudhary, P.; Akopova, I.; Nguyen, P.M.; Hare, R.J.; Gryczynski, I.; Vishwanatha, J.K. Exosomal Annexin II promotes angiogenesis and breast cancer metastasis. Mol. Cancer Res. 2017, 15, 93–105. [Google Scholar] [CrossRef] [PubMed]
- Bystricky, B.; Cierna, Z.; Sieberova, G.; Janega, P.; Karaba, M.; Minarik, G.; Benca, J.; Sedlackova, T.; Jurisova, S.; Gronesova, P.; et al. Relationship between circulating tumor cells and annexin A2 in early breast cancer patients. Anticancer Res. 2017, 37, 2727–2734. [Google Scholar] [CrossRef] [PubMed]
- Pi, F.; Zhang, H.; Li, H.; Thiviyanathan, V.; Gorenstein, D.G.; Sood, A.K.; Guo, P. RNA nanoparticles harboring annexin A2 aptamer can target ovarian cancer for tumor-specific doxorubicin delivery. Nanomed. Nanotechnol. Biol. Med. 2017, 13, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhao, Z.; Bai, H.; Fu, T.; Yang, C.; Hu, X.; Liu, Q.; Champanhac, C.; Teng, I.-T.; Ye, M.; et al. DNA Aptamer selected against pancreatic ductal adenocarcinoma for in vivo imaging and clinical tissue recognition. Theranostics 2015, 5, 985–994. [Google Scholar] [CrossRef] [PubMed]
- Bardeesy, N.; DePinho, R.A. Pancreatic cancer biology and genetics. Nat. Rev. Cancer 2002, 2, 897–909. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-X.; Knyazev, P.G.; Cheburkin, Y.V.; Sharma, K.; Knyazev, Y.P.; Orfi, L.; Szabadkai, I.; Daub, H.; Keri, G.; Ullrich, A. AXL is a potential target for therapeutic intervention in breast cancer progression. Cancer Res. 2008, 68, 1905–1915. [Google Scholar] [CrossRef] [PubMed]
- Shinh, Y.-S.; Lai, C.-Y.; Kao, Y.-R.; Shiah, S.-G.; Chu, Y.-W.; Lee, H.-S.; Wu, C.-W. Expression of AXL in lung adenocarcinoma and correlation with tumor progression. Neoplasia 2005, 7, 1058–1064. [Google Scholar] [CrossRef]
- Sun, W.; Fujimoto, J.; Tamaya, T. Coexpression of Gas6/AXL in human ovarian cancers. Oncology 2004, 66, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Koorstra, J.-B.M.; Karikari, C.A.; Feldmann, G.; Bisht, S.; Rojas, P.L.; Offerhaus, G.J.A.; Alvarez, H.; Maitra, A. The AXL receptor tyrosine kinase confers an adverse prognostic influence in pancreatic cancer and represents a new therapeutic target. Cancer Boil. Ther. 2009, 8, 618–626. [Google Scholar] [CrossRef]
- Sainaghi, P.P.; Castello, L.; Bergamasco, L.; Galletti, M.; Bellosta, P.; Avanzi, G.C. Gas6 induces proliferation in prostate carcinoma cell lines expressing the AXL receptor. J. Cell. Physiol. 2005, 204, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Sinha, S.; Boysen, J.; Nelson, M.; Secreto, C.; Warner, S.L.; Bearss, D.J.; Lesnick, C.; Shanafelt, T.D.; Kay, N.E.; Ghosh, A.K. Targeted AXL inhibition primes chronic lymphocytic leukemia b cells to apoptosis and shows synergistic/additive effects in combination with BTK inhibitors. Clin. Cancer Res. 2015, 21, 2115–2126. [Google Scholar] [CrossRef] [PubMed]
- Cerchia, L.; Esposito, C.L.; Camorani, S.; Rienzo, A.; Stasio, L.; Insabato, L.; Affuso, A.; de Franciscis, V. Targeting AXL with an high-affinity inhibitory aptamer. Mol. Ther. 2012, 20, 2291–2303. [Google Scholar] [CrossRef] [PubMed]
- Antony, J.; Tan, T.Z.; Kelly, Z.; Low, J.; Choolani, M.; Recchi, C.; Gabra, H.; Thiery, J.P.; Huang, R.Y.-J. The GAS6-AXL signaling network is a mesenchymal (Mes) molecular subtype-specific therapeutic target for ovarian cancer. Sci. Signal. 2016, 9, ra97. [Google Scholar] [CrossRef] [PubMed]
