MSC1 Cells Suppress Colorectal Cancer Cell Growth via Metabolic Reprogramming, Laminin–Integrin Adhesion Signaling, Oxidative Stress Resistance, and a Tumor-Suppressive Secretome
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition of WJ-MSC Expressed Genes Dataset
2.2. PPI Network Construction and Hub Analysis
2.3. Functional Enrichment of PPI Network Clusters
2.4. Subnetwork Analysis of PPI Network
2.5. Identification and Classification of TF Target Genes
- (a)
- ECM;
- (b)
- Cell membrane;
- (c)
- Cytoplasm;
- (d)
- Endosome;
- (e)
- Endoplasmic reticulum (ER).
- (a)
- Cell death and growth inhibition;
- (b)
- Cell contact;
- (c)
- Glucose and insulin signaling;
- (d)
- Metabolism;
- (e)
- Laminins, integrins, and kinases (PKA, PKC, CKII).
- ECM & Cell Death and Growth Inhibition;
- Cell Membrane & Cell Contact and Cell Death and Growth Inhibition;
- ECM and Cell Membrane & Laminins, integrins, and kinases (PKA, PKC, CKII);
- Cell Membrane and Cytoplasm and Endosome and ER & Glucose and insulin signaling;
- Cell Membrane and Cytoplasm and Endosome and ER & Metabolism.
2.6. Transcriptional Enrichment Analysis of TF Target Genes
2.7. WJ-MSC Isolation
2.8. WJ-MSC and RKO Cell Cultures
2.9. MSC1 Cell Polarization via TLR4 Activation with LPS
2.10. MTT Assays of MSC1 and RKO Co-Cultures
2.11. Manual Curation of Anti-Cancer Ligands and Receptors from TF Target Genes
- (a)
- ECM & Cell Death and Growth Inhibition, and
- (b)
- Cell Membrane and Cell Contact & Cell Death and Growth Inhibition
2.12. Data Availability
3. Results
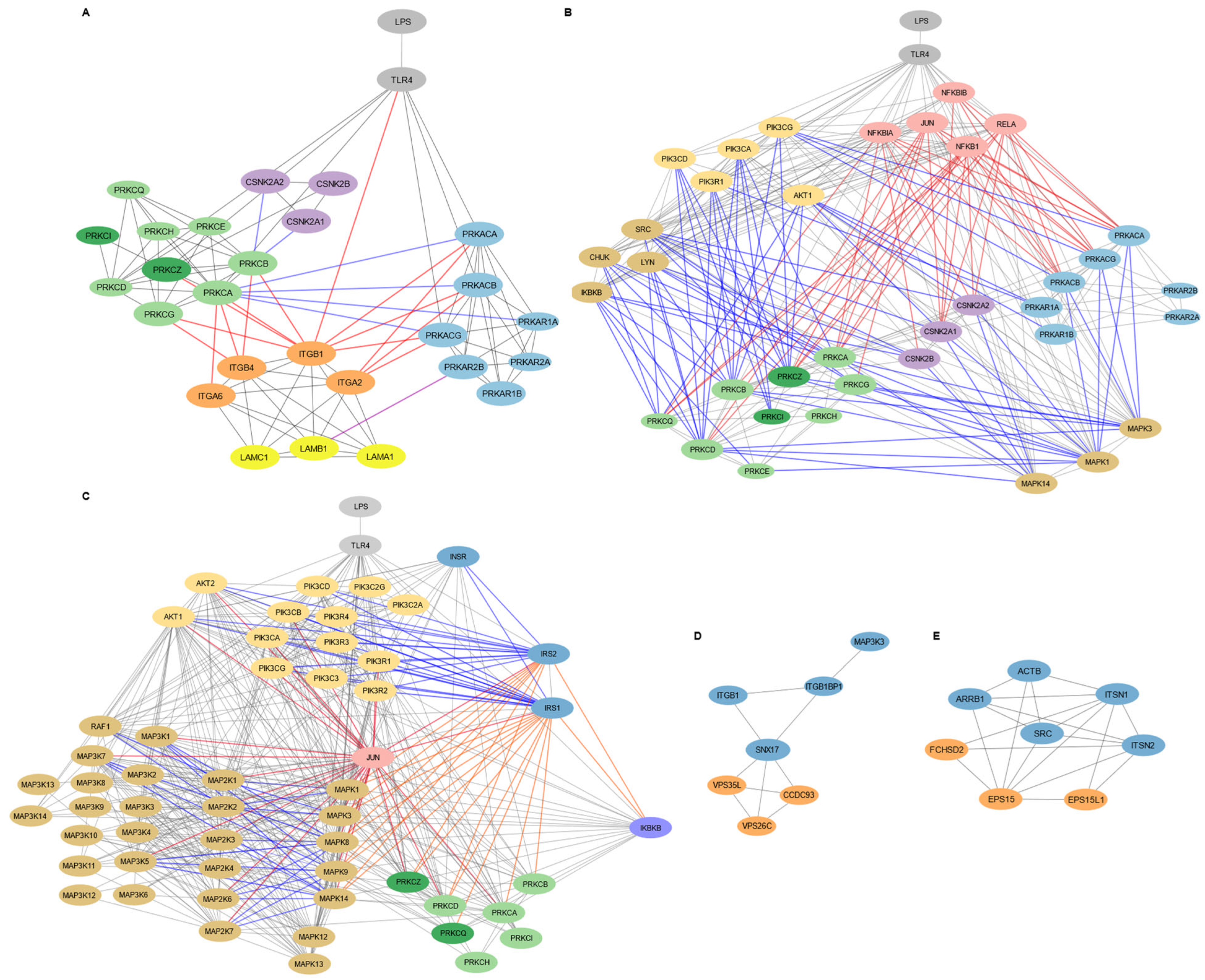
3.1. Core Network Hubs in MSC1 Cells Include SRC, AKT1, ITGB1, and NFKB1
3.2. MSC1 Polarization Activates Kinase Signaling, Immune Modulation, Stress Resistance, ECM Remodeling, and Metabolic Adaptation
3.2.1. Activation of MAPKs, PI3K/AKT, and Kinase Signaling Modules
3.2.2. Immunomodulation via Balanced Inflammatory and Anti-Inflammatory Signaling and Transcription
3.2.3. ECM and Cytoskeletal Remodeling and Integrin Signaling
3.2.4. Stress Resistance and Survival Mechanisms via Balanced Proliferation, Survival, Apoptosis, Stemness, and Epigenetic Remodeling
3.2.5. Metabolic Adaptation: Glucose, Lipid, and Energy Homeostasis
3.3. Subnetwork Mapping Identifies Laminin–Integrin–Kinase Mediated Adhesion, Insulin Signaling, and Membrane Dynamics in MSC1 Cells
3.3.1. Laminin-111/Integrin-Mediated Cell Adhesion and Migration in MSC1 Cells Is Regulated by PKA, PKC, and CKII Signaling
3.3.2. Insulin Signaling Pathway in MSC1 Cells Intersects with MAPK and Inflammatory Networks
3.3.3. Endocytosis and Membrane Localization Subnetworks in MSC1 Cells
3.4. Functional Enrichment of MSC1 Cells TF Target Genes Highlights Metabolic Reprogramming and Tumor-Suppressive Programs
3.4.1. Balanced Regulation of Growth, Apoptosis, and Survival in MSC1 Cells via TGF-β, Kinase, and Growth Factor Signaling
3.4.2. MSC1-Mediated Proliferation Suppression and Immune Modulation by Secretion of TNFs, NO, Cytokines, and Direct Cell–Cell Contact
3.4.3. ECM Remodeling Promotes Tumor-Suppressive Matrix Stabilization via Integrins and MSC1 Cell Migration via Cytoskeletal Reorganization
3.4.4. Stress Resistance via ROS Signaling, Antioxidant Adaptation, and Epigenetic Modification
