Single-Cell Transcriptomics Unveils the Mechanistic Role of FOSL1 in Cutaneous Wound Healing
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection and Batch Correction
2.2. Identification of Differentially Expressed Genes (DEGs)
2.3. TRRUST–STRING Integrated Gene–PPI Analysis
2.4. Feature Selection with Machine Learning Algorithms
2.5. Animal Study Design
2.6. Western Blot
2.7. Single-Cell RNA Sequencing Data Preprocessing and Pseudotime Analysis
2.8. Enrichment Analysis
2.9. Statistical Framework and Visualization
3. Results
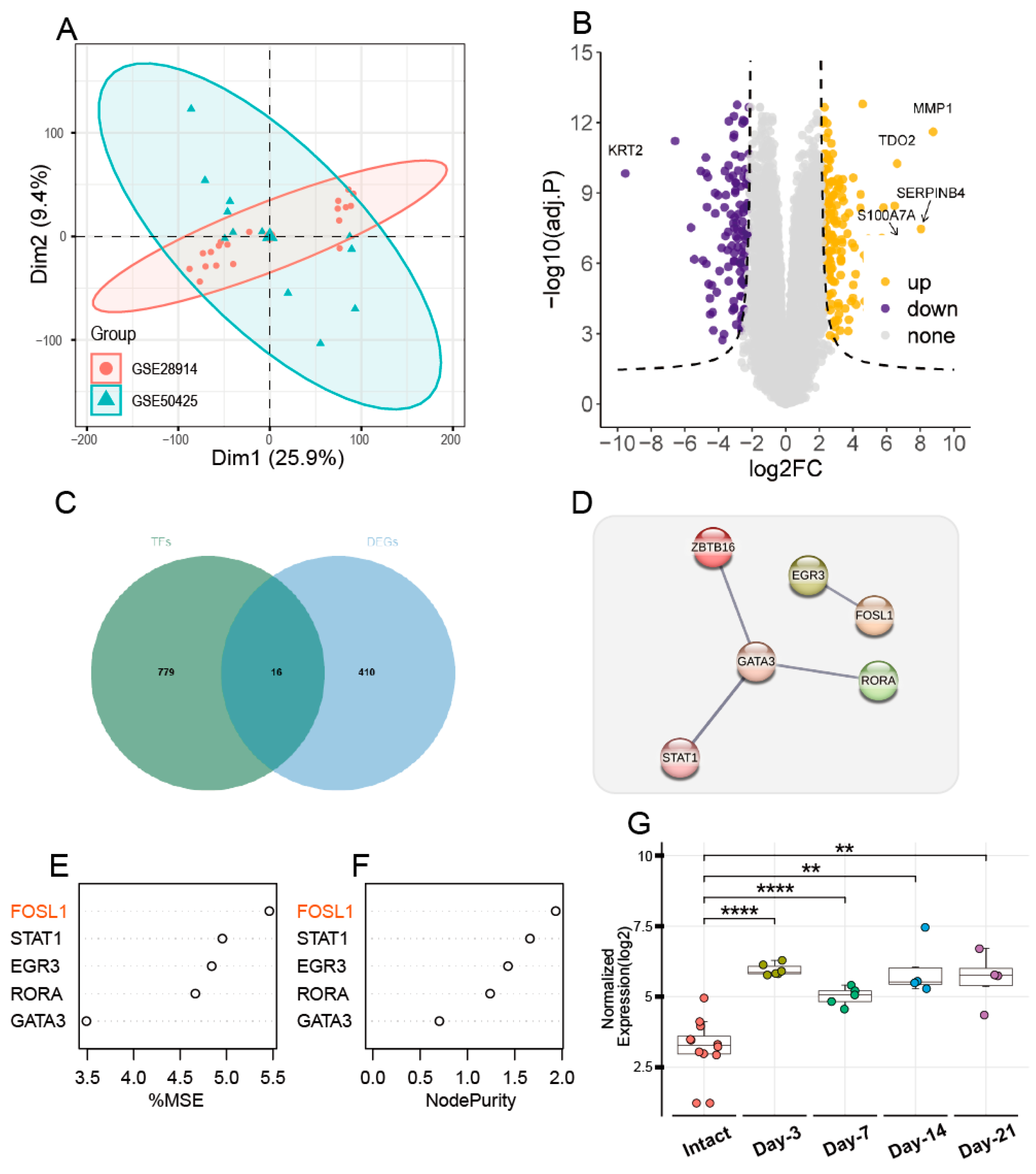
3.1. FOSL1 Identification Through Comprehensive Data Analysis
3.2. Animal Experiments
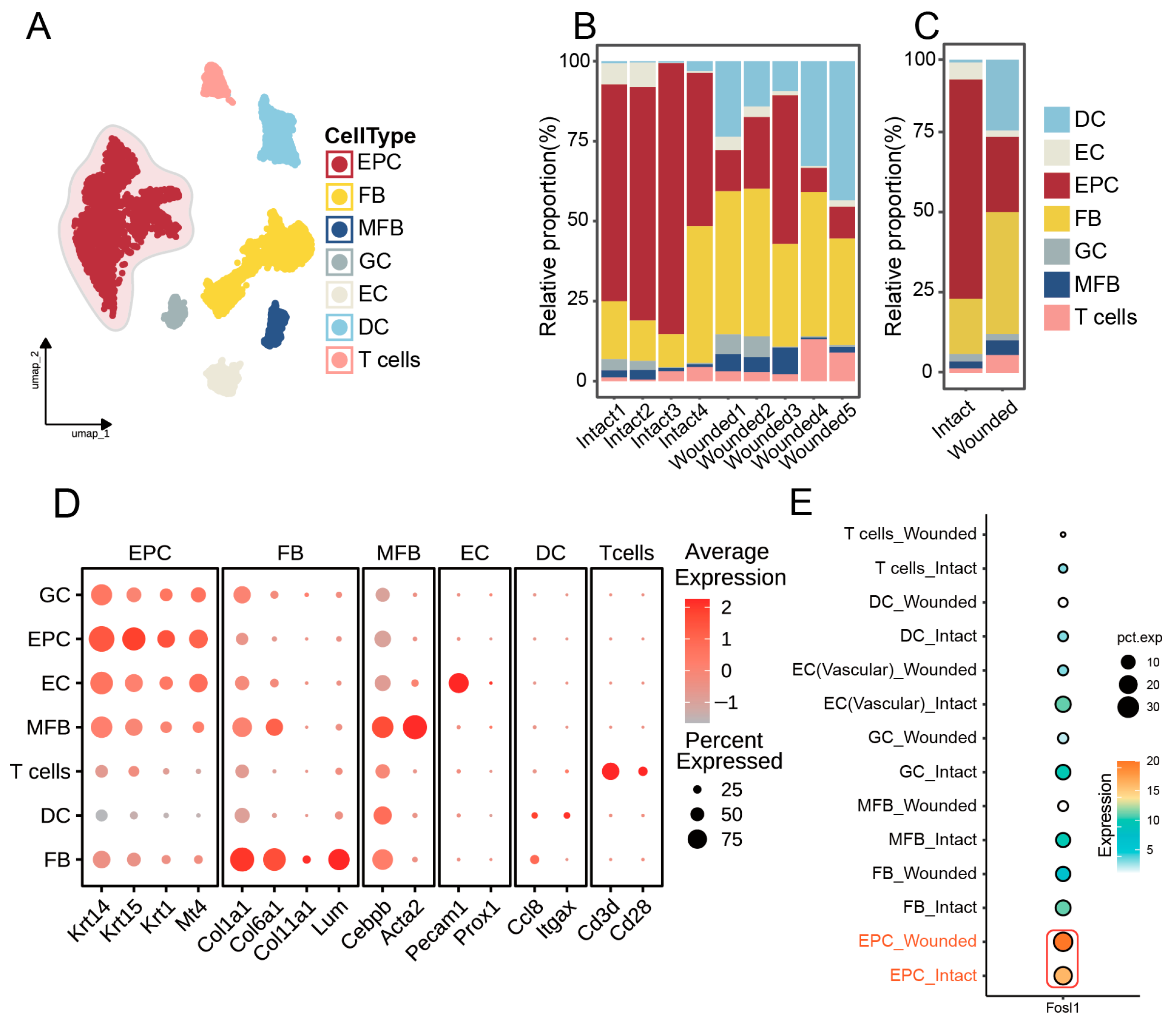
3.3. Identification of Cellular Composition in Wounded and Intact Samples
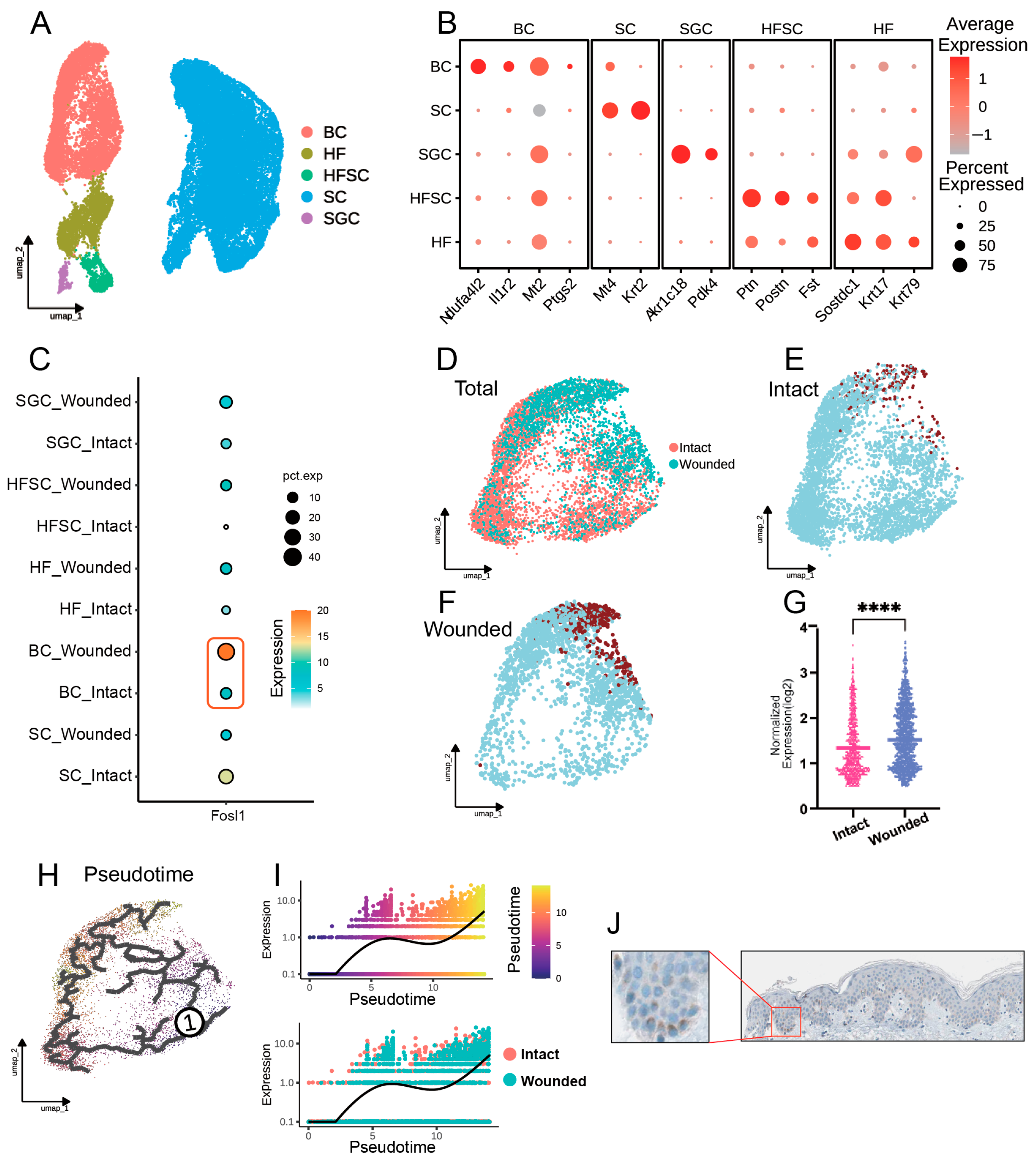
3.4. FOSL1 Expression and Cellular Dynamics in Basal Cells During Wound Repair
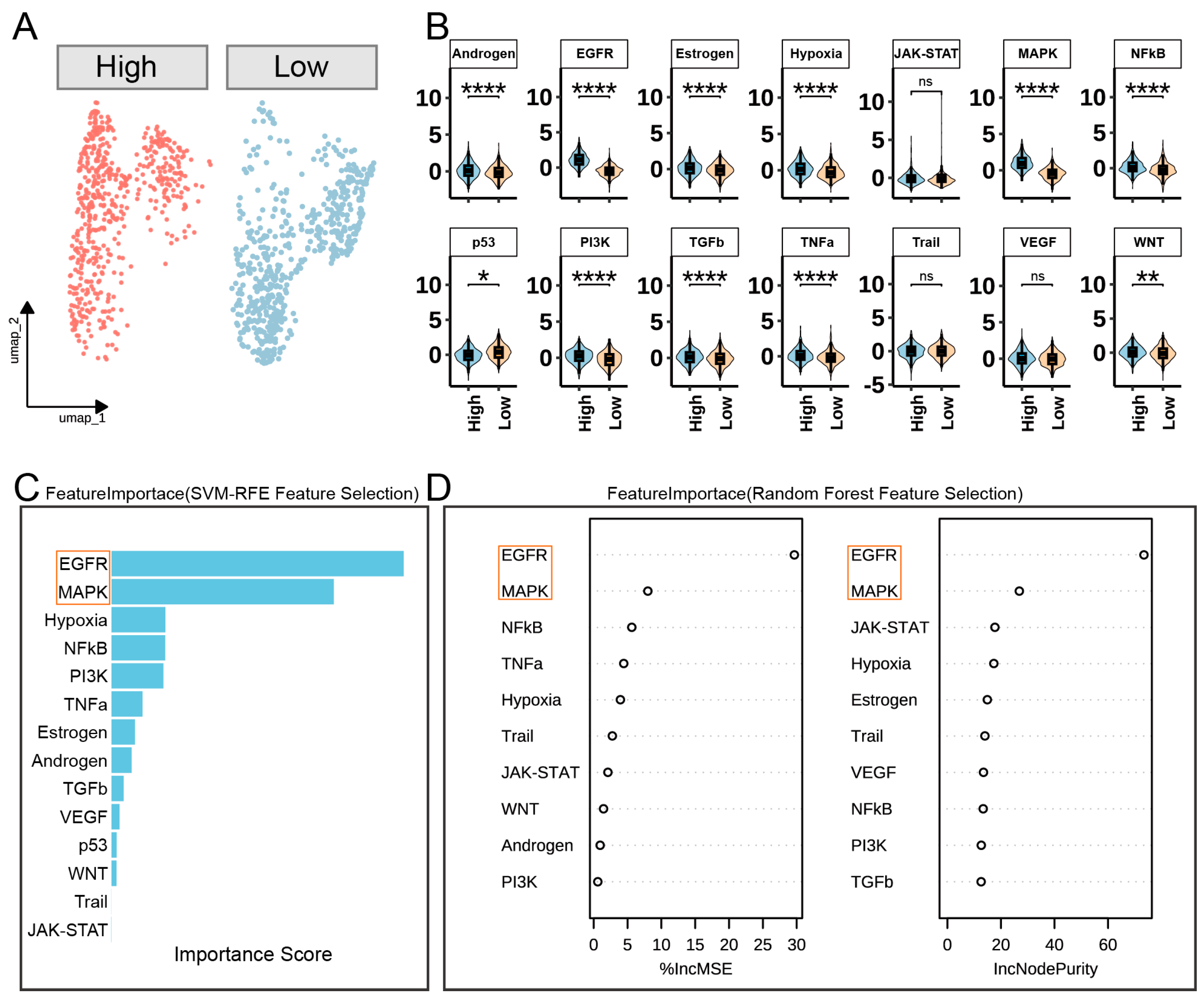
3.5. FOSL1-Mediated Signaling Pathways in Basal Cells During Wound Healing
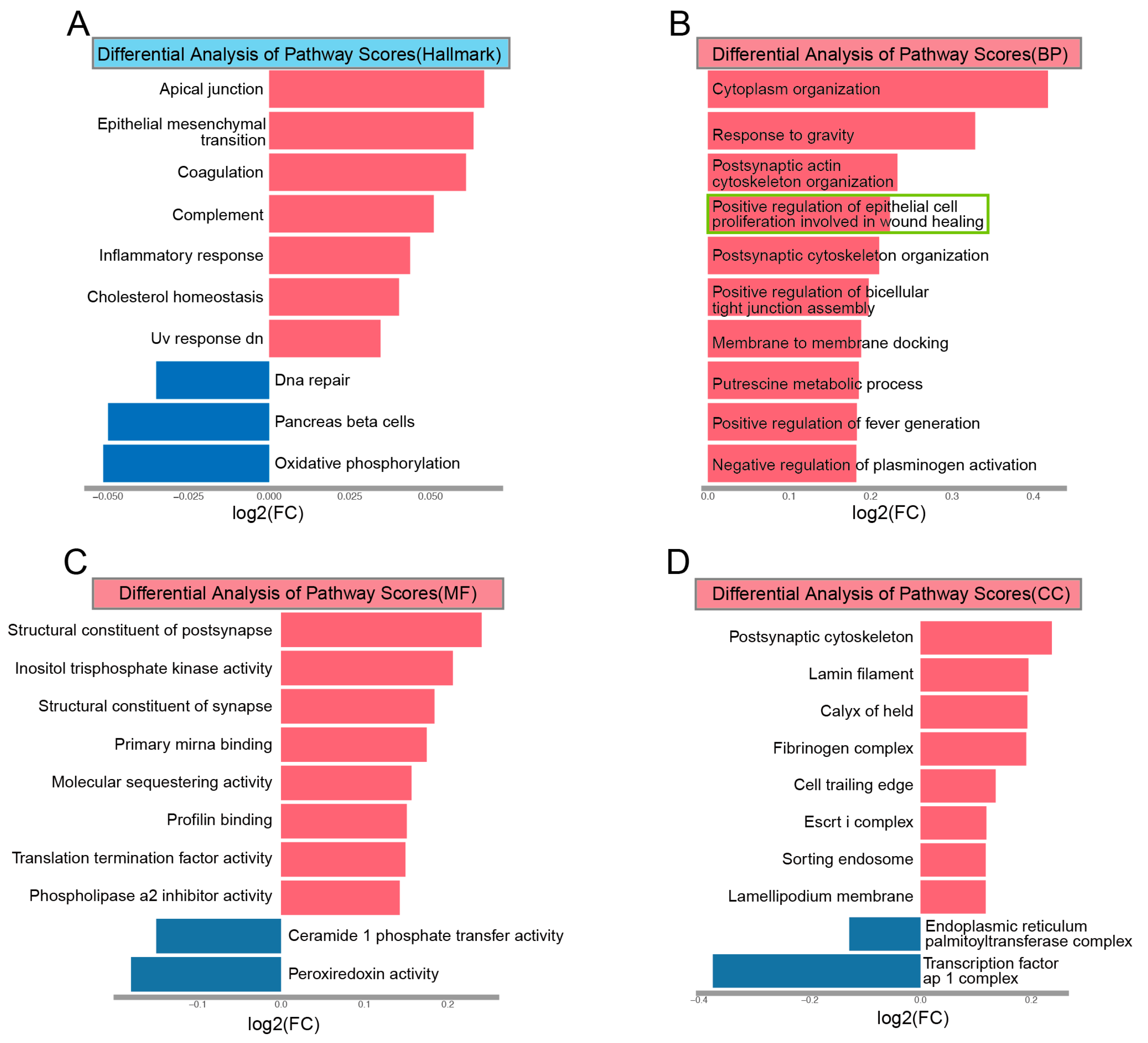
