Genetic Variants and Soluble Isoforms of PD-1/PD-L1 as Novel Biomarkers for Pancreatic Ductal Adenocarcinoma (PDAC) Susceptibility and Prognosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Population
2.2. Genotyping of PD-1/PD-L1 Variants
2.3. Measurement of sPD-1/sPD-L1
2.4. Retrieving Genes Correlated with PDAC
2.5. Protein–Protein Interaction
2.6. Mapping of Kaplan–Meier Survival Curve of Mutant Genes and Screening of Prognostic Biomarkers
2.7. Statistical Analysis
3. Results
3.1. Study Population
3.2. Genotyping of PD-1/PD-L1 Variants
3.3. Measurements of sPD-1/sPD-L1
3.4. Genes Correlated with PDAC
3.5. Protein–Protein Interactions
- (1)
- Co-expression: This indicates that PD-1 and PD-L1 show similar expression patterns across multiple datasets or conditions, suggesting they may functionally cooperate.
- (2)
- Text mining: Associations are identified from published scientific literature, where PD-1 and PD-L1 are frequently mentioned together, implying a functional or biological relationship.
- (3)
- Experimentally determined: This evidence comes from laboratory studies that directly demonstrate the interaction between PD-1 and PD-L1, such as binding assays, co-immunoprecipitation, or crystallography.
- (4)
- Curated databases: Information is drawn from established biological databases that manually collect and verify known protein–protein interactions reported in the literature.
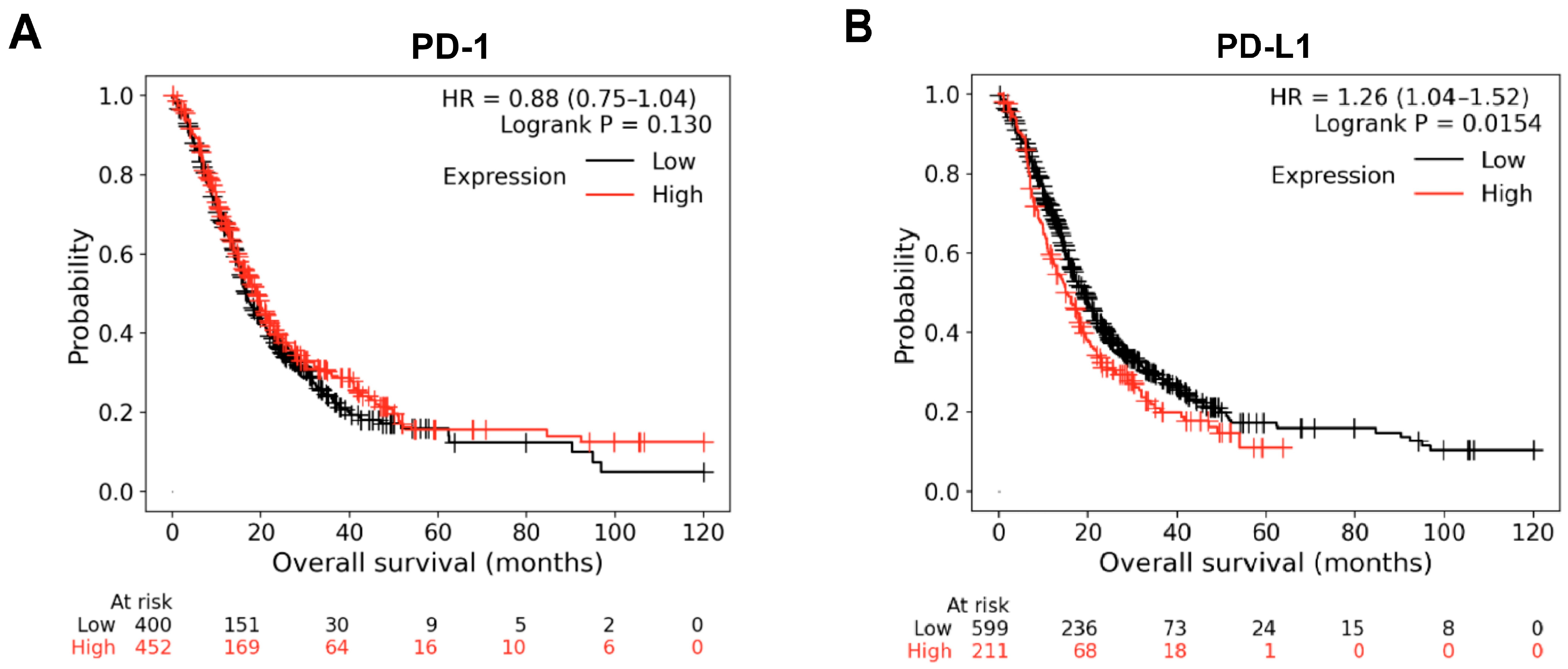
3.6. Kaplan–Meier Survival Curve Analysis of PD-1 and PD-L1 Genes and Screening of Prognostic Biomarkers
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| 3’UTR | 3′-untranslated region |
| CA19-9 | Carbohydrate antigen 19-9 |
| CEA | Carcinoembryonic antigen |
| HCC | Hepatocellular carcinoma |
| NSCLC | Non-small cell lung carcinoma |
| OR | Odds ratio |
| OS | Overall Survival |
| PD-1 | Programmed cell death protein-1 |
| PDAC | Pancreatic ductal adenocarcinoma |
| PD-L1 | Programmed cell death protein ligand 1 |
| SNPs | Single-nucleotide polymorphisms |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer Statistics, 2024. CA. Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef]
- Mukund, A.; Afridi, M.A.; Karolak, A.; Park, M.A.; Permuth, J.B.; Rasool, G. Pancreatic Ductal Adenocarcinoma (PDAC): A Review of Recent Advancements Enabled by Artificial Intelligence. Cancers 2024, 16, 2240. [Google Scholar] [CrossRef]
- Resell, M.; Rabben, H.L.; Sharma, A.; Hagen, L.; Hoang, L.; Skogaker, N.T.; Aarvik, A.; Bjåstad, E.K.; Svensson, M.K.; Amrutkar, M.; et al. Proteomics Profiling of Research Models for Studying Pancreatic Ductal Adenocarcinoma. Sci. Data 2025, 12, 266. [Google Scholar] [CrossRef]
