Prognostic Impact of KRAS and SMARCA4 Mutations and Co-Mutations on Survival in Non-Small Cell Lung Cancer: Insights from the AACR GENIE BPC Dataset
Abstract
1. Introduction
2. Materials and Methods
Statistical Methods
3. Results
3.1. Clinical and Genomic Characteristics
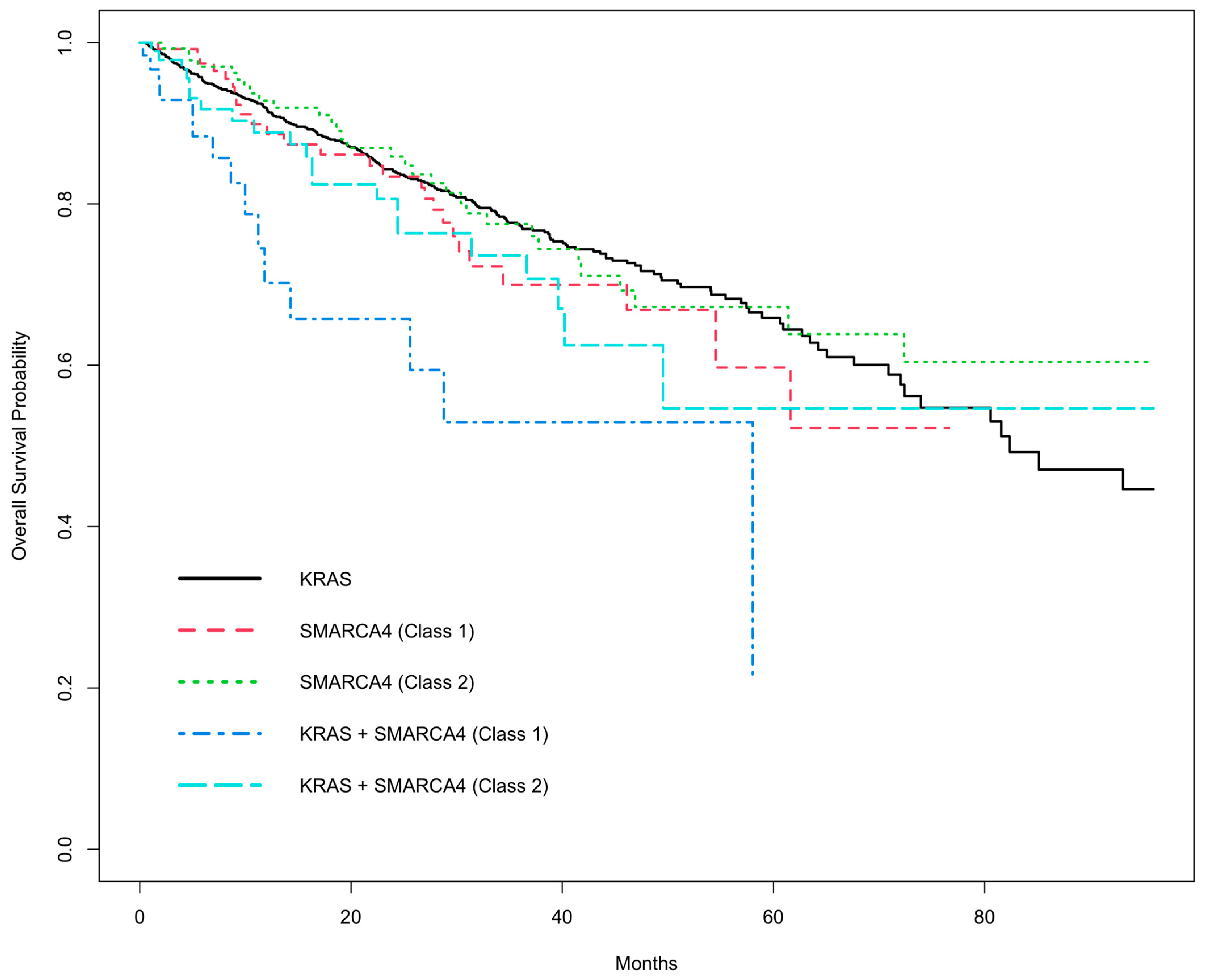
3.2. Univariate OS Analysis
3.3. Multivariate Analysis
4. Discussion
4.1. Biological Insights
4.2. Emerging Therapeutic Strategies for SMARCA4-Mutated NSCLC
4.3. Clinical Implications
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Health Organization. Lung Cancer. Available online: https://www.who.int/news-room/fact-sheets/detail/lung-cancer (accessed on 13 November 2024).
