FAM111B Overexpression and Immune Cell Infiltration: Implications for Ovarian Cancer Immunotherapy
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Microarray
2.2. Reagent
2.3. Multiplex Immunofluorescence and Immunohistochemistry
2.4. Digital Image Acquisition and Analysis
2.5. Bioinformatics Analysis
2.6. Statistical Analysis
3. Results
3.1. Multiplex Immunofluorescence Staining-Based Evaluation of FAM111B Expression and Immune Cell Subtypes in OC Tissues
3.2. FAM111B-Positive and Immune Cells Are Mainly Distributed in Tumor Tissues
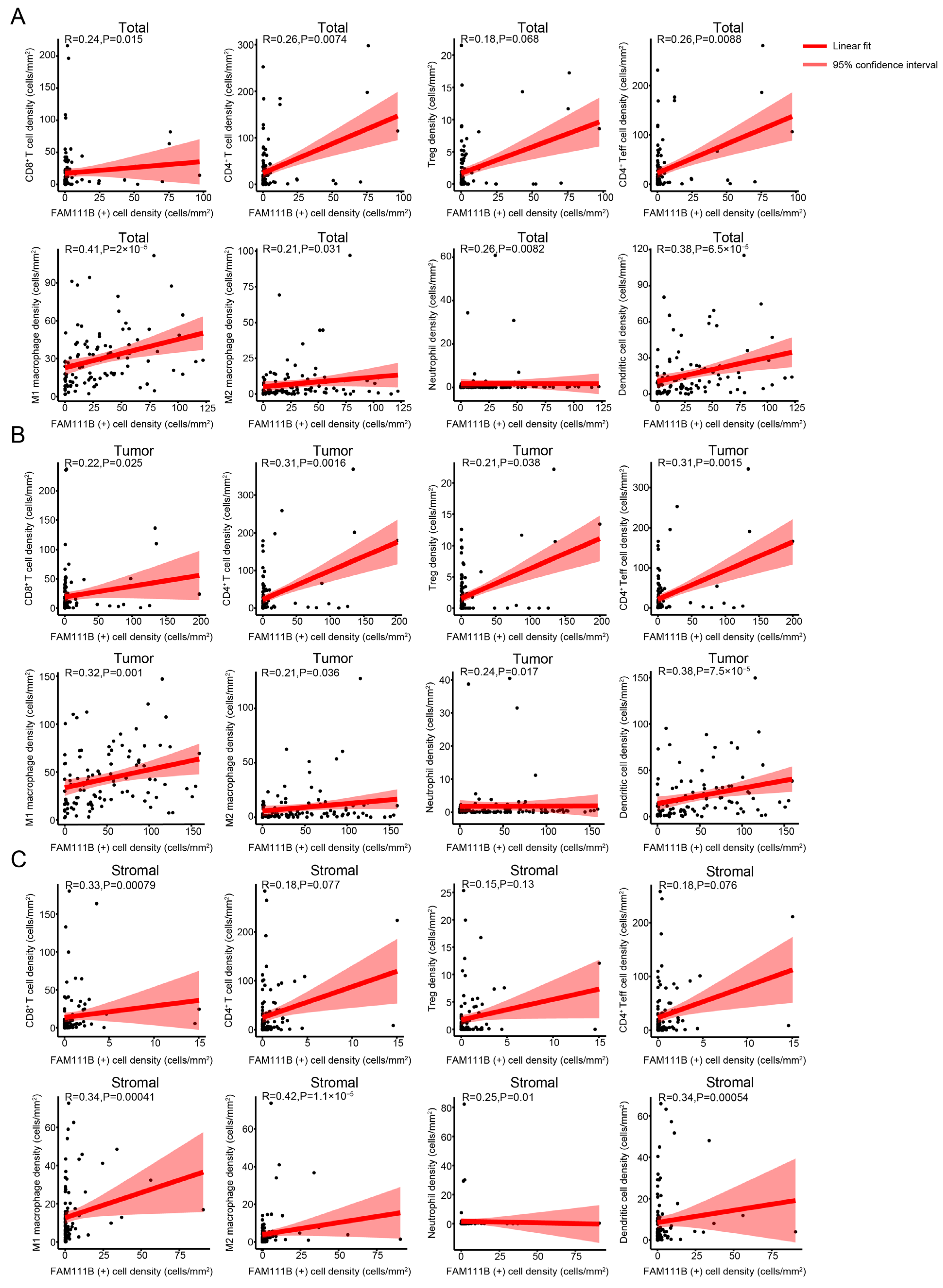
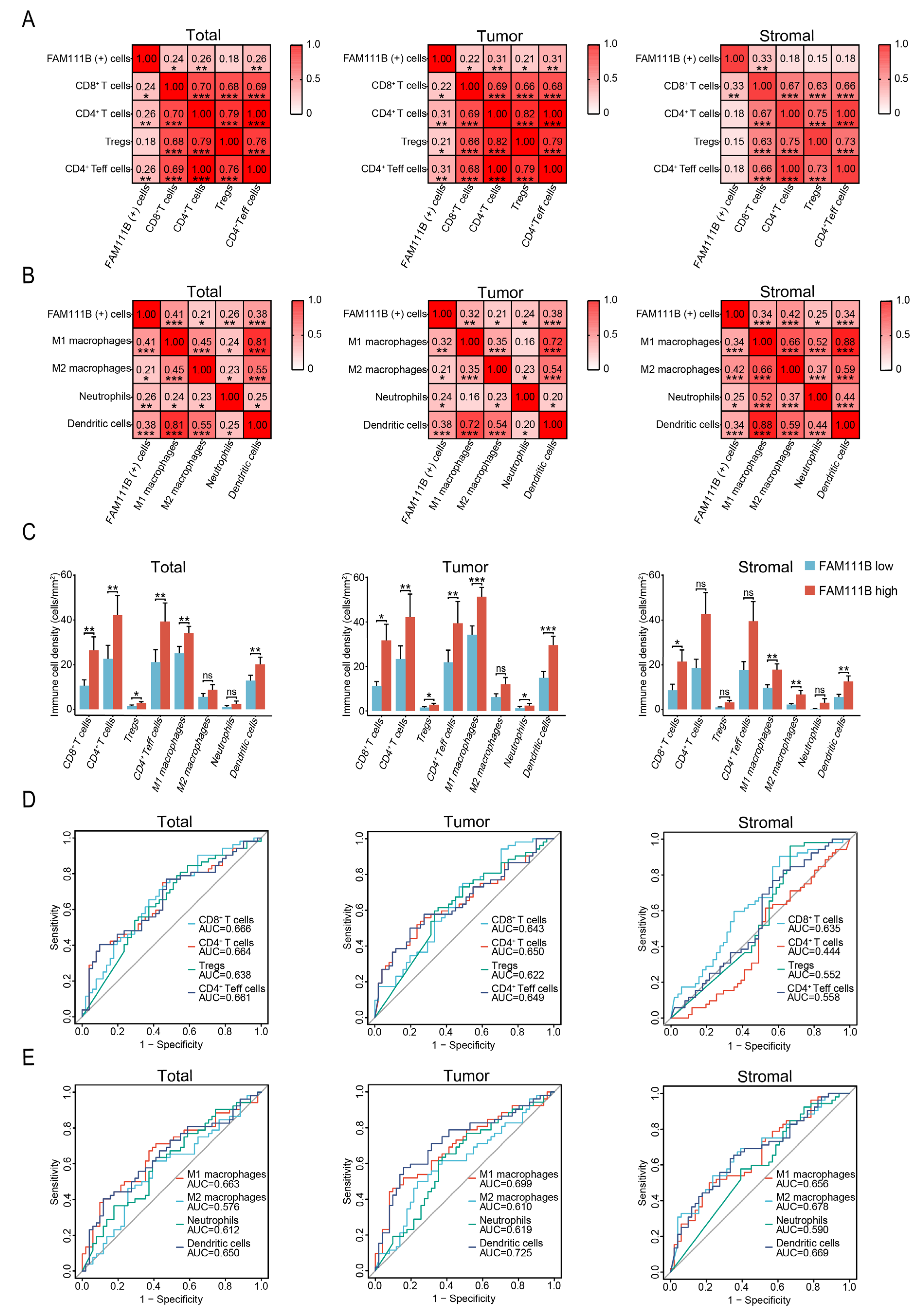