- Khayat, S.; Choudhary, K.; Claude Nshimiyimana, J.; Gurav, J.; Hneini, A.; Nazir, A.; Chaito, H.; Wojtara, M.; Uwishema, O. Pancreatic Cancer: From Early Detection to Personalized Treatment Approaches. Ann. Med. Surg. 2024, 86, 2866–2872. [Google Scholar] [CrossRef]
- Boekestijn, B.; Feshtali, S.; Vasen, H.; van Leerdam, M.E.; Bonsing, B.A.; Mieog, J.S.D.; Wasser, M.N. Screening for Pancreatic Cancer in High-Risk Individuals Using MRI: Optimization of Scan Techniques to Detect Small Lesions. Fam. Cancer 2024, 23, 295–308. [Google Scholar] [CrossRef]
- Samir, S.; El-Ashry, M.; Soliman, W.; Hassan, M. Urinary Biomarkers Analysis as a Diagnostic Tool for Early Detection of Pancreatic Adenocarcinoma: Molecular Quantification Approach. Comput. Biol. Chem. 2024, 112, 108171. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.; Nasr, S.M.; Elzallat, M. Effect of CD133 Polymorphisms on the Risk of Developing Liver Cirrhosis and Hepatocellular Carcinoma Induced by Viral Hepatitis. Virus Res. 2022, 312, 198714. [Google Scholar] [CrossRef]
- Yu, B.; Shao, S.; Ma, W. Frontiers in Pancreatic Cancer on Biomarkers, Microenvironment, and Immunotherapy. Cancer Lett. 2025, 610, 217350. [Google Scholar] [CrossRef] [PubMed]
- Rabi, L.T.; Valente, D.Z.; de Souza Teixeira, E.; Peres, K.C.; de Oliveira Almeida, M.; Bufalo, N.E.; Ward, L.S. Potential New Cancer Biomarkers Revealed by Quantum Chemistry Associated with Bioinformatics in the Study of Selectin Polymorphisms. Heliyon 2024, 10, e28830. [Google Scholar] [CrossRef]
- Hassan, M.; Elzallat, M.; Mohammed, D.M.; Balata, M.; El-Maadawy, W.H. Exploiting Regulatory T Cells (Tregs): Cutting-Edge Therapy for Autoimmune Diseases. Int. Immunopharmacol. 2025, 155, 114624. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Nishimura, S.; Goto, K. PD-1/PD-L1 Immune Checkpoint in Bone and Soft Tissue Tumors (Review). Mol. Clin. Oncol. 2025, 22, 31. [Google Scholar] [CrossRef] [PubMed]
- Suminaga, K.; Nomizo, T.; Yoshida, H.; Ozasa, H. The Impact of PD-L1 Polymorphisms on the Efficacy of Immune Checkpoint Inhibitors Depends on the Tumor Proportion Score: A Retrospective Study. J. Cancer Res. Clin. Oncol. 2025, 151, 61. [Google Scholar] [CrossRef] [PubMed]
- Nussinov, R.; Yavuz, B.R.; Jang, H. Molecular Principles Underlying Aggressive Cancers. Signal Transduct. Target. Ther. 2025, 10, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Chen, S.; Chen, Y.; Lin, J.; Lin, J.; Wang, Y.; Liu, C.; Kang, M. Programmed Death-1 Polymorphisms Is Associated with Risk of Esophagogastric Junction Adenocarcinoma in the Chinese Han Population: A Case-Control Study Involving 2, 740 Subjects. Oncotarget 2017, 8, 39198–39208. [Google Scholar] [CrossRef] [PubMed]