- Thandra, K.; Barsouk, A.; Saginala, K.; Aluru, J.S.; Barsouk, A. Epidemiology of lung cancer. Contemp. Oncol./Współczesna Onkol. 2021, 25, 45–52. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Ganti, A.K.; Klein, A.B.; Cotarla, I.; Seal, B.; Chou, E. Update of Incidence, Prevalence, Survival, and Initial Treatment in Patients With Non-Small Cell Lung Cancer in the US. JAMA Oncol. 2021, 7, 1824–1832. [Google Scholar] [CrossRef]
- Surveillance Research Program, National Cancer Institute. SEER*Explorer: An Interactive Website for SEER Cancer Statistics. Available online: https://seer.cancer.gov/statistics-network/explorer/ (accessed on 23 November 2024).
- Soria, J.C.; Ohe, Y.; Vansteenkiste, J.; Reungwetwattana, T.; Chewaskulyong, B.; Lee, K.H.; Dechaphunkul, A.; Imamura, F.; Nogami, N.; Kurata, T.; et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 378, 113–125. [Google Scholar] [CrossRef]
- Jänne, P.A.; Riely, G.J.; Gadgeel, S.M.; Heist, R.S.; Ou, S.I.; Pacheco, J.M.; Johnson, M.L.; Sabari, J.K.; Leventakos, K.; Yau, E.; et al. Adagrasib in Non-Small-Cell Lung Cancer Harboring a KRASG12C Mutation. N. Engl. J. Med. 2022, 387, 120–131. [Google Scholar] [CrossRef]
- Jordan, E.J.; Kim, H.R.; Arcila, M.E.; Barron, D.; Chakravarty, D.; Gao, J.; Chang, M.T.; Ni, A.; Kundra, R.; Jonsson, P.; et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017, 7, 596–609. [Google Scholar] [CrossRef]
- Pan, W.; Yang, Y.; Zhu, H.; Zhang, Y.; Zhou, R.; Sun, X. KRAS Mutation Is a Weak, but Valid Predictor for Poor Prognosis and Treatment Outcomes in NSCLC: A Meta-Analysis of 41 Studies. Oncotarget 2016, 7, 8373–8388. [Google Scholar] [CrossRef]
- Manolakos, P.; Ward, L.D. A Critical Review of the Prognostic and Predictive Implications of KRAS and STK11 Mutations and Co-Mutations in Metastatic Non-Small Lung Cancer. J. Pers. Med. 2023, 13, 1010. [Google Scholar] [CrossRef] [PubMed]
- Skoulidis, F.; Goldberg, M.E.; Greenawalt, D.M.; Hellmann, M.D.; Awad, M.M.; Gainor, J.F.; Schrock, A.B.; Hartmaier, R.J.; Trabucco, S.E.; Gay, L.; et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS-Mutant Lung Adenocarcinoma. Cancer Discov. 2018, 8, 822–835. [Google Scholar] [CrossRef] [PubMed]
- Dagogo-Jack, I.; Schrock, A.B.; Kem, M.; Jessop, N.; Lee, J.; Ali, S.M.; Ross, J.S.; Lennerz, J.K.; Shaw, A.T.; Mino-Kenudson, M.; et al. Clinicopathologic Characteristics of BRG1-Deficient NSCLC. J. Thorac. Oncol. 2020, 15, 766–776. [Google Scholar] [CrossRef]
- Schoenfeld, A.J.; Bandlamudi, C.; Lavery, J.A.; Montecalvo, J.; Namakydoust, A.; Rizvi, H.; Egger, J.; Concepcion, C.P.; Paul, S.; Arcila, M.E.; et al. The Genomic Landscape of SMARCA4 Alterations and Associations with Outcomes in Patients with Lung Cancer. Clin. Cancer Res. 2020, 26, 5701–5708. [Google Scholar] [CrossRef]
- Herzberg, B.; Gandhi, N.; Henick, B.; Xiu, J.; Vanderwalde, A.; Reuss, J.; Murray, J.; Concepcion-Crisol, C. Effects of Mutations in SWI/SNF Subunits on Context-Specific Prognosis in Driver Positive and Driver Negative NSCLC. In Proceedings of the 2023 American Society of Clinical Oncology Annual Meeting, Chicago, IL, USA, 2–6 June 2023. Abstract 9039. [Google Scholar]
