Implication of p16 Promoter Methylation, the BRAFV600E Mutation, and ETS1 Expression Determination on Papillary Thyroid Carcinoma Prognosis and High-Risk Patients’ Selection
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tissue Samples
2.2. RNA and DNA Isolation
2.3. p16 Methylation Analysis
2.4. The BRAFV600E Mutational Analysis
2.5. Reverse Transcription and Quantitative PCR of ETS1 mRNA
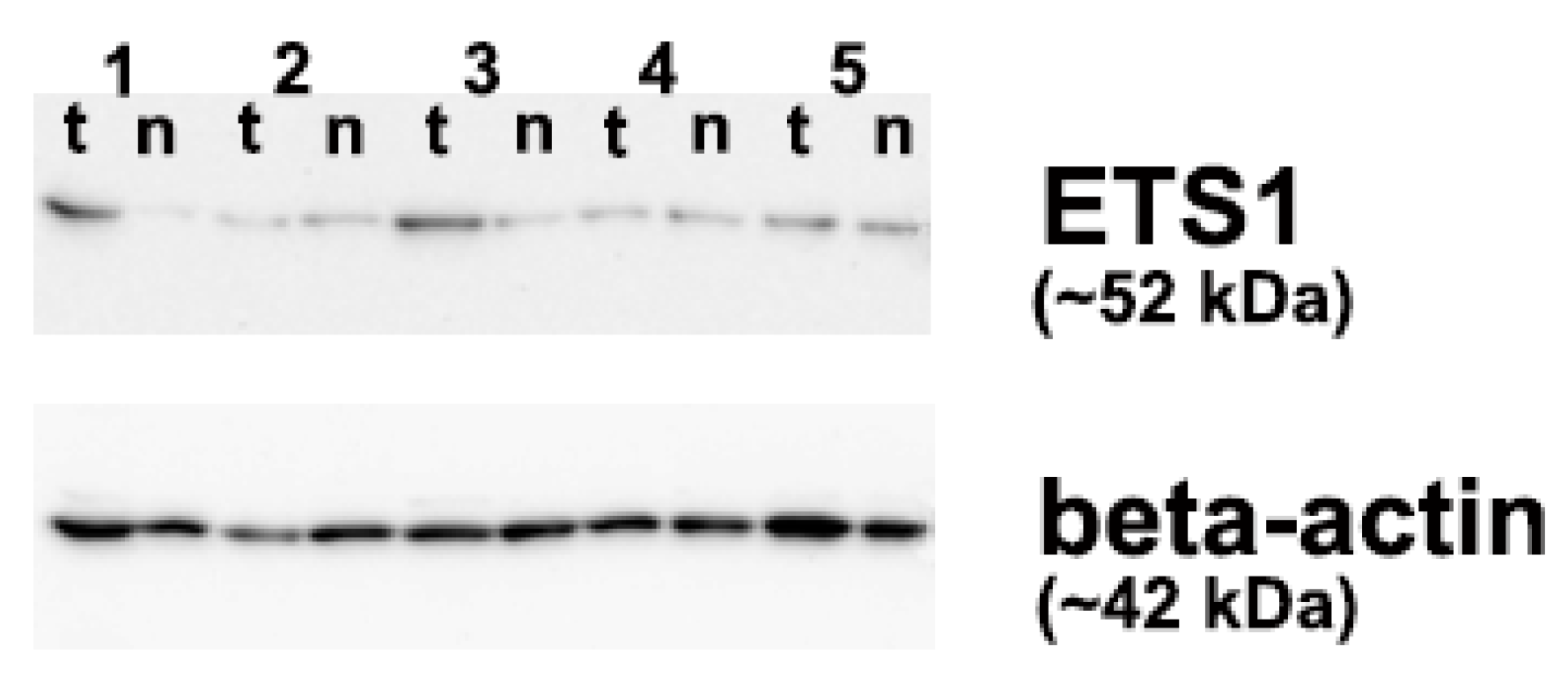
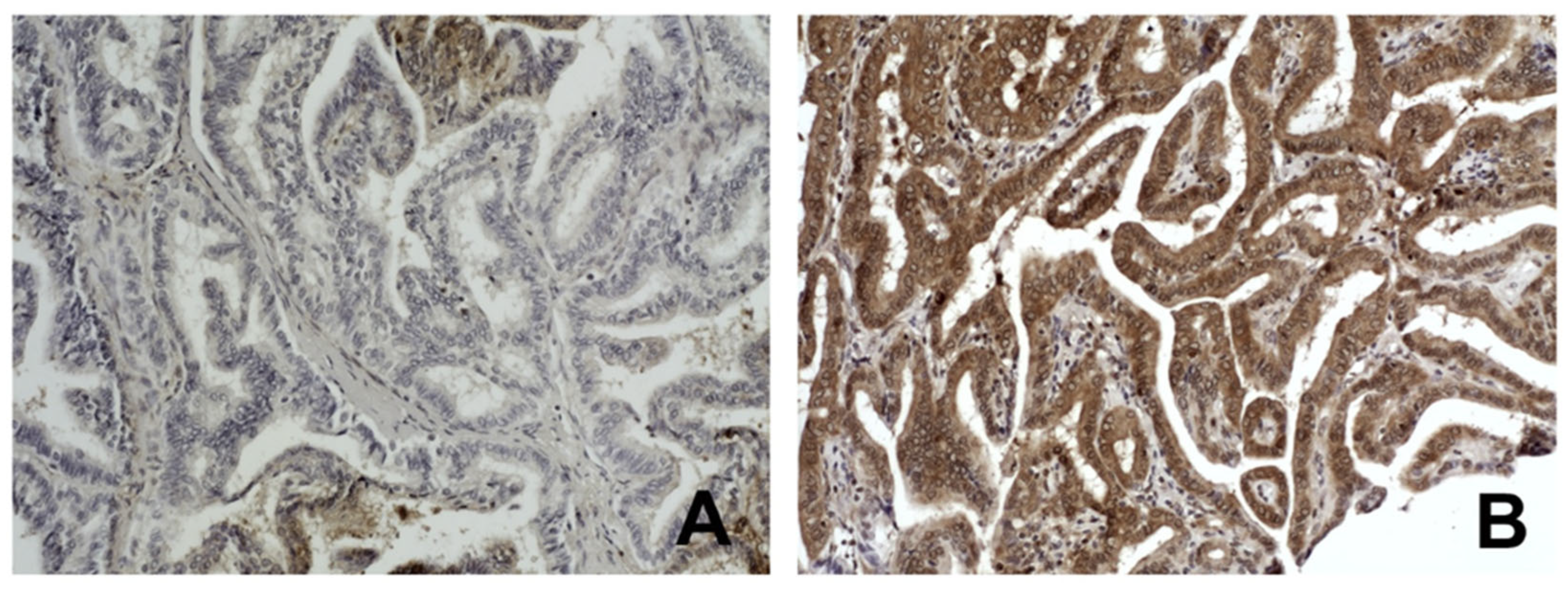
2.6. Quantification of ETS1 Protein Levels
2.7. Statistical Analysis
3. Results
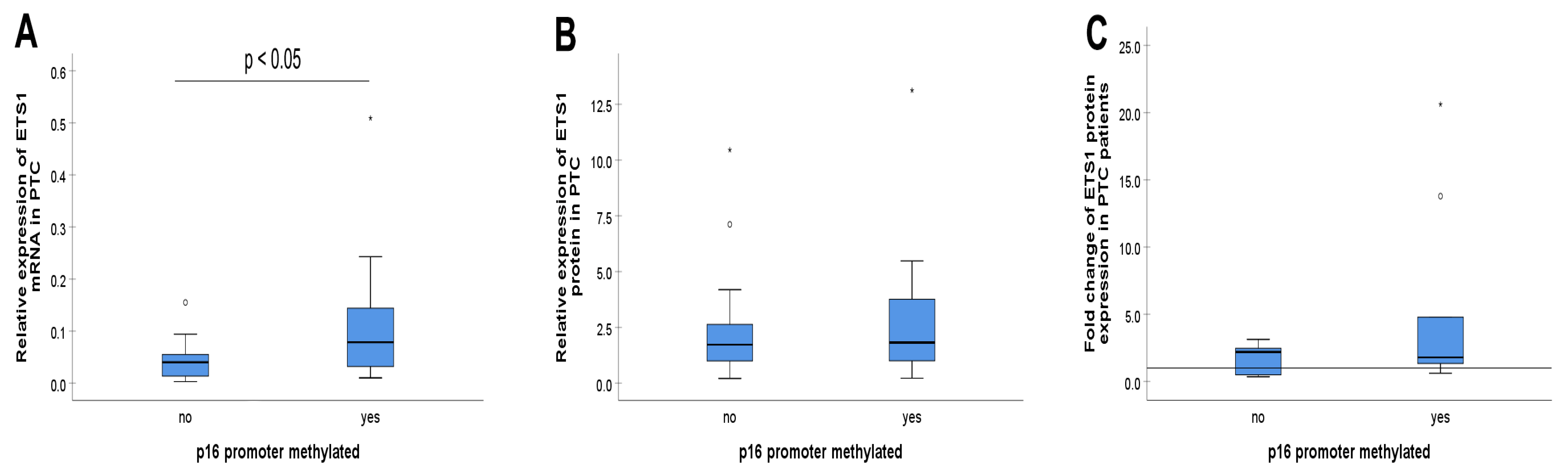
3.1. The p16 Promoter Methylation
3.2. The p16 Promoter Methylation and the BRAFV600E Mutation Status
3.3. The p16 Promoter Methylation and ETS1
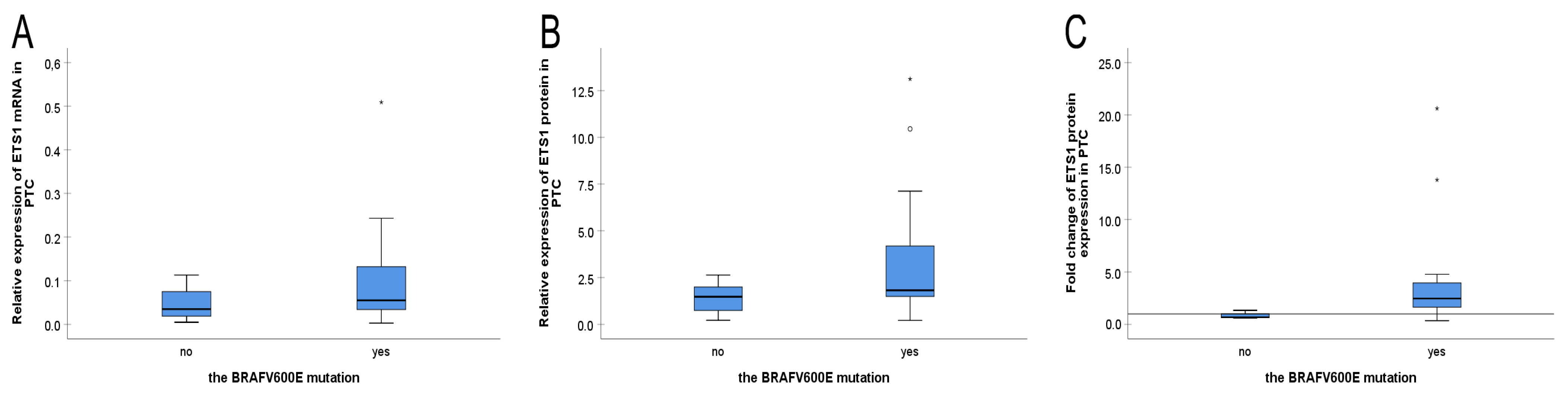
3.4. The BRAFV600E Mutation Status and ETS1
3.5. The Combination of the p16 Promoter Methylation, the BRAFV600E Mutation and ETS1 Levels
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| PTC | Papillary thyroid carcinoma |
| ETS1 | E26 transformation-specific |
| PCR | polymerase chain reaction |
| MSP | methylation-specific PCR |
| MASA | mutant allele-specific PCR amplification |
| qPCR | quantitative PCR |
| IHC | immunohistochemistry |
References
- Chmielik, E.; Rusinek, D.; Oczko-Wojciechowska, M.; Jarzab, M.; Krajewska, J.; Czarniecka, A.; Jarzab, B. Heterogeneity of Thyroid Cancer. Pathobiology 2018, 85, 117–129. [Google Scholar] [CrossRef] [PubMed]
- Bhaijee, F.; Nikiforov, Y.E. Molecular Analysis of Thyroid Tumors. Endocr. Pathol. 2011, 22, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Su, Y.; Qin Dai, R.; Zhou, F.; Shen, C.; Su, Z.Y.; Yang, G.R. The Analysis of Clinical Biological and Pathological Features in Papillary Thyroid Carcinoma. Am. J. Biomed. Life Sci. 2021, 9, 292. [Google Scholar] [CrossRef]