- Ge, J.; Zhu, L.; Zhou, J.; Li, G.; Li, Y.; Li, S.; Wu, Z.; Rong, J.; Yuan, H.; Liu, Y.; et al. Association between Co-Inhibitory Molecule Gene Tagging Single Nucleotide Polymorphisms and the Risk of Colorectal Cancer in Chinese. J. Cancer Res. Clin. Oncol. 2015, 141, 1533–1544. [Google Scholar] [CrossRef]
- Li, F.; Fan, X.; Wang, X.; Deng, H.; Zhang, X.; Zhang, K.; Li, N.; Han, Q.; Lv, Y.; Liu, Z. Genetic Association and Interaction of PD1 and TIM3 Polymorphisms in Susceptibility of Chronic Hepatitis B Virus Infection and Hepatocarcinogenesis. Discov. Med. 2019, 27, 79–92. [Google Scholar]
- Huang, K.; Hu, E.; Li, W.; Lv, J.; He, Y.; Deng, G.; Xiao, J.; Yang, C.; Zhao, X.; Chen, L.; et al. Association of PD-1 Polymorphisms with the Risk and Prognosis of Lung Adenocarcinoma in the Northeastern Chinese Han Population. BMC Med. Genet. 2019, 20, 177. [Google Scholar] [CrossRef]
- Zou, J.; Wu, D.; Li, T.; Wang, X.; Liu, Y.; Tan, S. Association of PD-L1 Gene Rs4143815 C>G Polymorphism and Human Cancer Susceptibility: A Systematic Review and Meta-Analysis. Pathol. Res. Pract. 2019, 215, 229–234. [Google Scholar] [CrossRef]
- Tan, D.; Sheng, L.; Yi, Q.H. Correlation of PD-1/PD-L1 Polymorphisms and Expressions with Clinicopathologic Features and Prognosis of Ovarian Cancer. Cancer Biomark. 2018, 21, 287–297. [Google Scholar] [CrossRef]
- Gong, Q.; Qie, H.L.; Dong, S.Y.; Jiang, H.T. Implication of PD-L1 Polymorphisms Rs2297136 on Clinical Outcomes of Patients with Advanced NSCLC Who Received PD-1 Blockades: A Retrospective Exploratory Study. Oncol. Lett. 2024, 27, 144. [Google Scholar] [CrossRef]
- Dragu, D.; Necula, L.G.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Soluble PD-L1: From Immune Evasion to Cancer Therapy. Life 2025, 15, 626. [Google Scholar] [CrossRef]
- Ćeriman Krstić, V.; Jovanović, D.; Samardžić, N.; Gajić, M.; Kotur Stevuljević, J.; Klisic, A.; Soldatović, I.; Radončić, D.; Roksandić Milenković, M.; Šeha, B.; et al. The Potential Role of SPD-L1 as a Predictive Biomarker in EGFR-Positive Non-Small-Cell Lung Cancer. Curr. Issues Mol. Biol. 2025, 47, 45. [Google Scholar] [CrossRef] [PubMed]
- Abu Hejleh, T.; Furqan, M.; Ballas, Z.; Clamon, G. The Clinical Significance of Soluble PD-1 and PD-L1 in Lung Cancer. Crit. Rev. Oncol. Hematol. 2019, 143, 148–152. [Google Scholar] [CrossRef]