- Halmos, B.; Haura, E.B. New twists in the AXL(e) of tumor progression. Sci. Signal. 2016, 9, fs14. [Google Scholar] [CrossRef] [PubMed]
- Willis, S.; Villalobos, V.M.; Gevaert, O.; Abramovitz, M.; Williams, C.; Sikic, B.I.; Leyland-Jones, B. Single gene prognostic biomarkers in ovarian cancer: A meta-analysis. PLoS ONE 2016, 11, e0149183. [Google Scholar] [CrossRef] [PubMed]
- Rea, K.; Pinciroli, P.; Sensi, M.; Alciato, F.; Bisaro, B.; Lozneanu, L.; Raspagliesi, F.; Centritto, F.; Cabodi, S.; Defilippi, P.; et al. Novel AXL-driven signaling pathway and molecular signature characterize high-grade ovarian cancer patients with poor clinical outcome. Oncotarget 2015, 6, 30859. [Google Scholar] [PubMed]
- Kanlikilicer, P.; Ozpolat, B.; Aslan, B.; Bayraktar, R.; Gurbuz, N.; Rodriguez-Aguayo, C.; Bayraktar, E.; Denizli, M.; Gonzalez-Villasana, V.; Ivan, C.; et al. Hampering AXL tyrosine kinase mediated ovarian cancer metastasis via serum stable DNA-aptamer therapeutics. Mol. Ther. Nucleic Acids 2017, in press. [Google Scholar] [CrossRef]
- Liu, J.; Liu, H.; Sefah, K.; Liu, B.; Pu, Y.; van Simaeys, D.; Tan, W. Selection of Aptamers specific for adipose tissue. PLoS ONE 2012, 7, e37789. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Liu, J.; Tong, G.; Liu, B.; Wang, G.; Liu, H. Adipo8, a high-affinity DNA aptamer, can differentiate among adipocytes and inhibit intracellular lipid accumulation in vitro. Sci. China Chem. 2015, 58, 1612–1620. [Google Scholar] [CrossRef]
- Higashimoto, Y.; Matsui, T.; Nishino, Y.; Taira, J.; Inoue, H.; Takeuchi, M.; Yamagishi, S. Blockade by phosphorothioate aptamers of advanced glycation end products-induced damage in cultured pericytes and endothelial cells. Microvasc. Res. 2013, 90, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Byun, K.; Yoo, Y.; Son, M.; Lee, J.; Jeong, G.-B.; Park, Y.M.; Salekdeh, G.H.; Lee, B. Advanced glycation end-products produced systemically and by macrophages: A common contributor to inflammation and degenerative diseases. Pharmacol. Ther. 2017. [Google Scholar] [CrossRef] [PubMed]
- Maeda, S.; Matsui, T.; Ojima, A.; Suematsu, M.; Kaseda, K.; Higashimoto, Y.; Yamakawa, R.; Yamagishi, S. DNA aptamer raised against advanced glycation end products prevents abnormalities in electroretinograms of experimental diabetic retinopathy. Ophthalmic Res. 2015, 54, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Higashimoto, Y.; Yamagishi, S.; Nakamura, K.; Matsui, T.; Takeuchi, M.; Noguchi, M.; Inoue, H. In vitro selection of DNA aptamers that block toxic effects of AGE on cultured retinal pericytes. Microvasc. Res. 2007, 74, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Han, X.; Wang, Z.; Wu, S.; Zheng, R. Thioaptamer conjugated single-wall carbon nanotubes in human breast cancer targeted photothermal therapy in vivo in vivo and in vitro in vitro. Int. J. Clin. Exp. Med. 2016, 9, 58–64. [Google Scholar]