- Liang, X.; Gao, X.; Wang, F.; Li, S.; Zhou, Y.; Guo, P.; Meng, Y.; Lu, T. Clinical Characteristics and Prognostic Analysis of SMARCA4-Deficient Non-Small Cell Lung Cancer. Cancer Med. 2023, 12, 14171–14182. [Google Scholar] [CrossRef]
- Manolakos, P.; Boccuto, L.; Ivankovic, D.S. A Critical Review of the Impact of SMARCA4 Mutations on Survival Outcomes in Non-Small Cell Lung Cancer. J. Pers. Med. 2024, 14, 684. [Google Scholar] [CrossRef]
- Hodges, H.; Stanton, B.; Cermakova, K.; Chang, C.; Miller, E.; Kirkland, J.; Ku, W.; Veverka, V.; Zhao, K.; Crabtree, G. Dominant-Negative SMARCA4 Mutants Alter the Accessibility Landscape of Tissue-Unrestricted Enhancers. Nat. Struct. Mol. Biol. 2018, 25, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Mardinian, K.; Adashek, J.J.; Botta, G.P.; Kato, S.; Kurzrock, R. SMARCA4: Implications of an Altered Chromatin-Remodeling Gene for Cancer Development and Therapy. Mol. Cancer Ther. 2021, 20, 2341–2351. [Google Scholar] [CrossRef] [PubMed]
- Fernando, T.M.; Piskol, R.; Bainer, R.; Sokol, E.S.; Trabucco, S.E.; Zhang, Q.; Trinh, H.; Maund, S.; Kschonsak, M.; Chaudhuri, S.; et al. Functional Characterization of SMARCA4 Variants Identified by Targeted Exome-Sequencing of 131,668 Cancer Patients. Nat. Commun. 2020, 11, 5551. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, H.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef]
- Alessi, J.V.; Ricciuti, B.; Spurr, L.F.; Gupta, H.; Li, Y.Y.; Glass, C.; Nishino, M.; Cherniack, A.D.; Lindsay, J.; Sharma, B.; et al. SMARCA4 and Other SWItch/Sucrose Non-Fermentable Family Genomic Alterations in NSCLC: Clinicopathologic Characteristics and Outcomes to Immune Checkpoint Inhibition. J. Thorac. Oncol. 2021, 16, 1176–1187. [Google Scholar] [CrossRef]
- Alessi, J.V.; Elkrief, A.; Ricciuti, B.; Wang, X.; Cortellini, A.; Vaz, V.R.; Lamberti, G.; Frias, R.L.; Venkatraman, D.; Fulgenzi, C.A.; et al. Clinicopathologic and Genomic Factors Impacting Efficacy of First-Line Chemoimmunotherapy in Advanced NSCLC. J. Thorac. Oncol. 2023, 18, 731–743. [Google Scholar] [CrossRef]
- Negrao, M.V.; Araujo, H.A.; Lamberti, G.; Cooper, A.J.; Akhave, N.S.; Zhou, T.; Delasos, L.; Hicks, J.K.; Aldea, M.; Minuti, G.; et al. Comutations and KRASG12C Inhibitor Efficacy in Advanced NSCLC. Cancer Discov. 2023, 13, 1556–1571. [Google Scholar] [CrossRef]
- Boiarsky, D.; Lydon, C.A.; Chambers, E.S.; Sholl, L.M.; Nishino, M.; Skoulidis, F.; Heymach, J.; Luo, J.; Awad, M.; Janne, P.; et al. Molecular Markers of Metastatic Disease in KRAS-Mutant Lung Adenocarcinoma. Ann. Oncol. 2023, 34, 589–604. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ahmed, T.; Petty, W.J.; Grant, S.; Ruiz, J.; Lycan, T.W.; Topaloglu, U.; Chou, P.; Miller, L.D.; Hawkins, G.A.; et al. SMARCA4 Mutations in KRAS-Mutant Lung Adenocarcinoma: A Multi-Cohort Analysis. Mol. Oncol. 2021, 15, 462–472. [Google Scholar] [CrossRef]
- Choudhury, N.J.; Lavery, J.A.; Brown, S.; de Bruijn, I.; Jee, J.; Tran, T.N.; Rizvi, H.; Arbour, K.C.; Whiting, K.; Shen, R.; et al. The GENIE BPC NSCLC Cohort: A Real-World Repository Integrating Standardized Clinical and Genomic Data for 1,846 Patients with Non-Small Cell Lung Cancer. Clin. Cancer Res. 2023, 29, 3418–3428. [Google Scholar] [CrossRef]