- Zang, B.; Chen, C.; Zhao, J.Q. PD-1 Gene Rs10204525 and Rs7421861 Polymorphisms Are Associated with Increased Risk and Clinical Features of Esophageal Cancer in a Chinese Han Population. Aging 2020, 12, 3771–3790. [Google Scholar] [CrossRef]
- Safran, M.; Dalah, I.; Alexander, J.; Rosen, N.; Iny Stein, T.; Shmoish, M.; Nativ, N.; Bahir, I.; Doniger, T.; Krug, H.; et al. GeneCards Version 3: Tthe Human Gene Integrator. Database 2010, 2010, baq020. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING Database in 2023: Protein-Protein Association Networks and Functional Enrichment Analyses for Any Sequenced Genome of Interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Győrffy, B. Integrated Analysis of Public Datasets for the Discovery and Validation of Survival-Associated Genes in Solid Tumors. Innovation 2024, 5, 100625. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Khan, N.U.; Gu, W.; Lei, H.; Goel, A.; Chen, T. Informatics Strategies for Early Detection and Risk Mitigation in Pancreatic Cancer Patients. Neoplasia 2025, 60, 101129. [Google Scholar] [CrossRef]
- Tang, W.; Chen, Y.; Chen, S.; Sun, B.; Gu, H.; Kang, M. Programmed Death-1 (PD-1) Polymorphism Is Associated with Gastric Cardia Adenocarcinoma. Int. J. Clin. Exp. Med. 2015, 8, 8086–8093. [Google Scholar]
- Hashemi, M.; Karami, S.; Sarabandi, S.; Moazeni-Roodi, A.; Małecki, A.; Ghavami, S.; Wiechec, E. Association between PD-1 and PD-L1 Polymorphisms and the Risk of Cancer: A Meta-Analysis of Case-Control Studies. Cancers 2019, 11, 1150. [Google Scholar] [CrossRef] [PubMed]
- Kuol, N.; Yan, X.; Barriga, V.; Karakkat, J.; Vassilaros, S.; Fyssas, I.; Tsimpanis, A.; Fraser, S.; Nurgali, K.; Apostolopoulos, V. Pilot Study: Immune Checkpoints Polymorphisms in Greek Primary Breast Cancer Patients. Biomedicines 2022, 10, 1827. [Google Scholar] [CrossRef]
- Ren, H.T.; Li, Y.M.; Wang, X.J.; Kang, H.F.; Jin, T.B.; Ma, X.B.; Liu, X.H.; Wang, M.; Liu, K.; Xu, P.; et al. PD-1 Rs2227982 Polymorphism Is Associated with the Decreased Risk of Breast Cancer in Northwest Chinese Women. Medicine 2016, 95, e3760. [Google Scholar] [CrossRef]
- Du, W.; Zhu, J.; Chen, Y.; Zeng, Y.; Shen, D.; Zhang, N.; Ning, W.; Liu, Z.; Huang, J.A. Variant SNPs at the MicroRNA Complementary Site in the B7-H1 3′-Untranslated Region Increase the Risk of Non-Small Cell Lung Cancer. Mol. Med. Rep. 2017, 16, 2682–2690. [Google Scholar] [CrossRef]
- Wu, Y.; Zhao, T.; Jia, Z.; Cao, D.; Cao, X.; Pan, Y.; Zhao, D.; Zhang, B.; Jiang, J. Polymorphism of the Programmed Death-Ligand 1 Gene Is Associated with Its Protein Expression and Prognosis in Gastric Cancer. J. Gastroenterol. Hepatol. 2019, 34, 1201–1207. [Google Scholar] [CrossRef]