3.4.5. Metabolic Adaptation via Glucose, Lipid, and Amino Acid Pathways
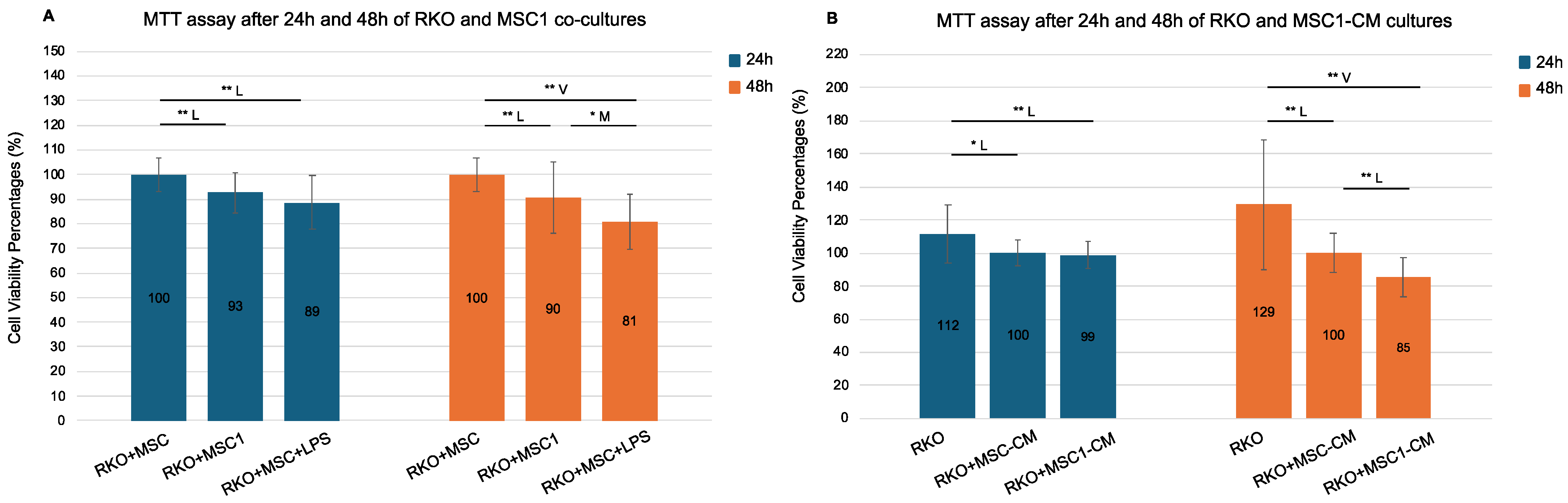
3.5. MSC1 Co-Culture and Secretome Reduce Colorectal Cancer Cell Viability by Suppressing Metabolic Activity
3.6. MSC1 Cells Express a Tumor-Suppressive Ligand and Receptor Profile
3.6.1. Apoptotic TNF Superfamily Members (TRAIL, LIGHT, TL1A, LTA) and Decoy Receptors
3.6.2. BMPs (BMP2, BMP4, BMP7) and Inhibins for Growth Suppression
3.6.3. Novel Putative Ligands and Receptors with Anti-Cancer Potential: SLIT3, CCN3, DCN, DNASE1L3, SULF1, THBS1, PTEN, OAS1, P2RX4, CDHR2, PTPRs, and DAB2IP
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| MSCs | Mesenchymal stem cells |
| MSC1 | Mesenchymal stem cells polarized to MSC1 phenotype |
| CRC | Colorectal cancer |
| LPS | Lipopolysaccharide |
| TLR4 | Toll-like receptor 4 |
| PPI | Protein–protein interaction |
| WJ-MSCs | Wharton’s jelly mesenchymal stem cells |
| EMT | Epithelial–mesenchymal transition |
| CAF | Cancer-associated fibroblast |
| ECM | Extracellular matrix |
| EPC | Edge percolated component |
| TFs | Transcription factors |
| TF targets | Transcription factor targets |
| TF target genes | Transcription factor target genes |
| GSEA | Gene set enrichment analysis |
| ORA | Over-representation analysis |
| qRT-PCR | Quantitative reverse transcription polymerase chain reaction |
| MSC1-CM | MSC1-conditioned medium |
| MSC-CM | MSC-conditioned medium |
| OD | Raw optical density |
| LDHA | Lactate dehydrogenase A |
| OXPHOS | oxidative phosphorylation |
| NO | Nitric oxide |
| ROS | Reactive oxygen species |
| IRS | Integrated stress response |
| TNF | Tumor necrosis factor |
| ecDNA | Extracellular DNA |
| TME | Tumor microenvironment |
| BMPs | Bone marrow proteins |
| BMPRs | Bone marrow protein receptors |
References
- Hass, R.; Kasper, C.; Böhm, S.; Jacobs, R. Different populations and sources of human mesenchymal stem cells (MSC): A comparison of adult and neonatal tissue-derived MSC. Cell Commun. Signal. 2011, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- El Omar, R.; Beroud, J.; Stoltz, J.F.; Menu, P.; Velot, É.; Decot, V. Umbilical cord mesenchymal stem cells: The new gold standard for mesenchymal stem cell-based therapies? Tissue Eng. Part B Rev. 2014, 20, 523–544. [Google Scholar] [CrossRef] [PubMed]
- Stuckey, D.W.; Shah, K. Stem cell-based therapies for cancer treatment: Separating hope from hype. Nat. Rev. Cancer 2014, 14, 683–691. [Google Scholar] [CrossRef] [PubMed]
- Minev, T.; Balbuena, S.; Gill, J.M.; Marincola, F.M.; Kesari, S.; Lin, F. Mesenchymal stem cells—the secret agents of cancer immunotherapy: Promises, challenges, and surprising twists. Oncotarget 2024, 15, 793–805. [Google Scholar] [CrossRef]
- O’Malley, G.; Heijltjes, M.; Houston, A.M.; Rani, S.; Ritter, T.; Egan, L.J.; Ryan, A.E. Mesenchymal stromal cells (MSCs) and colorectal cancer: A troublesome twosome for the anti-tumour immune response? Oncotarget 2016, 7, 60752–60774. [Google Scholar] [CrossRef]
- Jahangiri, B.; Khalaj-Kondori, M.; Asadollahi, E.; Purrafee Dizaj, L.; Sadeghizadeh, M. MSC-derived exosomes suppress colorectal cancer cell proliferation and metastasis via miR-100/mTOR/miR-143 pathway. Int. J. Pharm. 2022, 627, 122214. [Google Scholar] [CrossRef]