- Pelizzo, M.R.; Boschin, I.M.; Toniato, A.; Piotto, A.; Pagetta, C.; Gross, M.D.; Al-Nahhas, A.; Rubello, D. Papillary Thyroid Carcinoma: 35-Year Outcome and Prognostic Factors in 1858 Patients. Clin. Nucl. Med. 2007, 32, 440–444. [Google Scholar] [CrossRef]
- Ito, Y.; Kudo, T.; Kobayashi, K.; Miya, A.; Ichihara, K.; Miyauchi, A. Prognostic Factors for Recurrence of Papillary Thyroid Carcinoma in the Lymph Nodes, Lung, and Bone: Analysis of 5,768 Patients with Average 10-year Follow-up. World J. surg. 2012, 36, 1274–1278. [Google Scholar] [CrossRef]
- Ito, Y.; Miyauchi, A.; Kihara, M.; Fukushima, M.; Higashiyama, T.; Miya, A. Overall Survival of Papillary Thyroid Carcinoma Patients: A Single-Institution Long-Term Follow-Up of 5897 Patients. World J. Surg. 2018, 42, 615–622. [Google Scholar] [CrossRef]
- Vu-Phan, D.; Koenig, R.J. Genetics and epigenetics of sporadic thyroid cancer. Mol. Cell. Endocrinol. 2014, 386, 55–66. [Google Scholar] [CrossRef]
- Asa, S.L.; Ezzat, S. The epigenetic landscape of differentiated thyroid cancer. Mol. Cell. Endocrinol. 2018, 469, 3–10. [Google Scholar] [CrossRef]
- Rodríguez-Rodero, S.; Fernández, A.F.; Fernández-Morera, J.L.; Castro-Santos, P.; Bayon, G.F.; Ferrero, C.; Urdinguio, R.G.; Gonzalez-Marquez, R.; Suarez, C.; Fernández-Vega, I.; et al. DNA Methylation Signatures Identify Biologically Distinct Thyroid Cancer Subtypes. J. Clin. Endocrinol. Metab. 2013, 98, 2811–2821. [Google Scholar] [CrossRef]
- Wu, W.; Zhang, L.; Lin, J.; Huang, H.; Shi, B.; Lin, X.; Huang, Z.; Wang, C.; Qiu, J.; Wei, X. Hypermethylation of the HIC1 Promoter and Aberrant Expression of HIC1/SIRT1 Contribute to the Development of Thyroid Papillary Carcinoma. Oncotarget 2016, 7, 84416–84427. [Google Scholar] [CrossRef]