- Catalano, C.; da Silva Filho, M.I.; Frank, C.; Jiraskova, K.; Vymetalkova, V.; Levy, M.; Liska, V.; Vycital, O.; Naccarati, A.; Vodickova, L.; et al. Investigation of Single and Synergic Effects of NLRC5 and PD-L1 Variants on the Risk of Colorectal Cancer. PLoS ONE 2018, 13, e0192385. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, F.; Mao, Y.; Zhou, H.; Sun, J.; Li, R.; Liu, C.; Chen, W.; Hua, D.; Zhang, X. A MiR-570 Binding Site Polymorphism in the B7-H1 Gene Is Associated with the Risk of Gastric Adenocarcinoma. Hum. Genet. 2013, 132, 641–648. [Google Scholar] [CrossRef]
- Xie, Q.; Chen, Z.; Xia, L.; Zhao, Q.; Yu, H.; Yang, Z. Correlations of PD-L1 Gene Polymorphisms with Susceptibility and Prognosis in Hepatocellular Carcinoma in a Chinese Han Population. Gene 2018, 674, 188–194. [Google Scholar] [CrossRef]
- Hassan, M.; Attia, M.S.; Ali-Eldin, Z.; El Attar, G.; Elzallat, M.; Saad, H.H.K.; Isaac, A. Programmed Death-Ligand 1 (PD-L1) Polymorphisms as Predictive Biomarkers for the Development of Liver Cirrhosis and Hepatocellular Carcinoma in HCV Egyptian Patients. Tumour Virus Res. 2022, 14, 200249. [Google Scholar] [CrossRef]
- Qian, C.; Guo, H.; Chen, X.; Shi, A.; Li, S.; Wang, X.; Pan, J.; Fang, C. Association of PD-1 and PD-L1 Genetic Polymorphyisms with Type 1 Diabetes Susceptibility. J. Diabetes Res. 2018, 2018, 1614683. [Google Scholar] [CrossRef] [PubMed]
- Karami, S.; Sattarifard, H.; Kiumarsi, M.; Sarabandi, S.; Taheri, M.; Hashemi, M.; Bahari, G.; Ghavami, S. Evaluating the Possible Association between PD-1 (Rs11568821, Rs2227981, Rs2227982) and PD-L1 (Rs4143815, Rs2890658) Polymorphisms and Susceptibility to Breast Cancer in a Sample of Southeast Iranian Women. Asian Pac. J. Cancer Prev. 2020, 21, 3115–3123. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.M.; Li, Y.; Liu, J.H.; Wang, N.; Huang, X.; Cao, S.R.; Shan, B.E. Programmed Death-1 Ligand-1 Gene Rs2890658 Polymorphism Associated with the Risk of Esophageal Squamous Cell Carcinoma in Smokers. Cancer Biomark. 2017, 21, 65–71. [Google Scholar] [CrossRef]
- Shao, W.; Xu, Y.; Lin, S.; Gao, J.; Gao, J.; Wang, H. The Potential of Soluble Programmed Death-Ligand 1 (SPD-L1) as a Diagnosis Marker for Colorectal Cancer. Front. Oncol. 2022, 12, 988567. [Google Scholar] [CrossRef]
- He, Y.; Zhang, X.; Zhu, M.; He, W.; Hua, H.; Ye, F.; Zhou, X.; Chen, N.; Li, Y.; Zhong, W.; et al. Soluble PD-L1: A Potential Dynamic Predictive Biomarker for Immunotherapy in Patients with Proficient Mismatch Repair Colorectal Cancer. J. Transl. Med. 2023, 21, 1–13. [Google Scholar] [CrossRef]