3.6. GSVA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baum, C.L.; Arpey, C.J. Normal cutaneous wound healing: Clinical correlation with cellular and molecular events. Dermatol. Surg. 2005, 31, 674–686. [Google Scholar] [CrossRef]
- Gilliver, S.C.; Ashworth, J.J.; Ashcroft, G.S. The hormonal regulation of cutaneous wound healing. Clin. Dermatol. 2007, 25, 56–62. [Google Scholar] [CrossRef]
- Zhang, M.; Liang, L.; He, J.; He, Z.; Yue, C.; Jin, X.; Gao, M.; Xiao, S.; Zhou, Y. Fra-1 inhibits cell growth and the Warburg effect in cervical cancer cells via STAT1 regulation of the p53 signaling pathway. Front. Cell Dev. Biol. 2020, 8, 579629. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Bian, X.; Luo, L.; Bjorklund, A.K.; Li, L.; Zhang, L.; Chen, Y.; Guo, L.; Gao, J.; Cao, C.; et al. Spatiotemporal single-cell roadmap of human skin wound healing. Cell Stem Cell 2025, 32, 479–498.e8. [Google Scholar] [CrossRef]
- Escuin-Ordinas, H.; Li, S.; Xie, M.W.; Sun, L.; Hugo, W.; Huang, R.R.; Jiao, J.; de-Faria, F.M.; Realegeno, S.; Krystofinski, P. Cutaneous wound healing through paradoxical MAPK activation by BRAF inhibitors. Nat. Commun. 2016, 7, 12348. [Google Scholar] [CrossRef] [PubMed]