- Salimi, F.; Asadikaram, G.; Ashrafi, M.R.; Zeynali Nejad, H.; Abolhassani, M.; Abbasi-Jorjandi, M.; Sanjari, M. Organochlorine Pesticides and Epigenetic Alterations in Thyroid Tumors. Front. Endocrinol. 2023, 14, 1130794. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.-L.; Tian, G.-L.; Chen, S.-J.; Xu, L.; Wang, H.-Q. Promoter Methylation of P16 and RASSF1A Genes May Contribute to the Risk of Papillary Thyroid Cancer: A Meta-Analysis. Exp. Ther. Med. 2015, 10, 1549–1555. [Google Scholar] [CrossRef]
- Mohammadi-asl, J.; Larijani, B.; Khorgami, Z.; Tavangar, S.M.; Haghpanah, V.; Kheirollahi, M.; Mehdipour, P. Qualitative and Quantitative Promoter Hypermethylation Patterns of the P16, TSHR, RASSF1A and RARβ2 Genes in Papillary Thyroid Carcinoma. Med. Oncol. 2011, 28, 1123–1128. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Pei, R.; Lu, Z.; Rao, X.; Liu, B. Methylation of P16 CpG Islands Correlated with Metastasis and Aggressiveness in Papillary Thyroid Carcinoma. J. Chin. Med. Assoc. 2013, 76, 135–139. [Google Scholar] [CrossRef] [PubMed]
- Xing, M. BRAF Mutation in Thyroid Cancer. Endocr. Relat. Cancer 2005, 12, 245–262. [Google Scholar] [CrossRef]
- Smith, R.A.; Salajegheh, A.; Weinstein, S.; Nassiri, M.; Lam, A.K. Correlation between BRAF Mutation and the Clinicopathological Parameters in Papillary Thyroid Carcinoma with Particular Reference to Follicular Variant. Hum. Pathol. 2011, 42, 500–506. [Google Scholar] [CrossRef][Green Version]
- Brait, M.; Loyo, M.; Rosenbaum, E.; Ostrow, K.L.; Markova, A.; Papagerakis, S.; Zahurak, M.; Goodman, S.M.; Zeiger, M.; Sidransky, D.; et al. Correlation between BRAF Mutation and Promoter Methylation of TIMP3, RARβ2 and RASSF1A in Thyroid Cancer. Epigenetics 2012, 7, 710–719. [Google Scholar] [CrossRef]