3.3. FAM111B Expression Is Positively Correlated with the Infiltration Levels of Different Immune Cells in OC Tissues
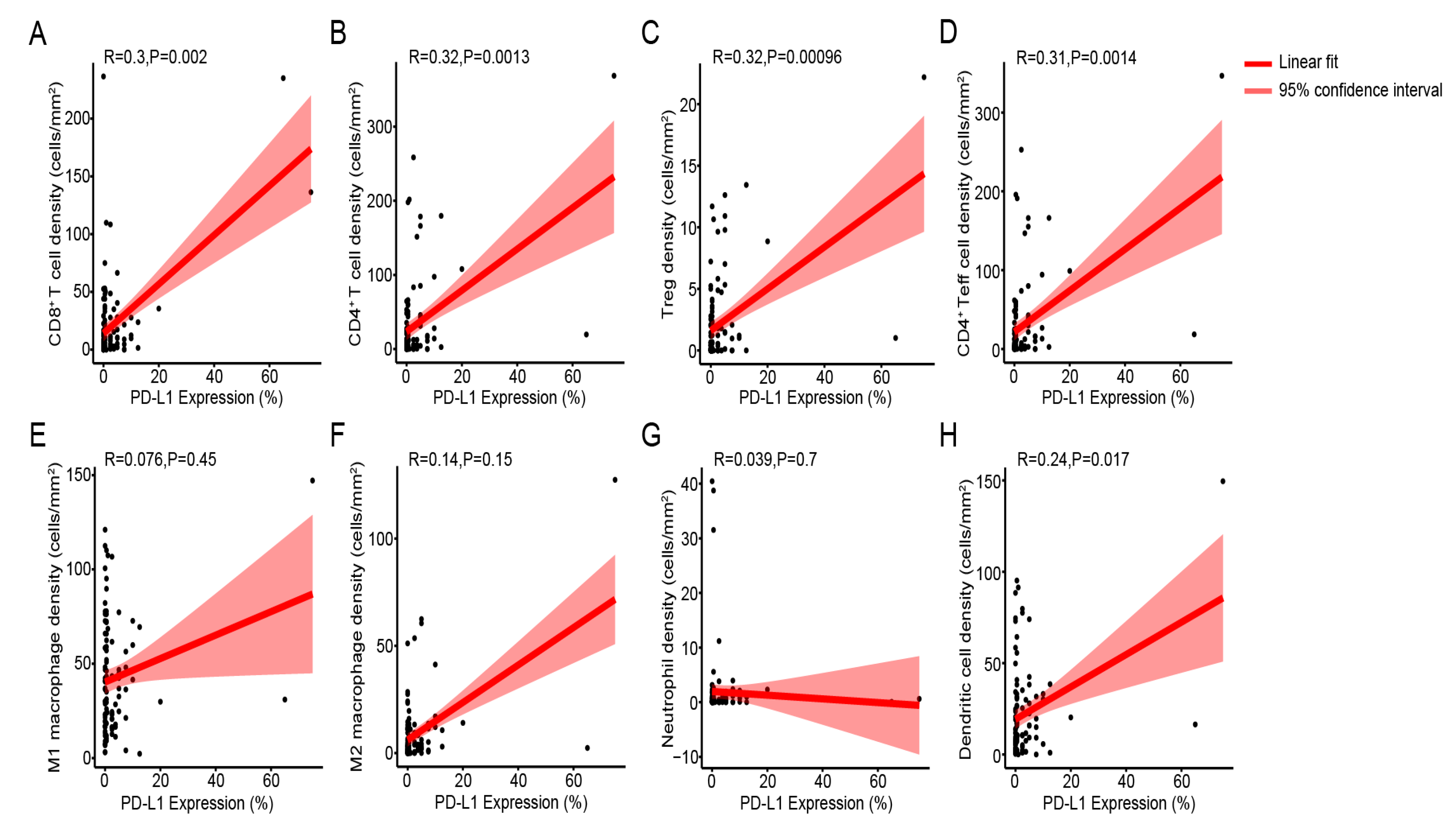
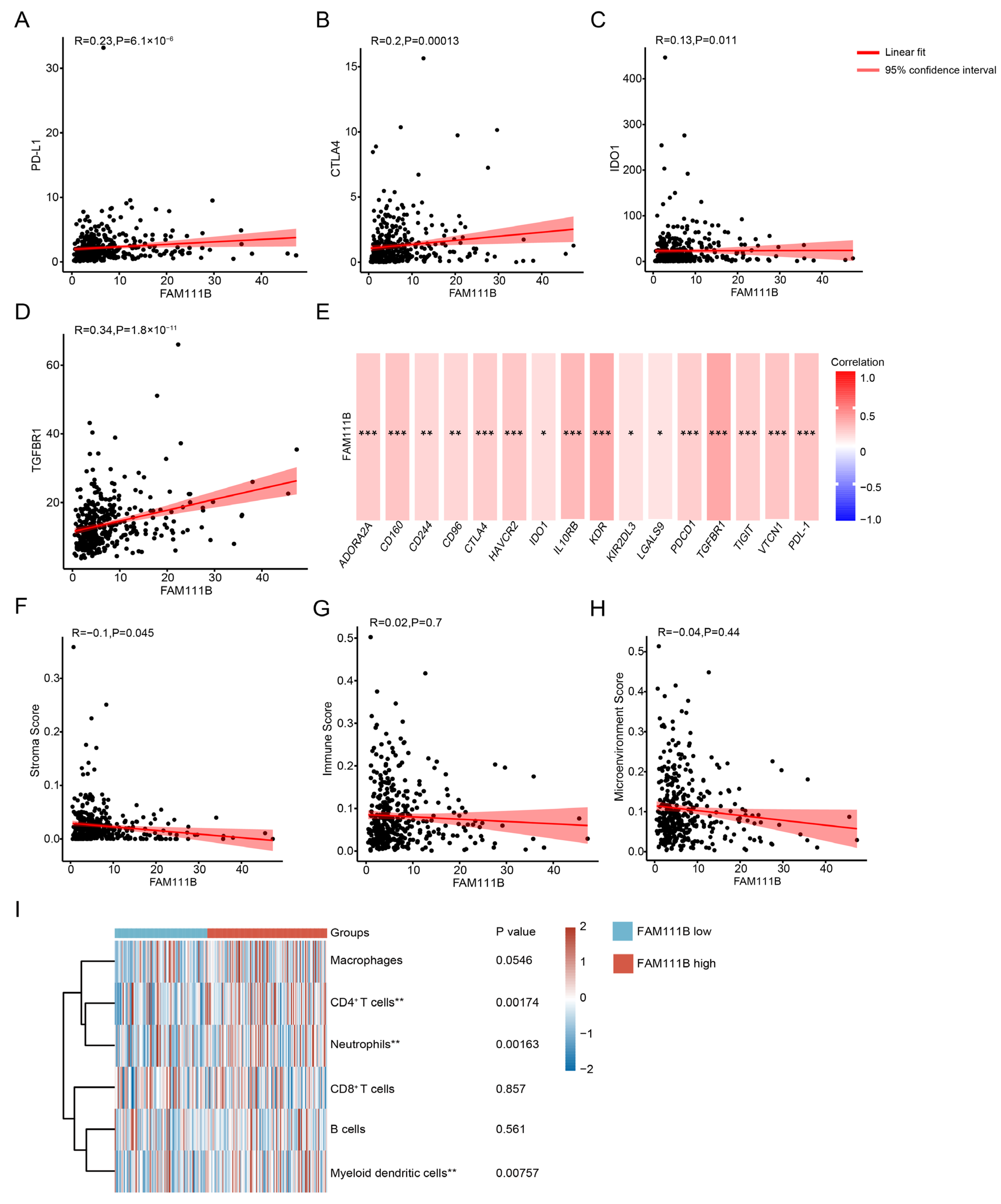