- Shinagawa, K.; Kitadai, Y.; Tanaka, M.; Sumida, T.; Kodama, M.; Higashi, Y.; Tanaka, S.; Yasui, W.; Chayama, K. Mesenchymal stem cells enhance growth and metastasis of colon cancer. Int. J. Cancer 2010, 127, 2323–2333. [Google Scholar] [CrossRef]
- Takigawa, H.; Kitadai, Y.; Shinagawa, K.; Yuge, R.; Higashi, Y.; Tanaka, S.; Yasui, W.; Chayama, K. Mesenchymal stem cells induce epithelial to mesenchymal transition in colon cancer cells through direct cell-to-cell contact. Neoplasia 2017, 19, 429–438. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Spaeth, E.L.; Dembinski, J.L.; Sasser, A.K.; Watson, K.; Klopp, A.; Hall, B.; Andreeff, M.; Marini, F.C. Mesenchymal stem cell transition to tumor-associated fibroblasts contributes to fibrovascular network expansion and tumor progression. PLoS ONE 2009, 4, e4992. [Google Scholar] [CrossRef]
- Lu, P.; Weaver, V.M.; Werb, Z. The extracellular matrix: A dynamic niche in cancer progression. J. Cell Biol. 2012, 196, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Stipp, C.S. Laminin-binding integrins and their tetraspanin partners as potential antimetastatic targets. Expert Rev. Mol. Med. 2010, 12, e3. [Google Scholar] [CrossRef]
- Hoxhaj, G.; Manning, B.D. The PI3K–AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even Warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Waterman, R.S.; Tomchuck, S.L.; Henkle, S.L.; Betancourt, A.M. A new mesenchymal stem cell (MSC) paradigm: Polarization into a pro-inflammatory MSC1 or an immunosuppressive MSC2 phenotype. PLoS ONE 2010, 5, e10088. [Google Scholar] [CrossRef]
- Waterman, R.S.; Henkle, S.L.; Betancourt, A.M. Mesenchymal stem cell 1 (MSC1)-based therapy attenuates tumor growth whereas MSC2-treatment promotes tumor growth and metastasis. PLoS ONE 2012, 7, e45590. [Google Scholar] [CrossRef]
- Choi, J.; Pacheco, C.M.; Mosbergen, R.; Korn, O.; Chen, T.; Nagpal, I.; Englart, S.; Angel, P.W.; Wells, C.A. Stemformatics: Visualize and download curated stem cell data. Nucleic Acids Res. 2019, 47, D841–D846. [Google Scholar] [CrossRef]
- Martins, J.P.; Santos, J.M.; de Almeida, J.M.; Filipe, M.A.; de Almeida, M.V.; Almeida, S.C.; Água-Doce, A.; Varela, A.; Gilljam, M.; Stellan, B.; et al. Towards an advanced therapy medicinal product based on mesenchymal stromal cells isolated from the umbilical cord tissue: Quality and safety data6. Stem Cell Res. Ther. 2014, 5, 9. [Google Scholar] [CrossRef]
- Kolberg, L.; Raudvere, U.; Kuzmin, I.; Adler, P.; Vilo, J.; Peterson, H. g:Profiler—Interoperable web service for functional enrichment analysis and gene identifier mapping (2023 update). Nucleic Acids Res. 2023, 51, W207–W212. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Franz, M.; Lopes, C.T.; Huck, G.; Dong, Y.; Sumer, O.; Bader, G.D. Cytoscape.js: A graph theory library for visualisation and analysis. Bioinformatics 2016, 32, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W.V. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics 2003, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2025. Nucleic Acids Res. 2025, 53, D609–D617. [Google Scholar] [CrossRef]
- Broad Institute. GSEA (Gene Set Enrichment Analysis) Software, Version 4.3.3 [Build 4/2/2024]. Broad Institute of MIT and Harvard 2024. Available online: https://www.gsea-msigdb.org/ (accessed on 5 June 2025).