- Chang, B.; Huang, T.; Wei, H.; Shen, L.; Zhu, D.; He, W.; Chen, Q.; Zhang, H.; Li, Y.; Huang, R.; et al. The Correlation and Prognostic Value of Serum Levels of Soluble Programmed Death Protein 1 (SPD-1) and Soluble Programmed Death-Ligand 1 (SPD-L1) in Patients with Hepatocellular Carcinoma. Cancer Immunol. Immunother. 2019, 68, 353–363. [Google Scholar] [CrossRef]
- Ugurel, S.; Schadendorf, D.; Horny, K.; Sucker, A.; Schramm, S.; Utikal, J.; Pföhler, C.; Herbst, R.; Schilling, B.; Blank, C.; et al. Elevated Baseline Serum PD-1 or PD-L1 Predicts Poor Outcome of PD-1 Inhibition Therapy in Metastatic Melanoma. Ann. Oncol. 2020, 31, 144–152. [Google Scholar] [CrossRef]
- Costantini, A.; Julie, C.; Dumenil, C.; Hélias-Rodzewicz, Z.; Tisserand, J.; Dumoulin, J.; Giraud, V.; Labrune, S.; Chinet, T.; Emile, J.F.; et al. Predictive Role of Plasmatic Biomarkers in Advanced Non-Small Cell Lung Cancer Treated by Nivolumab. Oncoimmunology 2018, 7, e1452581. [Google Scholar] [CrossRef]
- Mahoney, K.M.; Ross-Macdonald, P.; Yuan, L.; Song, L.; Veras, E.; Wind-Rotolo, M.; Mcdermott, D.F.; Stephen Hodi, F.; Choueiri, T.K.; Freeman, G.J. Soluble PD-L1 as an Early Marker of Progressive Disease on Nivolumab. J. Immunother. Cancer 2022, 10, e003527. [Google Scholar] [CrossRef]
- Cheng, H.Y.; Kang, P.J.; Chuang, Y.H.; Wang, Y.H.; Jan, M.C.; Wu, C.F.; Lin, C.L.; Liu, C.J.; Liaw, Y.F.; Lin, S.M.; et al. Circulating Programmed Death-1 as a Marker for Sustained High Hepatitis b Viral Load and Risk of Hepatocellular Carcinoma E95870. PLoS ONE 2014, 9, e95870. [Google Scholar] [CrossRef] [PubMed]
- Sorensen, S.F.; Demuth, C.; Weber, B.; Sorensen, B.S.; Meldgaard, P. Increase in Soluble PD-1 Is Associated with Prolonged Survival in Patients with Advanced EGFR-Mutated Non-Small Cell Lung Cancer Treated with Erlotinib. Lung Cancer 2016, 100, 77–84. [Google Scholar] [CrossRef] [PubMed]




| Control (n = 150) | PDAC (n = 150) | p Value | |
|---|---|---|---|
| Age (year) | 58.38 ± 0.60 | 59.39 ± 0.92 | 0.18 |
| Gender | |||
| Males | 91 (60.70%) | 100 (66.70%) | 0.28 |
| Females | 59 (39.30%) | 50 (33.30%) | |
| Smoking | |||
| No | 75 (50.00%) | 84 (56.00%) | 0.30 |
| Yes | 75 (50.00%) | 66 (44.00%) | |
| Routine Lab. | |||
| ALT (IU/L) | 22.78 ± 1.12 | 84.61 ± 9.93 | <0.001 |
| AST (IU/L) | 25.61 ± 0.98 | 71.57 ± 5.29 | <0.001 |
| Total bilirubin (mg/dL) | 0.60 ± 0.02 | 8.06 ± 0.47 | <0.001 |
| Albumin (g/dL) | 3.91 ± 0.03 | 3.71 ± 0.22 | <0.001 |