- AACR Project GENIE Consortium. AACR Project GENIE: Powering Precision Medicine Through an International Consortium. Cancer Discov. 2017, 7, 818–831. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Lavery, J.A.; Lepisto, E.M.; Brown, S.; Rizvi, H.; McCarthy, C.; LeNoue-Newton, M.; Yu, C.; Lee, J.; Guo, X.; Yu, T.; et al. A Scalable Quality Assurance Process for Curating Oncology Electronic Health Records: The Project GENIE Biopharma Collaborative Approach. JCO Clin. Cancer Inform. 2022, 6, e2100105. [Google Scholar] [CrossRef]
- Wankhede, D.; Grover, S.; Hofman, P. SMARCA4 Alterations in Non-Small Cell Lung Cancer: A Systematic Review and Meta-Analysis. J. Clin. Pathol. 2024, 77, 457–463. [Google Scholar] [CrossRef]
- Alver, B.H.; Kim, K.H.; Lu, P.; Wang, X.; Manchester, H.E.; Wang, W.; Haswell, J.R.; Park, P.J.; Roberts, C.W. The SWI/SNF Chromatin Remodeling Complex Is Required for Maintenance of Lineage Specific Enhancers. Nat. Commun. 2017, 8, 14648. [Google Scholar] [CrossRef] [PubMed]
- Ito, K.; Hulse, M.; Agarwal, A.; Carter, J.; Sivakumar, M.; Vykuntam, K.; Fultang, N.; Schwab, A.; Wang, M.; Coward, M.; et al. Discovery of PRT3789, a Selective SMARCA2 Degrader. In Proceedings of the AACR-NCI-EORTC International Conference on Molecular Targets and Cancer Therapeutics, Hynes Convention Center, Boston, MA, USA, 11–15 October 2023. [Google Scholar]
- Dagogo-Jack, I.; Dowlati, A.; Guo, R.; Awad, M.M.; Swalduz, A.; Calvo, E.; Moreno Garcia, V.; Adjei, A.A.; Lorusso, P.; Punekar, S.; et al. A Phase 1 Study of PRT3789, a Potent and Selective Degrader of SMARCA2 in Patients with Advanced or Metastatic Solid Tumors and a SMARCA4 Mutation. In Proceedings of the Annual European Society for Medical Oncology Congress (ESMO), Madrid, Spain, 20–24 October 2023. [Google Scholar]
- Yap, T.A.; Tolcher, A.W.; Plummer, R.; Mukker, J.K.; Enderlin, M.; Hicking, C.; Grombacher, T.; Locatelli, G.; Szucs, Z.; Gounaris, I.; et al. First-in-Human Study of the Ataxia Telangiectasia and Rad3-Related (ATR) Inhibitor Tuvusertib (M1774) as Monotherapy in Patients with Solid Tumors. Clin. Cancer Res. 2024, 30, 2057–2067. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Cappuzzo, F.; Yamamoto, N.; Vokes, N.; Gray, J.E.E.; Owonikoko, T.K.; Ariyasu, R.; Ishii, H.; Kang, J.H.; Lee, S.; et al. 104TiP—Phase Ib/IIa Study of ATR Inhibitor Tuvusertib + Anti-PD-1 Cemiplimab in Patients with Advanced Non-Squamous Non-Small Cell Lung Cancer (NSCLC) That Has Progressed on Prior Anti-PD-(L)1 and Platinum-Based Therapies. Ann. Oncol. 2024, 9 (Suppl. 3), 102683. [Google Scholar] [CrossRef]
| Clinical | N = 659, n (%) |
|---|---|
| Age at NGS sequencing (mean years) | 67.4 |
| Sex | |
| Female | 397 (60%) |
| Male | 262 (40%) |
| Race | |
| Non-White/Other | 59 (9%) |
| White | 600 (91%) |
| Stage | |
| I–III (early and locally advanced) | 392 (59%) |
| IV (metastatic) | 267 (41%) |
| Smoking History | |
| Never | 50 (8%) |
| Former | 496 (75%) |