- Xie, Z.; Bailey, A.; Kuleshov, M.V.; Clarke, D.J.B.; Evangelista, J.E.; Jenkins, S.L.; Lachmann, A.; Wojciechowicz, M.L.; Kropiwnicki, E.; Jagodnik, K.M.; et al. Gene set knowledge discovery with Enrichr. Curr. Protoc. 2021, 1, e90. [Google Scholar] [CrossRef]
- Galliou, P.A. Supplementary Datasets for “MSC1 Cells Suppress Colorectal Cancer Cell Growth via Metabolic Reprogramming, Laminin–Integrin Adhesion Signaling, Oxidative Stress Resistance, and a Tumor-Suppressive Secretome” (Version 1) [Data Set]. Zenodo 2025. Available online: https://zenodo.org/records/15540018 (accessed on 27 April 2025).
- Ashkenazi, A.; Herbst, R.S. To kill a tumor cell: The potential of proapoptotic receptor agonists. J. Clin. Investig. 2008, 118, 1979–1990. [Google Scholar] [CrossRef]
- Fan, Z.; Yu, P.; Wang, Y.; Wang, Y.; Fu, M.L.; Liu, W.; Sun, Y.; Fu, Y.X. NK-cell activation by LIGHT triggers tumor-specific CD8+ T-cell immunity to reject established tumors. Blood 2006, 107, 1342–1351. [Google Scholar] [CrossRef]
- Han, M.; Sun, Y.; Zhao, W.; Feng, L.; Zhang, H.; Zhang, S.; Liu, Z.; Gao, C.; Sun, H.; Li, J.; et al. Comprehensive characterization of TNFSF14/LIGHT with implications in prognosis and immunotherapy of human gliomas. Front. Immunol. 2022, 13, 1025286. [Google Scholar] [CrossRef] [PubMed]
- Migone, T.S.; Zhang, J.; Luo, X.; Zhuang, L.; Chen, C.; Hu, B.; Hong, J.S.; Perry, J.W.; Chen, S.-F.; Zhou, J.X.H.; et al. TL1A Is a TNF-like Ligand for DR3 and TR6/DcR3 and Functions as a T Cell Costimulator. Immunity 2002, 16, 479–492. [Google Scholar] [CrossRef]
- Zhao, C.; Han, Q.; Ying, H.Y.; Liu, Y.; Wang, X.; Wang, X.Y. TNFSF15 facilitates differentiation and polarization of macrophages toward M1 phenotype to inhibit tumor growth. OncoImmunology 2022, 11, 2032918. [Google Scholar] [CrossRef]
- Niu, W.; Liu, Q.; Huo, X.; Luo, Y.; Zhang, X. TL1A promotes metastasis and EMT process of colorectal cancer. Heliyon 2024, 10, e24392. [Google Scholar] [CrossRef]
- Upadhyay, V.; Fu, Y.X. Lymphotoxin signalling in immune homeostasis and the control of microorganisms. Nat. Rev. Immunol. 2013, 13, 270–279. [Google Scholar] [CrossRef]
- Hu, X.; Zimmerman, M.A.; Bardhan, K.; Yang, D.; Waller, J.L.; Liles, G.B.; Lee, J.R.; Pollock, R.; Lev, D.; Ware, C.F.; et al. Lymphotoxin β receptor mediates caspase-dependent tumor cell apoptosis in vitro and tumor suppression in vivo despite induction of NF-κB activation. Carcinogenesis 2013, 34, 1105–1114. [Google Scholar] [CrossRef] [PubMed]
- Hardwick, J.C.H.; Kodach, L.L.; Offerhaus, G.J.A.; van den Brink, G.R. Bone morphogenetic protein signalling in colorectal cancer. Nat. Rev. Cancer 2008, 8, 806–812. [Google Scholar] [CrossRef]
- Liu, R.X.; Ren, W.Y.; Ma, Y.; Liao, Y.P.; Wang, H.; Zhu, J.H.; Jiang, H.T.; Wu, K.; He, B.C.; Sun, W.J.; et al. BMP7 mediates the anticancer effect of honokiol by upregulating p53 in HCT116 cells. Int. J. Oncol. 2017, 51, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Vishnubalaji, R.; Yue, S.; Alfayez, M.; Kassem, M.; Liu, F.F.; Aldahmash, A.; Alajez, N.M. Bone morphogenetic protein 2 (BMP2) induces growth suppression and enhances chemosensitivity of human colon cancer cells. Cancer Cell Int. 2016, 16, 77. [Google Scholar] [CrossRef]
- Risbridger, G.P.; Schmitt, J.F.; Robertson, D.M. Activins and inhibins in endocrine and other tumors. Endocr. Rev. 2001, 22, 836–858. [Google Scholar] [CrossRef]
- Tong, M.; Jun, T.; Nie, Y.; Hao, J.; Fan, D. The role of the Slit/Robo signaling pathway. J. Cancer 2019, 10, 2694–2705. [Google Scholar] [CrossRef]