- Agrawal, N.; Akbani, R.; Aksoy, B.A.; Ally, A.; Arachchi, H.; Asa, S.L.; Auman, J.T.; Balasundaram, M.; Balu, S.; Baylin, S.B.; et al. Integrated Genomic Characterization of Papillary Thyroid Carcinoma. Cell 2014, 159, 676–690. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Tsuji, E.; Yagi, K.; Matsusaka, K.; Tsuji, S.; Kurebayashi, J.; Ogawa, T.; Aburatani, H.; Kaneda, A. Aberrantly Methylated Genes in Human Papillary Thyroid Cancer and Their Association with BRAF/RAS Mutation. Front. Genet. 2013, 4, 271. [Google Scholar] [CrossRef]
- Nakayama, T.; Ito, M.; Ohtsuru, A.; Naito, S.; Nakashima, M.; Sekine, I. Expression of the Ets-1 Proto-Oncogene in Human Thyroid Tumor. Mod. Pathol. 1999, 12, 61–68. [Google Scholar]
- de Nigris, F.; Mega, T.; Berger, N.; Barone, M.V.; Santoro, M.; Viglietto, G.; Verde, P.; Fusco, A. Induction of ETS-1 and ETS-2 Transcription Factors Is Required for Thyroid Cell Transformation. Cancer Res. 2001, 61, 2267–2275. [Google Scholar] [PubMed]
- Plotnik, J.P.; Budka, J.A.; Ferris, M.W.; Hollenhorst, P.C. ETS1 Is a Genome-Wide Effector of RAS/ERK Signaling in Epithelial Cells. Nucleic Acids Res. 2014, 42, 11928–11940. [Google Scholar] [CrossRef] [PubMed]
- Dittmer, J. The biology of the Ets1 proto-oncogene. Mol Cancer. Mol. Cancer 2003, 2, 29. [Google Scholar] [CrossRef] [PubMed]
- Kar, A.; Gutierrez-Hartmann, A. Molecular Mechanisms of ETS Transcription Factor-Mediated Tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 522–543. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, Z.; Sun, M.; Huang, W.; Xia, L. ETS Transcription Factors: Multifaceted Players from Cancer Progression to Tumor Immunity. Biochim. Biophys. Acta (BBA) Rev. Cancer 2023, 1878, 188872. [Google Scholar] [CrossRef]
- Novković, S.S.; Šelemetjev, S.; Miljuš, J.J.; Živaljević, V.; Dunđerović, D.; Milinković, M.; Denčić, T.I. ETS1 Protein Expression May Be Altered by the Complementarity of ETS1 mRNA Sequences with miR-203a-3p and miR-204-3p in Papillary Thyroid Carcinoma. Int. J. Mol. Sci. 2025, 26, 1253. [Google Scholar] [CrossRef]