| Urea (mg/dL) | 26.15 ± 0.82 | 40.03 ± 1.91 | <0.001 |
| Creatinine (mg/dL) | 0.94 ± 0.02 | 1.20 ± 0.04 | <0.001 |
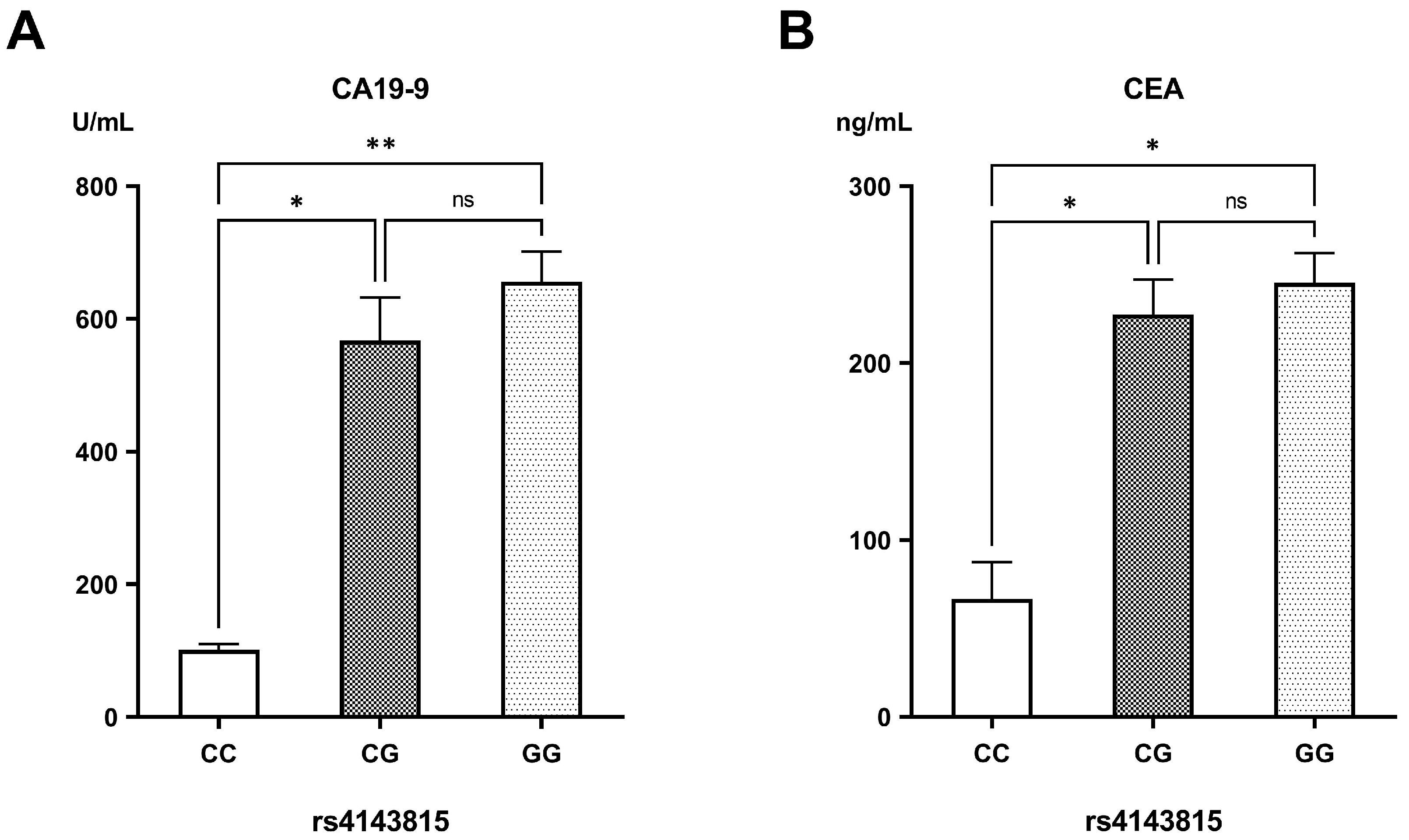
| CA19-9 (U/mL) | 18.44 ± 0.95 | 618.43 ± 35.83 | <0.001 |
| CEA (ng/mL) | 2.75 ± 0.17 | 229.01 ± 12.73 | <0.001 |
| Radiological grading | |||
| Grade 1 | 36 (24.00%) | ||
| Grade 2 | 46 (30.70%) | ||
| Grade 3 | 68 (45.30%) | ||
| Clinical Grading of PDAC | p Value | |||
|---|---|---|---|---|
| Grade 1 | Grade 2 | Grade 3 | ||
| CA19-9 (U/mL) | 508.53 ± 56.43 | 567.75 ± 49.37 | 690.28 ± 66.34 | 0.113 |
| CEA (ng/mL) | 201.72 ± 26.93 | 231.89 ± 20.65 | 251.67 ± 19.97 | 0.305 |
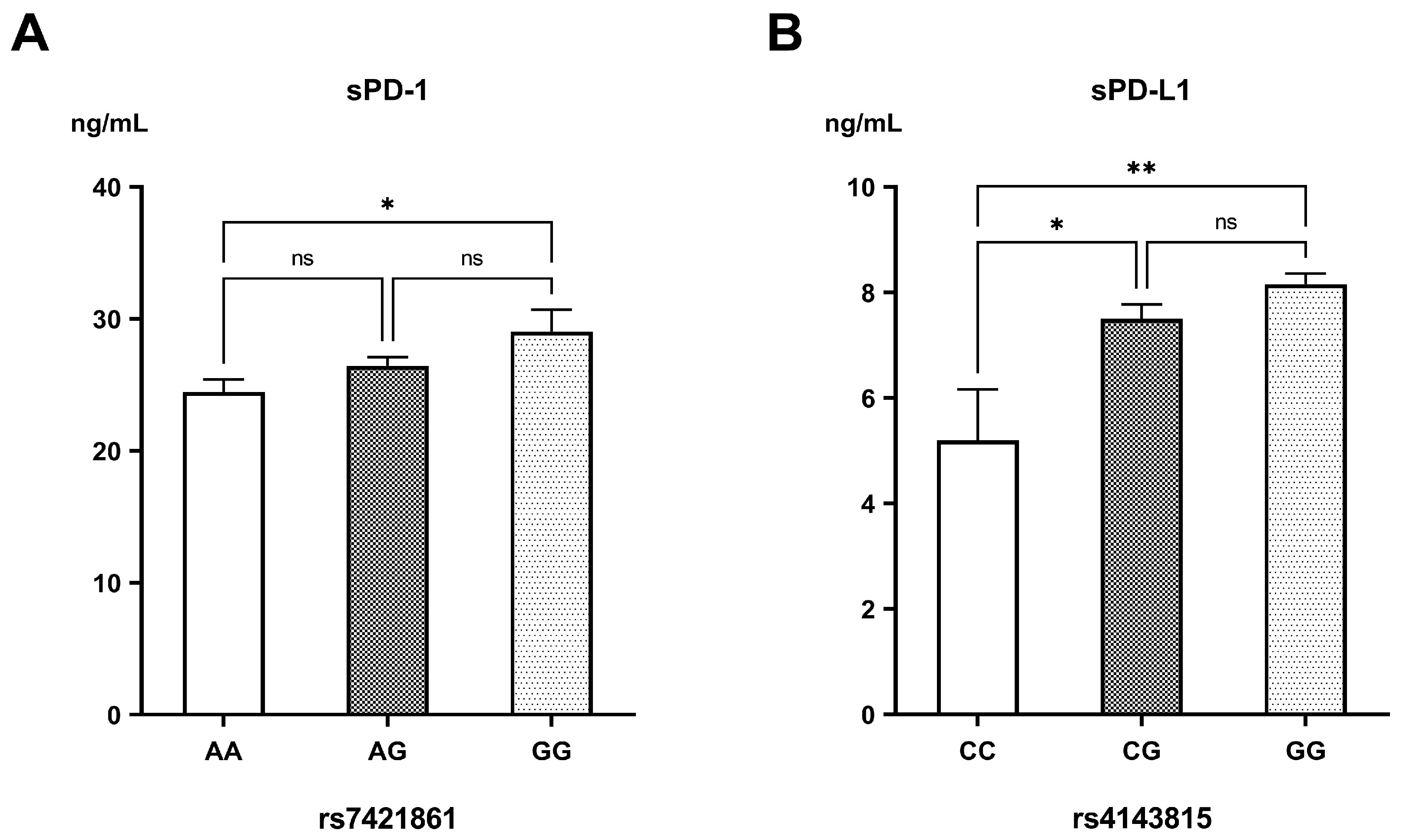
| PD-1 rs7421861 | ||||
| AA (reference) | 21 (58.30%) a | 7 (15.20%) b | 16 (23.50%) b | <0.001 |
| AG (heterozygous) | 13 (36.10%) a | 33 (71.70%) b | 41 (60.30%) ab | |
| GG (variant) | 2 (5.60%) a | 6 (13.00%) a | 11 (16.20%) a | |
| A | 55 (76.40%) a | 47 (51.10%) b | 73 (53.70%) b | 0.002 |
| G | 17 (23.60%) a | 45 (48.90%) b | 63 (46.30%) b | |
| PD-L1 rs2297136 | ||||
| AA (reference) | 12 (33.30%) a | 11 (23.90%) a | 22 (32.40%) a | 0.697 |
| AG (heterozygous) | 24 (26.70%) a | 34 (73.90%) a | 44 (64.70%) a | |
| GG (variant) | 0 (0.00%) a | 1 (2.20%) a | 2 (2.90%) a | |
| A | 48 (66.70%) a | 56 (60.90%) a | 88 (64.70%) a | 0.725 |
| G | 24 (33.30%) a | 36 (39.10%) a | 48 (35.30%) a | |
| PD-L1 rs4143815 | ||||
| CC (reference) | 4 (11.10%) a | 1 (2.20%) ab | 0 (0.00%) b | <0.001 |
| CG (heterozygous) | 17 (47.20%) a | 19 (41.30%) a | 12 (17.60%) b | |
| GG (variant) | 15 (41.70%) a | 26 (56.50%) a | 56 (82.40%) b | |
| C | 25 (34.70%) a | 21 (22.80%) a | 12 (8.80%) b | <0.001 |
| G | 47 (65.30%) a | 71 (77.20%) a | 124 (91.20%) b | |
| Control (n = 150) | PDAC (n = 150) | p Value | Odds Ratio | Confidence Interval | |
|---|---|---|---|---|---|
| Genotype AA (reference) AG (heterozygous) GG (variant) | 82 (54.70%) 59 (39.30%) 9 (6.00%) | 44 (29.30%) 87 (58.0%) 19 (12.70%) | <0.001 | ||
| Genotype AA AG + GG | 82 (54.70%) 68 (45.30%) | 44 (29.30%) 106 (70.70%) | <0.001 | 0.34 | 0.21–0.55 |
| Genotype AG AA + GG | 59 (39.30%) 91 (60.70%) | 87 (58.0%) 63 (42.0%) | 0.001 | 2.130 | 1.34–3.38 |
| Genotype GG AA + AG | 9 (6.0%) 141 (94.0%) | 19 (12.70%) 131 (87.30%) | 0.047 | 2.27 | 0.99–5.20 |
| Allele A (reference) G (variant) | 223 (74.30%) 77 (25.70%) | 175 (58.30%) 125 (41.70%) | <0.001 | 0.48 2.07 | 0.34–0.68 1.46–2.92 |
| Control (n = 150) | PDAC (n = 150) | p Value | Odds Ratio | Confidence Interval | |
|---|---|---|---|---|---|