- Li, J.; Ye, L.; Sun, P.; Zheng, F.; Ruge, F.; Satherley, L.K.; Feng, Y.; Zhao, H.; Du, G.; Wang, T.; et al. Reduced NOV expression correlates with disease progression in colorectal cancer and is associated with survival, invasion, and chemoresistance of cancer cells. Oncotarget 2017, 8, 26231–26244. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Mondal, D.K.; Ulas, M.; Neill, T.; Iozzo, R.V. Oncosuppressive roles of decorin through regulation of multiple receptors and diverse signaling pathways. Am. J. Physiol. Cell Physiol. 2022, 322, C764–C776. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Nakano, H.; Fan, W.; Li, Y.; Sil, P.; Nakano, K.; Junqueira, C.; Ritter, J.; Yang, Y.; Hernandez, C.; et al. DNASE1L3 enhances antitumor immunity and suppresses tumor progression in colon cancer. JCI Insight 2023, 8, e168161. [Google Scholar] [CrossRef]
- Lui, C.T.; Zhu, S.T.; Li, P. SULF1 inhibits proliferation and invasion of esophageal squamous cell carcinoma cells by decreasing heparin-binding growth factor signaling. Dig. Dis. Sci. 2013, 58, 1256–1263. [Google Scholar] [CrossRef]
- Kaur, S.; Bronson, S.M.; Pal-Nath, D.; Miller, T.W.; Soto-Pantoja, D.R.; Roberts, D.D. Functions of Thrombospondin-1 in the tumor microenvironment. Int. J. Mol. Sci. 2021, 22, 4570. [Google Scholar] [CrossRef]
- Naderali, E.; Khaki, A.A.; Rad, J.S.; Ali-Hemmati, A.; Rahmati, M.; Charoudeh, H.N.A. Regulation and modulation of PTEN activity. Mol. Biol. Rep. 2018, 45, 2869–2881. [Google Scholar] [CrossRef]
- Jiang, S.; Deng, X.; Luo, M.; Zhou, L.; Chai, J.J.; Tian, C.; Yan, Y.; Luo, Z. Pan-cancer analysis identified OAS1 as a potential prognostic biomarker for multiple tumor types. Front. Oncol. 2023, 13, 1234567. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.H.; Hao, Y.; Mousawi, F.; Peng, H.; Yang, X. Expression of P2 purinergic receptors in mesenchymal stem cells and their roles in extracellular nucleotide regulation of cell functions. J. Cell. Physiol. 2017, 232, 287–297. [Google Scholar] [CrossRef]
- Bloch, E.; Sikorski, E.L.; Pontoriero, D.; Day, E.K.; Berger, B.W.; Lazzara, M.J.; Thévenin, D. Disrupting the transmembrane domain-mediated oligomerization of protein tyrosine phosphatase receptor J inhibits EGFR-driven cancer cell phenotypes. J. Biol. Chem. 2019, 294, 18796–18806. [Google Scholar] [CrossRef]
- Pittenger, M.F.; Discher, D.E.; Péault, B.M.; Phinney, D.G.; Hare, J.M.; Caplan, A.I. Mesenchymal stem cell perspective: Cell biology to clinical progress. NPJ Regen. Med. 2019, 4, 22. [Google Scholar] [CrossRef]
- Kawasaki, T.; Kawai, T. Toll-like receptor signaling pathways. Front. Immunol. 2014, 5, 461. [Google Scholar] [CrossRef]
- Pickup, M.W.; Mouw, J.K.; Weaver, V.M. The extracellular matrix modulates the hallmarks of cancer. EMBO Rep. 2014, 15, 1243–1253. [Google Scholar] [CrossRef]
- Koliakos, G.; Kouzi-Koliakos, K.; Triantos, A.; Dimitriadou, A.; Papageorgiou, A. Laminin-1 phosphorylation by protein kinase A: Effect on self-assembly and heparin binding. J. Biochem. Mol. Biol. 2000, 33, 370–378. [Google Scholar]
- Koliakos, G.; Trachana, V.; Gaitatzi, M.; Dimitriadou, A.; Papageorgiou, A. Phosphorylation of laminin-1 by protein kinase C. Mol. Cells 2001, 11, 179–185. [Google Scholar] [CrossRef]
- Bohana-Kashtan, O.; Pinna, L.A.; Fishelson, Z. Extracellular phosphorylation of C9 by protein kinase CK2 regulates complement-mediated lysis. Eur. J. Immunol. 2005, 35, 1939–1948. [Google Scholar] [CrossRef]
- Galliou, P.A.; Verrou, K.M.; Koliakos, G. Phosphorylation mapping of laminin α1-chain: Kinases in association with active sites. Comput. Biol. Chem. 2019, 80, 480–497. [Google Scholar] [CrossRef]
- Verrou, K.M.; Galliou, P.A.; Papaioannou, M.; Koliakos, G. Phosphorylation mapping of laminin β1-chain: Kinases in association with active sites. J. Biosci. 2019, 44, 55. [Google Scholar] [CrossRef]
- Galliou, P.A.; Verrou, K.M.; Papanikolaou, N.A.; Koliakos, G. Phosphorylation mapping of laminin γ1-chain: Kinases, functional interaction sequences, and phosphorylation-interfering cancer mutations. J. Biosci. 2024, 49, 85. [Google Scholar] [CrossRef]