- Peng, D.; Li, W.; Zhang, B.; Liu, X. Overexpression of lncRNA SLC26A4-AS1 inhibits papillary thyroid carcinoma progression through recruiting ETS1 to promote ITPR1-mediated autophagy. J. Cell. Mol. Med. 2021, 25, 8148–8158. [Google Scholar] [CrossRef]
- Peyret, V.; Nazar, M.; Martín, M.; Quintar, A.A.; Fernandez, E.A.; Geysels, R.C.; Fuziwara, C.S.; Montesinos, M.M.; Maldonado, C.A.; Santisteban, P.; et al. Functional Toll-like Receptor 4 Overexpression in Papillary Thyroid Cancer by MAPK/ERK–Induced ETS1 Transcriptional Activity. Mol. Cancer Res. 2018, 16, 833–845. [Google Scholar] [CrossRef]
- Wan, S.-M.; Peng, P.; Guan, T. Ets-1 Regulates Its Target Genes Mainly by DNA Methylation in Human Ovarian Cancer. J. Obstet. Gynaecol. 2013, 33, 877–881. [Google Scholar] [CrossRef]
- Stojanović, S.; Šelemetjev, S.; Đorić, I.; Janković Miljuš, J.; Tatić, S.; Živaljević, V.; Išić Denčić, T. BRAFV600E, BANCR, miR-203a-3p and miR-204-3p in Risk Stratification of PTC Patients. Biomedicines 2023, 11, 3338. [Google Scholar] [CrossRef]
- DeLellis, R.A.; Lloyd, R.V.; Heitz, P.U.; Eng, C. (Eds.) Pathology and Genetics of Tumours of Endocrine Organs: WHO Classification of Tumours, 3rd ed.; IARC Press: Lyon, France, 2004; ISBN 978-92-832-2416-7. [Google Scholar]
- Edge, S.B.; Compton, C.C. The American Joint Committee on Cancer: The 7th Edition of the AJCC Cancer Staging Manual and the Future of TNM. Ann. Surg. Oncol. 2010, 17, 1471–1474. [Google Scholar] [CrossRef] [PubMed]
- Basolo, F.; Torregrossa, L.; Giannini, R.; Miccoli, M.; Lupi, C.; Sensi, E.; Berti, P.; Elisei, R.; Vitti, P.; Baggiani, A.; et al. Correlation between the BRAF V600E Mutation and Tumor Invasiveness in Papillary Thyroid Carcinomas Smaller than 20 Millimeters: Analysis of 1060 Cases. J. Clin. Endocrinol. Metab. 2010, 95, 4197–4205. [Google Scholar] [CrossRef] [PubMed]