- Sarate, R.M.; Hochstetter, J.; Valet, M.; Hallou, A.; Song, Y.; Bansaccal, N.; Ligare, M.; Aragona, M.; Engelman, D.; Bauduin, A. Dynamic regulation of tissue fluidity controls skin repair during wound healing. Cell 2024, 187, 5298–5315.e19. [Google Scholar] [CrossRef] [PubMed]
- Lê, S.; Josse, J.; Husson, F. FactoMineR: An R package for multivariate analysis. J. Stat. Softw. 2008, 25, 1–18. [Google Scholar] [CrossRef]
- Han, H.; Cho, J.-W.; Lee, S.; Yun, A.; Kim, H.; Bae, D.; Yang, S.; Kim, C.Y.; Lee, M.; Kim, E. TRRUST v2: An expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018, 46, D380–D386. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudie, F.; Djemiel, C.; Klopp, C. jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Speiser, J.L.; Miller, M.E.; Tooze, J.; Ip, E. A comparison of random forest variable selection methods for classification prediction modeling. Expert Syst. Appl. 2019, 134, 93–101. [Google Scholar] [CrossRef]
- Kuhn, M. Building predictive models in R using the caret package. J. Stat. Softw. 2008, 28, 1–26. [Google Scholar] [CrossRef]
- Chandrasekar, V.; Mohammad, S.; Aboumarzouk, O.; Singh, A.V.; Dakua, S.P. Quantitative prediction of toxicological points of departure using two-stage machine learning models: A new approach methodology (NAM) for chemical risk assessment. J. Hazard. Mater. 2025, 487, 137071. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Stuart, T.; Kowalski, M.H.; Choudhary, S.; Hoffman, P.; Hartman, A.; Srivastava, A.; Molla, G.; Madad, S.; Fernandez-Granda, C. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024, 42, 293–304. [Google Scholar] [CrossRef]
- Korsunsky, I.; Millard, N.; Fan, J.; Slowikowski, K.; Zhang, F.; Wei, K.; Baglaenko, Y.; Brenner, M.; Loh, P.-r.; Raychaudhuri, S. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods 2019, 16, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Spielmann, M.; Qiu, X.; Huang, X.; Ibrahim, D.M.; Hill, A.J.; Zhang, F.; Mundlos, S.; Christiansen, L.; Steemers, F.J. The single-cell transcriptional landscape of mammalian organogenesis. Nature 2019, 566, 496–502. [Google Scholar] [CrossRef]