- Grafe, I.; Alexander, S.; Peterson, J.R.; Snider, T.N.; Levi, B.; Lee, B.; Mishina, Y. TGF-β family signaling in mesenchymal differentiation. Cold Spring Harb. Perspect. Biol. 2018, 10, a022202. [Google Scholar] [CrossRef]
- Pala, R.; Cruciani, S.; Manca, A.; Garroni, G.; El Faqir, M.A.; Lentini, V.; Capobianco, G.; Pantaleo, A.; Maioli, M. Mesenchymal stem cell behavior under microgravity: From stress response to a premature senescence. Int. J. Mol. Sci. 2023, 24, 7753. [Google Scholar] [CrossRef]
- Olesen, K.; Moruzzi, N.; Bulatovic, I.; Folmes, C.; Jeon, R.; Felldin, U.; Terzic, A.; Simonson, O.E.; Le Blanc, K.; Österholm, C.; et al. Diversity of respiratory parameters and metabolic adaptation to low oxygen tension in mesenchymal stromal cells. Metab. Open 2022, 13, 100167. [Google Scholar] [CrossRef]
- Wen, F.; Liu, Y.; Wang, W.; Li, M.; Guo, F.; Sang, Y.; Zhuang, J.; Chen, Y.; Ma, L.; Wang, X.; et al. Adenomatous polyposis coli genotype-dependent toll-like receptor 4 activity in colon cancer. Oncotarget 2016, 7, 7761–7772. [Google Scholar] [CrossRef]
- Kuo, W.T.; Lee, T.C.; Yang, H.Y.; Chen, C.Y.; Au, Y.C.; Lu, Y.Z.; Wu, L.L.; Wei, S.C.; Ni, Y.H.; Lin, B.R.; et al. LPS receptor subunits have antagonistic roles in epithelial apoptosis and colonic carcinogenesis. Cell Death Differ. 2015, 22, 1590–1604. [Google Scholar] [CrossRef]
- Barrett, T.; Wilhite, S.E.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; Marshall, K.A.; Phillippy, K.H.; Sherman, P.M.; Holko, M.; et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 2012, 41, D991–D995. [Google Scholar] [CrossRef]
- Edgar, R. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef]
- Kim, S.H.; Das, A.; Chai, J.C.; Binas, B.; Choi, M.R.; Park, K.S.; Lee, Y.S.; Jung, K.H.; Chai, Y.G. Transcriptome sequencing–wide functional analysis of human mesenchymal stem cells in response to TLR4 ligand. Sci. Rep. 2016, 6, 30311. [Google Scholar] [CrossRef]
- Kim, S.H.; Choi, H.I.; Choi, M.R.; An, G.Y.; Binas, B.; Jung, K.H.; Chai, Y.G. Epigenetic regulation of IFITM1 expression in lipopolysaccharide-stimulated human mesenchymal stromal cells. Stem Cell Res. Ther. 2020, 11, 16. [Google Scholar] [CrossRef]
- Ge, S.X.; Son, E.W.; Yao, R. iDEP: An integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 2018, 19, 534. [Google Scholar] [CrossRef]


| Enriched Pathway | Size | Nes | FDR q-Value | Leading Edge Genes |
|---|---|---|---|---|
| Kinase Signaling and Second Messenger Regulation | ||||
| Diacylglycerol-Dependent Serine Threonine Kinase Activity | 9 | 2.86 | 0.00 | PRKACA, PRKACB, PRKCB, PRKCD, ATF4, PLA2G5, PLA2G6, PLCB3, PLCB4, PLCB2, ADCY4, ADCY8, EGFR |
| Phosphatidylinositol Signaling System | 22 | 2.50 | 0.00 | PIP4K2A, PIP4K2B, PRKCB, PLCG2, PIP5K1A, PLCG1, ITPK1, PLCB3, PLCB4, PLCB2, PLCD1, DGKA, DGKZ, DGKI, DGKH, ITPR1, PTEN, PIK3C2A |
| Immune Modulation and Inflammation Regulation | ||||
| Fc Epsilon RI Signaling Pathway | 19 | 2.08 | 0.02 | PRKACB, PRKCB, PLCG2, PLCG1 |
| Fc Gamma R-Mediated Phagocytosis | 26 | 1.90 | 0.04 | LYN, ITGA2B, ITGA11, ITGB1, ITGB4, ITGAL, ITGB8, ITGB7, ITGB6, ITGA3, ITGA2, ITGA1, ITGA7, ITGA6, ITGA5 |
| Metabolic Adaptation | ||||
| Inositol Phosphate Metabolism | 13 | 2.39 | 0.00 | PIP4K2A, PIP4K2B, PLCG2, PIP5K1A, PLCG1, ITPK1, PLCB3, PLCB4, PLCB2, PLCD1, PTEN, PIK3C2A |
| Chrebp Pathway | 5 | 2.30 | 0.01 | PRKACA, PRKACB, PLCB2, ADCY4, ADCY8, KCNB1 |
| ECM remodeling and Differentiation | ||||
| Integrin Complex | 16 | 2.49 | 0.02 | GJA1, ITGA2B, ITGA11, ITGB1, ITGB4, ITGB8, ITGB7, ITGB6, DSP, ITGA3, ITGA2, ITGA1, ITGA7, ITGA6, ITGA5 |
| Regulation of Extracellular Matrix Organization | 14 | −2.36 | 0.05 | TGFB2, TGFB1, TGFB3, BMP7, BMP4, BMP2, GSK3B, TGFBR1, TGFBR2 |
| Regulation of Epithelial to Mesenchymal Transition Involved in Endocardial Cushion Formation | 5 | −2.37 | 0.05 | FGFR4, FGFR3, FGFR2, FGFR1, THBS1 |