- Išić Denčić, T.; Bartolome, A.; Šelemetjev, S.; Đorić, I.; Tatić, S.; Živaljević, V.; Cvejić, D. High Expression and Localization of β-Catenin and Epidermal Growth Factor Receptor Identify High Risk Papillary Thyroid Carcinoma Patients. Exp. Mol. Pathol. 2018, 105, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Bettington, M.; Brown, I.S.; Rosty, C. Serrated Lesions of the Appendix in Serrated Polyposis Patients. Pathology 2016, 48, 30–34. [Google Scholar] [CrossRef]
- Boissière-Michot, F.; Frugier, H.; Ho-Pun-Cheung, A.; Lopez-Crapez, E.; Duffour, J.; Bibeau, F. Immunohistochemical Staining for P16 and BRAFV600E Is Useful to Distinguish between Sporadic and Hereditary (Lynch Syndrome-Related) Microsatellite Instable Colorectal Carcinomas. Virchows Arch. 2016, 469, 135–144. [Google Scholar] [CrossRef]
- Louis, I.V.S.; Bohjanen, P.R. Post-Transcriptional Regulation of Cytokine and Growth Factor Signaling in Cancer. Cytokine Growth Factor. Rev. 2017, 33, 83–93. [Google Scholar] [CrossRef]
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723. [Google Scholar] [CrossRef]
- Beňačka, R.; Szabóová, D.; Guľašová, Z.; Hertelyová, Z.; Radoňak, J. Non-Coding RNAs in Human Cancer and Other Diseases: Overview of the Diagnostic Potential. Int. J. Mol. Sci. 2023, 24, 16213. [Google Scholar] [CrossRef]
- Ros, J.; Baraibar, I.; Sardo, E.; Mulet, N.; Salvà, F.; Argilés, G.; Martini, G.; Ciardiello, D.; Cuadra, J.L.; Tabernero, J.; et al. BRAF, MEK and EGFR Inhibition as Treatment Strategies in BRAF V600E Metastatic Colorectal Cancer. Ther. Adv. Med. Oncol. 2021, 13, 1758835921992974. [Google Scholar] [CrossRef]
- Schubert, L.; Mariko, M.L.; Clerc, J.; Huillard, O.; Groussin, L. MAPK Pathway Inhibitors in Thyroid Cancer: Preclinical and Clinical Data. Cancers 2023, 15, 710. [Google Scholar] [CrossRef]