- Schubert, M.; Klinger, B.; Klünemann, M.; Sieber, A.; Uhlitz, F.; Sauer, S.; Garnett, M.J.; Blüthgen, N.; Saez-Rodriguez, J. Perturbation-response genes reveal signaling footprints in cancer gene expression. Nat. Commun. 2018, 9, 20. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Balli, M.; Chui, J.S.-H.; Athanasouli, P.; Abreu de Oliveira, W.A.; El Laithy, Y.; Sampaolesi, M.; Lluis, F. Activator protein-1 transcriptional activity drives soluble micrograft-mediated cell migration and promotes the matrix remodeling machinery. Stem Cells Int. 2019, 2019, 6461580. [Google Scholar] [CrossRef] [PubMed]
- Thuraisingam, T.; Xu, Y.Z.; Eadie, K.; Heravi, M.; Guiot, M.-C.; Greemberg, R.; Gaestel, M.; Radzioch, D. MAPKAPK-2 signaling is critical for cutaneous wound healing. J. Investig. Dermatol. 2010, 130, 278–286. [Google Scholar] [CrossRef] [PubMed]
- Repertinger, S.K.; Campagnaro, E.; Fuhrman, J.; El-Abaseri, T.; Yuspa, S.H.; Hansen, L.A. EGFR enhances early healing after cutaneous incisional wounding. J. Investig. Dermatol. 2004, 123, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Wnorowski, A.M.; de Souza, A.; Chachoua, A.; Cohen, D.E. The management of EGFR inhibitor adverse events: A case series and treatment paradigm. Int. J. Dermatol. 2012, 51, 223–232. [Google Scholar] [CrossRef]
- Lacouture, M.E. Mechanisms of cutaneous toxicities to EGFR inhibitors. Nat. Rev. Cancer 2006, 6, 803–812. [Google Scholar] [CrossRef]

| Accession | Country | Sample | Organism | Wound Date | Data Type | Manufacturer | Platform |
|---|---|---|---|---|---|---|---|
| GSE28914 | Finland | Tissue | Homo sapiens | Intact, 3rd, 7th | Chip | Affymetrix | U133 Plus 2.0 Array |
| GSE50425 | Finland | Tissue | Homo sapiens | Intact, 14th, 21st | Chip | Illumina | HumanHT-12 |
| GSE142471 | USA | Tissue | Mus musculus | Intact, 4th | Single cell | Illumina | Hiseq 4000 |
| GSE245864 | USA | Tissue | Mus musculus | Intact, 3rd | Single cell | MGItech | DNBSEQ-G400 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, J.; Zheng, G.; Luo, Y.; Ge, L.; Liu, Z.; Huang, W.; Jin, M.; Kong, Y.; Xu, S.; Jin, Z.; et