- Mu, Q.; Annapragada, A.; Srivastava, M.; Li, X.; Wu, J.; Thiviyanathan, V.; Wang, H.; Williams, A.; Gorenstein, D.; Annapragada, A.; et al. Conjugate-SELEX: A high-throughput screening of thioaptamer-liposomal nanoparticle conjugates for targeted intracellular delivery of anticancer drugs. Mol. Ther. Nucleic Acids 2016, 5, e382. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Elizondo-Riojas, M.-A.; Li, X.; Lokesh, G.L.R.; Somasunderam, A.; Thiviyanathan, V.; Volk, D.E.; Durland, R.H.; Englehardt, J.; Cavasotto, C.N.; et al. X-Aptamers: A bead-based selection method for random incorporation of drug-like moieties onto next-generation aptamers for enhanced binding. Biochem. 2012, 51, 8321–8323. [Google Scholar] [CrossRef] [PubMed]
- Thiel, W.H.; Bair, T.; Peek, A.S.; Liu, X.; Dassie, J.; Stockdale, K.R.; Behlke, M.A.; Miller, F.J., Jr.; Giangrande, P.H. Rapid identification of cell-specific, internalizing RNA aptamers with bioinformatics analyses of a cell-based aptamer selection. PLoS ONE 2012, 7, e43836. [Google Scholar] [CrossRef] [PubMed]
- Lu, E.; Elizondo-Riojas, M.-A.; Chang, J.T.; Volk, D.E. Aptaligner: Automated software for aligning pseudo-random DNA X-aptamers from next-generation sequencing data. Biochemistry 2014, 53, 3523–3525. [Google Scholar] [CrossRef] [PubMed]
- Lipi, F.; Chen, S.; Chakravarthy, M.; Rakesh, S.; Veedu, R.N. In vitro evolution of chemically-modified nucleic acid aptamers: Pros and cons, and comprehensive selection strategies. RNA Biol. 2016, 13, 1232–1245. [Google Scholar] [CrossRef] [PubMed]
- Lokesh, G.L.; Wang, H.; Lam, C.H.; Thiviyanathan, V.; Ward, N.; Gorenstein, D.G.; Volk, D.E. X-Aptamer Selection and Validation. In RNA Nanostructures: Methods and Protocols; Bindewald, E., Shapiro, B., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1632, in press. [Google Scholar]
- Kraemer, S.; Vaught, J.D.; Bock, C.; Gold, L.; Katilius, E.; Keeney, T.R.; Kim, N.; Saccomano, N.A.; Wilcox, S.K.; Zichi, D.; et al. From SOMAmer-based biomarker discovery to diagnostic and clinical applications: A SOMAmer-based, streamlined multiplex proteomic assay. PLoS ONE 2011, 6, e26332. [Google Scholar] [CrossRef] [PubMed]
- Mercier, M.-C.; Dontenwill, M.; Choulier, L. Selection of nucleic acid aptamers targeting tumor cell-surface protein biomarkers. Cancers 2017, 9, 69. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.E.; Wu, H.; Niu, Y.; Cai, J. Improving the stability of aptamers by chemical modification. Curr. Med. Chem. 2011, 18, 4126–4138. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ren, X.; Schluesener, H.J.; Zhang, Z. Aptamers: Selection, modification and application to nervous system diseases. Curr. Med. Chem. 2011, 18, 4159–4168. [Google Scholar] [CrossRef] [PubMed]
- Meyer, C.; Hahn, U.; Rentmeister, A. Cell-specific aptamers as emerging therapeutics. J. Nucleic Acids 2011, 2011, 1–18. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Volk, D.E.; Lokesh, G.L.R. Development of Phosphorothioate DNA and DNA Thioaptamers. Biomedicines 2017, 5, 41. https://doi.org/10.3390/biomedicines5030041
Volk DE, Lokesh GLR. Development of Phosphorothioate DNA and DNA Thioaptamers. Biomedicines. 2017; 5(3):41. https://doi.org/10.3390/biomedicines5030041
Chicago/Turabian StyleVolk, David E., and Ganesh L. R. Lokesh. 2017. "Development of Phosphorothioate DNA and DNA Thioaptamers" Biomedicines 5, no. 3: 41. https://doi.org/10.3390/biomedicines5030041
APA StyleVolk, D. E., & Lokesh, G. L. R. (2017). Development of Phosphorothioate DNA and DNA Thioaptamers. Biomedicines, 5(3), 41. https://doi.org/10.3390/biomedicines5030041