| Cellular Component Disassembly | 56 | −2.47 | 0.05 | PLEKHA1, VEGFA, INPPL1, PDGFRA, PSEN1, TGFB1, TGFB3, BMP7, BMP4, DLG1, FGFR3, FGFR2, FGFR1, TGFBR1, TGFBR2, GREM1, LAMA5 |
| Anti-Cancer Mechanisms | ||||
| TGF Beta Signaling Pathway | 17 | −2.71 | 0.00 | TGFB2, TGFB1, TGFB3, INHBA, BMP7, BMP4, BMP2, DCN, TGFBR1, TGFBR2, THBS1, MAPK3 |
| Transforming Growth Factor Beta Receptor Binding | 5 | −2.38 | 0.02 | TGFB2, TGFB1, TGFB3, TGFBR1, TGFBR2 |
| TOB1 Pathway | 6 | −2.09 | 0.05 | GRB2, RAC1, HRAS, TGFB2, TGFB1, TGFB3, TGFBR1 |
| ALK Pathway | 12 | −2.53 | 0.00 | WNT4, TGFB1, PTCH1, BMP4, BMP2, DLG1, ILK, GREM1, LAMA5 |
| Fibroblast Growth Factor Binding | 5 | −2.43 | 0.03 | PIK3R4, PIK3R1, ZMPSTE24, CARMIL1, INSR, CTSS, CX3CL1, MAP1LC3A, PLAAT1, PLAAT3, SREBF2, ARHGEF2, DNASE1L3, CAMKK2, LIMA1, VMP1, FAP, IGF1R, TSC2, ADAM15, ADRB2 |
| P38 MAPK Pathway | 7 | −2.09 | 0.04 | TGFB2, TGFB1, TGFB3, TGFBR1, TGFBR2 |
| Term | Overlap | p-Value | Combined Score | Leading Edge Genes |
|---|---|---|---|---|
| ECM & Cell Death and Growth Inhibition | ||||
| TGF-β Signaling | ||||
| Positive Regulation of Pathway-Restricted SMAD Protein Phosphorylation | 10/49 | 0.00 | 2168 | BMP4, TGFB2, BMP2, TGFB1, GDF15, TGFB3, INHBA, INHA, BMP7, TGFBR2 |
| Apoptosis and Cell Death | ||||
| Regulation of Nitric Oxide-Mediated Signal Transduction | 3/5 | 0.00 | 5069 | THBS1, EGFR, VEGFA |
| TNFs Bind Their Physiological Receptors | 9/29 | 0.00 | 3886 | TNFRSF6B, TNFSF14, TNFSF15, TNFSF13, LTA, TNFSF11, TNFRSF11B, TNFRSF1B, TNFSF13B |
| Cell Growth Inhibition | ||||
| Negative Regulation of Smooth Muscle Cell Proliferation | 8/37 | 0.00 | 1868 | IL10, BMP4, BMP2, IGFBP5, TGFB3, PTEN, IL12B, APOE |
| Immune Modulation | ||||
| Immune Infiltration in Pancreatic Cancer WP5285 | 12/39 | 0.00 | 5279 | IL10, TGFB2, IL6, LGALS1, TGFB1, TGFB3, IL23A, CCL2, IL12B, LGALS9, MMP9, VEGFA |
| Tissue Repair and ECM Remodeling | ||||
| Platelet Mediated Interactions with Vascular and Circulating Cells | 5/17 | 0.00 | 1912 | TGFB2, TGFB1, TGFB3, CCL2, PF4 |
| Regulation of Collagen Biosynthetic Process | 5/18 | 0.00 | 1734 | BMP4, IL6, TGFB1, TGFB3, WNT4 |
| Cell Membrane & Cell Contact and Cell Death and Growth Inhibition | ||||
| Apoptosis and Cell Death | ||||
| Epithelial Cell Apoptotic Process | 2/10 | 0.00 | 1982 | BMPR2, DAB2IP |
| Negative Regulation of Anoikis | 2/16 | 0.00 | 1016 | ITGB1, SRC |
| Survival, Proliferation, and Growth Suppression | ||||
| Negative Regulation of Epidermal Growth Factor Receptor Signaling Pathway | 3/23 | 0.00 | 1639 | DAB2IP, PTPRJ, EGFR |
| Negative Regulation of Cell Growth | 7/125 | 0.00 | 1389 | DDX3X, BMPR2, CDHR2, RACK1, PTPRJ, ENO1, RTN4 |
| Negative Regulation of ERBB Signaling Pathway | 3/18 | 0.00 | 2319 | DAB2IP |
| TROP2 Regulatory Signaling | 4/45 | 0.00 | 1321 | ITGB1, SRC, RACK1, EGFR |
| Cell–Cell Contact Signaling | ||||
| Cadherin Binding | 24/319 | 0.00 | 75,601 | ITGB1, ACVR1, RAB1A, DDX3X, BMPR2, SRC, DAB2IP, PTPRJ, PSEN1, ENO1, PTPRH, FNBP1L, RTN4, EGFR, CD2AP, DLG1, P2RX4, HNRNPK, RUVBL1, RACK1, ITGA6, PKN2, MARK2, EIF4G2 |
| Contact Inhibition | 1/5 | 0.01 | 1006 | PTPRJ |
| Cytoskeletal Dynamics and Migration | ||||
| Contractile Actin Filament Bundle Assembly | 2/14 | 0.00 | 1224 | ITGB1, SRC |
| Regulation of Epithelial Cell Migration | 4/50 | 0.00 | 1143 | BMPR2, SRC, DAB2IP, RTN4 |
| Hypoxia | ||||
| Negative Regulation of Cellular Response to Hypoxia | 1/5 | 0.01 | 1006 | ENO1 |
| Cell Membrane and Cytoplasm and Endosome and ER & Metabolism | ||||
| Lipid Metabolism | ||||
| Regulation of Long-Chain Fatty Acid Import Across Plasma Membrane | 4/5 | 0.00 | 4686 | AKT2, ACSL5, IRS2, THBS1 |
| Lysosphingolipid and LPA Receptors | 8/14 | 0.00 | 2698 | PLPPR1, PLPPR2, LPAR1, S1PR1, LPAR2, S1PR2, S1PR5, S1PR4 |
| Arachidonate Production from DAG | 3/5 | 0.00 | 1191 | DAGLA, DAGLB, MGLL |