- Graves, B.J.; Petersen, J.M. Specificity within the Ets Family of Transcription Factors. Adv. Cancer Res. 1998, 75, 1–55. [Google Scholar] [CrossRef]
- Kozik, B.; Krajnovic, M.; Kokanov, N.; Jovanovic-Cupic, S.; Bozovic, A.; Todorovic, L.; Mandusic, V. Combined Analysis of KRAS Mutation and p16INK4a and p14ARF Methylation Status in Locally Advanced Rectal Carcinoma Treated with Preoperative Chemoradiotherapy. Arch. Biol. Sci. 2022, 74, 127–134. [Google Scholar] [CrossRef]

| Method | Primer | Sequence (5′-3′) |
|---|---|---|
| MASA | BRAF-f_wt (for wild type) | GTGATTTTGGTCTAGCTACAGT |
| BRAF-f_mut (for BRAFV600E) | GTGATTTTGGTCTAGCTACAGA | |
| BRAF-r | GGCCAAAAATTTAATCAGTGGA | |
| MSP | p16_U-f | TTATTAGAGGGTGGGGTGGATTGT |
| p16_U-r | CAACCCCAAACCACAACCATAA | |
| p16_M-f | TTATTAGAGGGTGGGGCGGATCGC | |
| p16_M-r | GACCCCGAACCGCGACCGTAA | |
| qPCR | ETS1-f | ACAGGACTCCATGGCAAACG |
| ETS1-r | ATGAAGAAAGCCTGGTGCAGT | |
| GAPDH-f | GAAGGTGAAGGTCGGAGT | |
| GAPDH-r | GAAGATGGTGATGGGATTTC |
| PTC | BRAFV600E | Total (n) | ||
|---|---|---|---|---|
| No | Yes | |||
| p16 methylated | no | 6 | 18 | 24 |
| yes | 11 | 22 | 33 | |
| Total (n) | 17 | 40 | 57 | |
| Clinicopathological Parameter | p16 Methylated and ETS1 Protein Levels High (n) | p-Value | ||
|---|---|---|---|---|
| None or One | Both | |||
| Gender | male | 13 | 2 | 0.949 |
| female | 29 | 10 | ||
| Intraglandular dissemination | no | 18 | 5 | 0.941 |
| yes | 24 | 7 | ||
| Lymph node metastasis | no | 33 | 10 | 0.718 |
| yes | 9 | 2 | ||
| Extrathyroid invasion | no | 28 | 10 | 0.265 |
| yes | 14 | 2 | ||
| Degree of tumor infiltration | 1 | 9 | 6 | 0.251 |
| 2 | 12 | 3 | ||
| 3 | 7 | 1 | ||
| 4 | 14 | 2 | ||
| pT | T1 | 8 | 1 | 0.032 |
| T2 | 14 | 9 | ||
| T3 | 19 | 1 | ||
| T4 | 1 | 1 | ||
| pTNM | I | 15 | 2 | 0.012 |
| II | 9 | 8 | ||