| Current | 113 (17%) |
| Mutation Type | |
| KRAS | 518 (79%) |
| SMARCA4 (class 1) | 41 (6%) |
| SMARCA4 (class 2) | 54 (8%) |
| KRAS + SMARCA4 (class 1) | 18 (3%) |
| KRAS + SMARCA4 (class 2) | 28 (4%) |
| OS Status from Cancer Diagnosis | Total | Living | Deceased | p-Value |
|---|---|---|---|---|
| Clinical | N = 659 | N = 311 | N = 348 | |
| Age at NGS sequencing (mean) | 66.7 | 68.0 | 0.085 | |
| Sex | 0.327 | |||
| Female | 397 | 194 | 203 | |
| Male | 262 | 117 | 145 | |
| Race | 0.713 | |||
| Non-White/Other | 59 | 26 | 33 | |
| White | 600 | 285 | 315 | |
| Stage | <0.001 | |||
| I–III (early and locally advanced) | 392 | 253 | 139 | |
| IV (metastatic) | 267 | 58 | 209 | |
| Smoking History | 0.901 | |||
| Never | 50 | 25 | 25 | |
| Former | 496 | 232 | 264 | |
| Current | 113 | 54 | 59 | |
| Genomic | N = 659 | |||
| Mutation Type | 0.001 | |||
| KRAS | 518 | 264 | 254 | |
| SMARCA4 (class 1) | 41 | 15 | 26 | |
| SMARCA4 (class 2) | 54 | 22 | 32 | |
| KRAS + SMARCA4 (class 1) | 18 | 3 | 15 | |
| KRAS + SMARCA4 (class 2) | 28 | 7 | 21 |
| Factor | HR | 95% CI | p-Value |
|---|---|---|---|
| Sex | |||
| (Reference group: Female) | |||
| Male | 1.18 | 0.94–1.47 | 0.153 |
| Age | 1.02 | 1.01–1.03 | 0.005 |
| Race | |||
| (Reference group: Non-White/Other) | |||
| White | 0.98 | 0.68–1.41 | 0.905 |
| Smoking History | |||
| (Reference group: Never smoker) | |||
| Former smoker | 1.08 | 0.70–1.66 | 0.732 |
| Current smoker | 1.21 | 0.75–1.98 | 0.437 |
| Stage | |||
| (Reference group: Stage I–III) | |||
| Stage IV | 4.01 | 3.21–5.01 | <0.001 |
| Genomic | |||
| (Reference group: KRAS) | |||
| SMARCA4 (class 1) | 1.18 | 0.78–1.78 | 0.438 |
| SMARCA4 (class 2) | 0.93 | 0.64–1.37 | 0.720 |
| KRAS + SMARCA4 (class 1) | 3.23 | 1.90–5.51 | <0.001 |
| KRAS + SMARCA4 (class 2) | 1.34 | 0.85–2.12 | 0.205 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manolakos, P.; Wang, Y.-B.; Withycombe, J.; Boccuto, L.; Ivankovic, D. Prognostic Impact of KRAS and SMARCA4 Mutations and Co-Mutations on Survival in Non-Small Cell Lung Cancer: Insights from the AACR GENIE BPC Dataset. Biomedicines 2025, 13, 2142. https://doi.org/10.3390/biomedicines13092142
Manolakos P, Wang Y-B, Withycombe J, Boccuto L, Ivankovic D. Prognostic Impact of KRAS and SMARCA4 Mutations and Co-Mutations on Survival in Non-Small Cell Lung Cancer: Insights from the AACR GENIE BPC Dataset. Biomedicines. 2025; 13(9):2142. https://doi.org/10.3390/biomedicines13092142
Chicago/Turabian StyleManolakos, Peter, Yu-Bo Wang, Janice Withycombe, Luigi Boccuto, and Diana Ivankovic. 2025. "Prognostic Impact of KRAS and SMARCA4 Mutations and Co-Mutations on Survival in Non-Small Cell Lung Cancer: Insights from the AACR GENIE BPC Dataset" Biomedicines 13, no. 9: 2142. https://doi.org/10.3390/biomedicines13092142
APA StyleManolakos, P., Wang, Y.-B., Withycombe, J., Boccuto, L., & Ivankovic, D. (2025). Prognostic Impact of KRAS and SMARCA4 Mutations and Co-Mutations on Survival in Non-Small Cell Lung Cancer: Insights from the AACR GENIE BPC Dataset. Biomedicines, 13(9), 2142. https://doi.org/10.3390/biomedicines13092142