| Glycerophospholipid Metabolic Process | 16/62 | 0.00 | 999 | PDGFRB, PLA2G4D, PLA2G4B, PLA2G4C, GDE1, PLAAT1, GDPD3, PLA2G6, PLCB3, PNPLA3, PIP5K1A, PLCG1, ENPP6, PLCB2, PLCD1, DGK |
| Lipid Signaling Pathways | ||||
| Positive Regulation of Phospholipase C Activity | 12/36 | 0.00 | 1206 | PDGFRB, PDGFRA, EDNRA, KIT, LPAR1, LPAR2, HRAS, S1PR4, ESR1, PLCB2, EGFR, FGFR1 |
| Inositol Lipid-Mediated Signaling | 10/33 | 0.00 | 838 | PDGFRB, PDGFRA, PLCB3, PLCB4, PLCG2, PLCG1, PLCB2, PLCD1, IGF1R, FGFR1 |
| Carbohydrate Metabolism | ||||
| Fructose Metabolism | 4/7 | 0.00 | 1370 | TKFC, ALDH1A1, SORD, ALDOB |
| Cellular Stress and Defense | ||||
| mRNA Protein and Metabolite Induction Pathway by Cyclosporin A | 4/7 | 0.00 | 1370 | SLC3A2, SLC7A11, NFE2L2, ATF4 |
| SOS-Mediated Signaling | 4/7 | 0.00 | 1370 | IRS1, GRB2, IRS2, HRAS |
| Positive Regulation of Protein Catabolic Process in The Vacuole | 3/5 | 0.00 | 1191 | LRP1, LRP2, LDL |
| Cell Membrane and Cytoplasm, and Endosome and ER & Glucose and Insulin | ||||
| Insulin Signaling Pathway | ||||
| Insulin Receptor Signaling Pathway | 22/47 | 0.00 | 11,689 | GSK3B, C2CD5, GSK3A, IRS1, INSR, GAB1, PIK3R3, IRS2, PIK3R2, IDE, PIK3R1, SORBS1, PIK3C2A, SLC39A14, IGF1R, FER, AKT2, GRB2, AP3S1, PTPN2, RHOQ, APPL1 |
| Negative Regulation of Insulin Receptor Signaling Pathway | 11/26 | 0.00 | 4338 | GSK3A, SOCS1, IRS1, PRKCB, PRKCD, PIP4K2A, TSC2, PIP4K2B, PRKCQ, TNS2, PTPN2 |
| IRS Activation | 3/5 | 0.00 | 2694 | IRS1, INSR, IRS2 |
| Glucose Metabolism | ||||
| Positive Regulation of Glucose Import | 11/25 | 0.00 | 4712 | PRKCI, OCLN, IRS1, CAPN10, AKT2, INSR, IRS2, PIK3R1, SORBS1, RHOQ, APPL1 |
| Glucose Transmembrane Transport | 8/22 | 0.00 | 2301 | SLC2A14, SORT1, SLC2A12, SLC2A1, SLC2A3, SLC2A4, SLC2A5, SLC5A3 |
| PI3K/AKT/mTOR Signaling Pathway | IRS1, INSR, AKT2, IRS2, SORBS1 | |||
| PI3K/AKT/mTOR Signaling | 8/105 | 0.00 | 170 | RAB10, SLC2A4 |
| Genes encoding proteins with anti-cancer effects on cancer cells | |
| Secreted | TNFSF10 (TRAIL), TNFSF14 (LIGHT), TNFSF15 (TL1A), LTA, BMP4, BMP7, BMP2, INHA, INHBA, SLIT3, CCN3, DCN, DNASE1L3, OAS1, SULF1, THBS1, PTEN |
| Receptors | TNFRSF1B (TNFR2), TNFRSF11B, TNFRSF6B (DcR3), BMPR2, ACVR1, P2RX4, CDHR2 |
| Genes encoding proteins that mediate MSC apoptosis and growth suppression | |
| Receptors | PTPRJ, PTPRH |
| Intracellular | DAB2IP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Galliou, P.-A.; Argyri, N.; Maria, P.; Koliakos, G.; Papanikolaou, N.A. MSC1 Cells Suppress Colorectal Cancer Cell Growth via Metabolic Reprogramming, Laminin–Integrin Adhesion Signaling, Oxidative Stress Resistance, and a Tumor-Suppressive Secretome. Biomedicines 2025, 13, 1503. https://doi.org/10.3390/biomedicines13061503
Galliou P-A, Argyri N, Maria P, Koliakos G, Papanikolaou NA. MSC1 Cells Suppress Colorectal Cancer Cell Growth via Metabolic Reprogramming, Laminin–Integrin Adhesion Signaling, Oxidative Stress Resistance, and a Tumor-Suppressive Secretome. Biomedicines. 2025; 13(6):1503. https://doi.org/10.3390/biomedicines13061503
Chicago/Turabian StyleGalliou, Panagiota-Angeliki, Niti Argyri, Papaioannou Maria, George Koliakos, and Nikolaos A. Papanikolaou. 2025. "MSC1 Cells Suppress Colorectal Cancer Cell Growth via Metabolic Reprogramming, Laminin–Integrin Adhesion Signaling, Oxidative Stress Resistance, and a Tumor-Suppressive Secretome" Biomedicines 13, no. 6: 1503. https://doi.org/10.3390/biomedicines13061503
APA StyleGalliou, P.-A., Argyri, N., Maria, P., Koliakos, G., & Papanikolaou, N. A. (2025). MSC1 Cells Suppress Colorectal Cancer Cell Growth via Metabolic Reprogramming, Laminin–Integrin Adhesion Signaling, Oxidative Stress Resistance, and a Tumor-Suppressive Secretome. Biomedicines, 13(6), 1503. https://doi.org/10.3390/biomedicines13061503