| III | 13 | 0 | ||
| IV | 5 | 2 | ||
| PTC | The Level of ETS1 Protein Expression | Total (n) | ||
|---|---|---|---|---|
| Low | High | |||
| p16 methylated/ BRAFV600E mutated | None or one | 12 | 7 | 19 |
| Both | 3 | 8 | 11 | |
| Total (n) | 15 | 15 | 30 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stojanović Novković, S.; Šelemetjev, S.; Krajnović, M.; Božović, A.; Kožik, B.; Prosenc Zmrzljak, U.; Išić Denčić, T. Implication of p16 Promoter Methylation, the BRAFV600E Mutation, and ETS1 Expression Determination on Papillary Thyroid Carcinoma Prognosis and High-Risk Patients’ Selection. Biomedicines 2025, 13, 1583. https://doi.org/10.3390/biomedicines13071583
Stojanović Novković S, Šelemetjev S, Krajnović M, Božović A, Kožik B, Prosenc Zmrzljak U, Išić Denčić T. Implication of p16 Promoter Methylation, the BRAFV600E Mutation, and ETS1 Expression Determination on Papillary Thyroid Carcinoma Prognosis and High-Risk Patients’ Selection. Biomedicines. 2025; 13(7):1583. https://doi.org/10.3390/biomedicines13071583
Chicago/Turabian StyleStojanović Novković, Stefana, Sonja Šelemetjev, Milena Krajnović, Ana Božović, Bojana Kožik, Uršula Prosenc Zmrzljak, and Tijana Išić Denčić. 2025. "Implication of p16 Promoter Methylation, the BRAFV600E Mutation, and ETS1 Expression Determination on Papillary Thyroid Carcinoma Prognosis and High-Risk Patients’ Selection" Biomedicines 13, no. 7: 1583. https://doi.org/10.3390/biomedicines13071583
APA StyleStojanović Novković, S., Šelemetjev, S., Krajnović, M., Božović, A., Kožik, B., Prosenc Zmrzljak, U., & Išić Denčić, T. (2025). Implication of p16 Promoter Methylation, the BRAFV600E Mutation, and ETS1 Expression Determination on Papillary Thyroid Carcinoma Prognosis and High-Risk Patients’ Selection. Biomedicines, 13(7), 1583. https://doi.org/10.3390/biomedicines13071583