| Genotype AA (reference) AG (heterozygous) GG (variant) | 35 (23.30%) 86 (57.30%) 29 (19.30%) | 45 (30.0%) 102 (68.0%) 3 (2.0%) | <0.001 | ||
| Genotype AA AG + GG | 35 (23.30%) 115 (76.70%) | 45 (30.0%) 105 (70.0%) | 0.192 | 1.4 | 0.84–2.36 |
| Genotype AG AA + GG | 86 (57.30%) 64 (42.70%) | 102 (68.0%) 48 (32.0%) | 0.056 | 1.58 | 0.987–2.53 |
| Genotype GG AA + AG | 29 (19.30%) 121 (80.70%) | 3 (2.0%) 147 (98.0%) | <0.001 | 0.09 | 0.03–0.29 |
| Allele A (reference) G (variant) | 156 (52.00%) 144 (48.00%) | 192 (64.00%) 108 (36.00%) | 0.003 | 1.64 0.61 | 1.18–2.28 0.44–0.85 |
| Control (n = 150) | PDAC (n = 150) | p Value | Odds Ratio | Confidence Interval | |
|---|---|---|---|---|---|
| Genotype CC (reference) CG (heterozygous) GG (variant) | 44 (29.30%) 67 (44.70%) 39 (26.00%) | 5 (3.30%) 48 (32.0%) 97 (64.70%) | <0.001 | ||
| Genotype CC CG + GG | 44 (29.30%) 106 (70.70%) | 5 (3.30%) 145 (96.70%) | <0.001 | 0.083 | 0.03–0.22 |
| Genotype CG CC + GG | 67 (44.70%) 83 (55.30%) | 48 (32.0%) 102 (68.0%) | 0.024 | 0.58 | 0.36–0.93 |
| Genotype GG CC + CG | 39 (26.00%) 111 (74.00%) | 97 (64.70%) 53 (35.30%) | <0.001 | 5.209 | 3.18–8.55 |
| Allele C (reference) G (variant) | 155 (51.70%) 145 (48.30%) | 58 (19.30%) 242 (80.70%) | <0.001 | 0.22 4.46 | 0.16–0.32 3.10–6.43 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, M.; El-Maadawy, W.H.; Elhusseny, Y.; Agamy, F.E.; Fahim, S.A.; Balata, M. Genetic Variants and Soluble Isoforms of PD-1/PD-L1 as Novel Biomarkers for Pancreatic Ductal Adenocarcinoma (PDAC) Susceptibility and Prognosis. Biomedicines 2025, 13, 2246. https://doi.org/10.3390/biomedicines13092246
Hassan M, El-Maadawy WH, Elhusseny Y, Agamy FE, Fahim SA, Balata M. Genetic Variants and Soluble Isoforms of PD-1/PD-L1 as Novel Biomarkers for Pancreatic Ductal Adenocarcinoma (PDAC) Susceptibility and Prognosis. Biomedicines. 2025; 13(9):2246. https://doi.org/10.3390/biomedicines13092246
Chicago/Turabian StyleHassan, Marwa, Walaa H. El-Maadawy, Yasmine Elhusseny, Fatma Elbatol Agamy, Sally A. Fahim, and Mahmoud Balata. 2025. "Genetic Variants and Soluble Isoforms of PD-1/PD-L1 as Novel Biomarkers for Pancreatic Ductal Adenocarcinoma (PDAC) Susceptibility and Prognosis" Biomedicines 13, no. 9: 2246. https://doi.org/10.3390/biomedicines13092246
APA StyleHassan, M., El-Maadawy, W. H., Elhusseny, Y., Agamy, F. E., Fahim, S. A., & Balata, M. (2025). Genetic Variants and Soluble Isoforms of PD-1/PD-L1 as Novel Biomarkers for Pancreatic Ductal Adenocarcinoma (PDAC) Susceptibility and Prognosis. Biomedicines, 13(9), 2246. https://doi.org/10.3390/biomedicines13092246