al. Single-Cell Transcriptomics Unveils the Mechanistic Role of FOSL1 in Cutaneous Wound Healing. Biomedicines 2025, 13, 1330. https://doi.org/10.3390/biomedicines13061330
Meng J, Zheng G, Luo Y, Ge L, Liu Z, Huang W, Jin M, Kong Y, Xu S, Jin Z, et al. Single-Cell Transcriptomics Unveils the Mechanistic Role of FOSL1 in Cutaneous Wound Healing. Biomedicines. 2025; 13(6):1330. https://doi.org/10.3390/biomedicines13061330
Chicago/Turabian StyleMeng, Jingbi, Ge Zheng, Yinli Luo, Ling Ge, Zhiqing Liu, Wenhua Huang, Meitong Jin, Yanli Kong, Shanhua Xu, Zhehu Jin, and et al. 2025. "Single-Cell Transcriptomics Unveils the Mechanistic Role of FOSL1 in Cutaneous Wound Healing" Biomedicines 13, no. 6: 1330. https://doi.org/10.3390/biomedicines13061330
APA StyleMeng, J., Zheng, G., Luo, Y., Ge, L., Liu, Z., Huang, W., Jin, M., Kong, Y., Xu, S., Jin, Z., & Pi, L. (2025). Single-Cell Transcriptomics Unveils the Mechanistic Role of FOSL1 in Cutaneous Wound Healing. Biomedicines, 13(6), 1330. https://doi.org/10.3390/biomedicines13061330