3.4. Immune Cell Density Levels Are Positively Correlated with PD-L1 Protein Expression
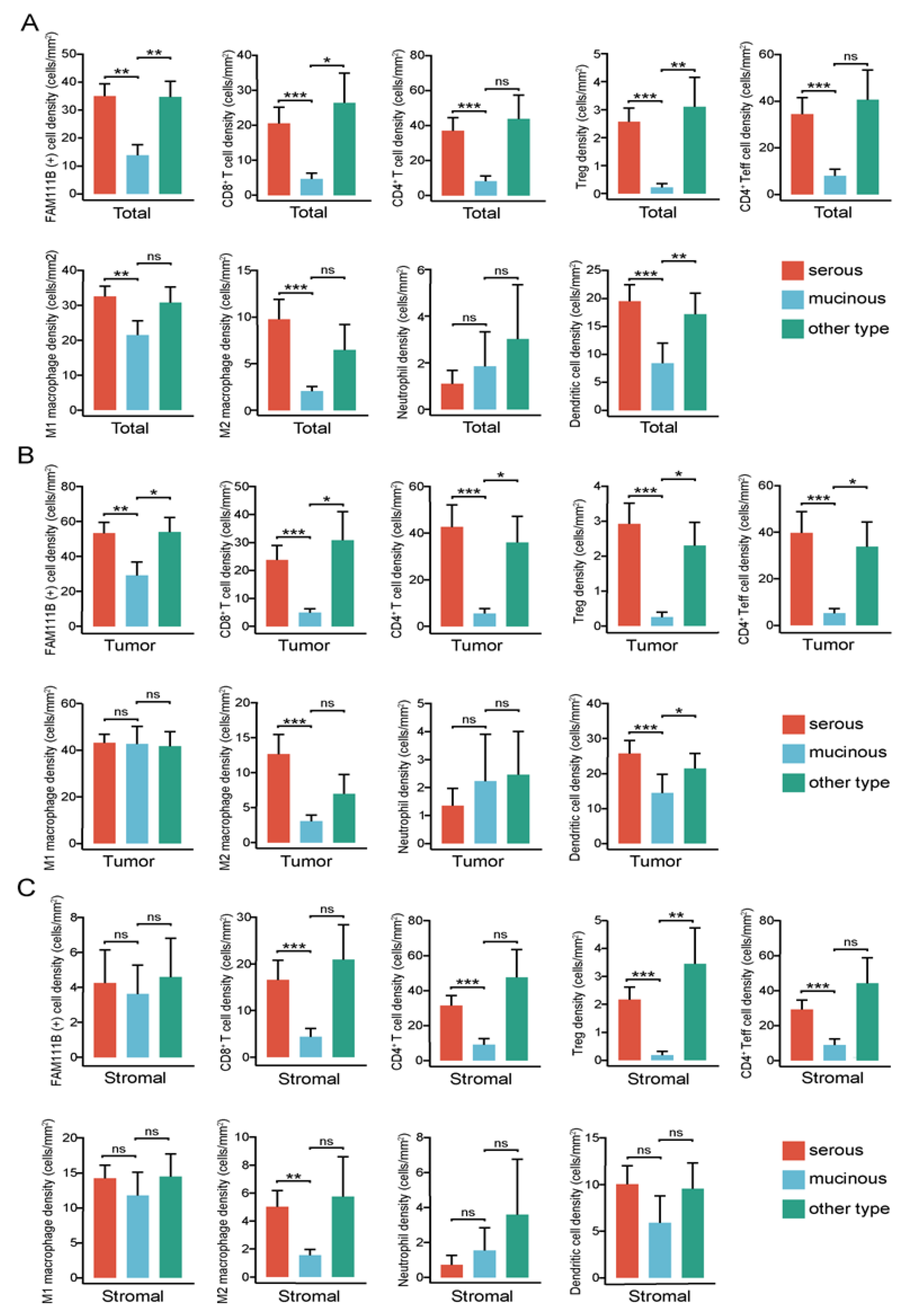
3.5. FAM111B-Positive and Immune Cell Density Levels Are Higher in Serous Carcinoma Relative to Mucinous Carcinoma
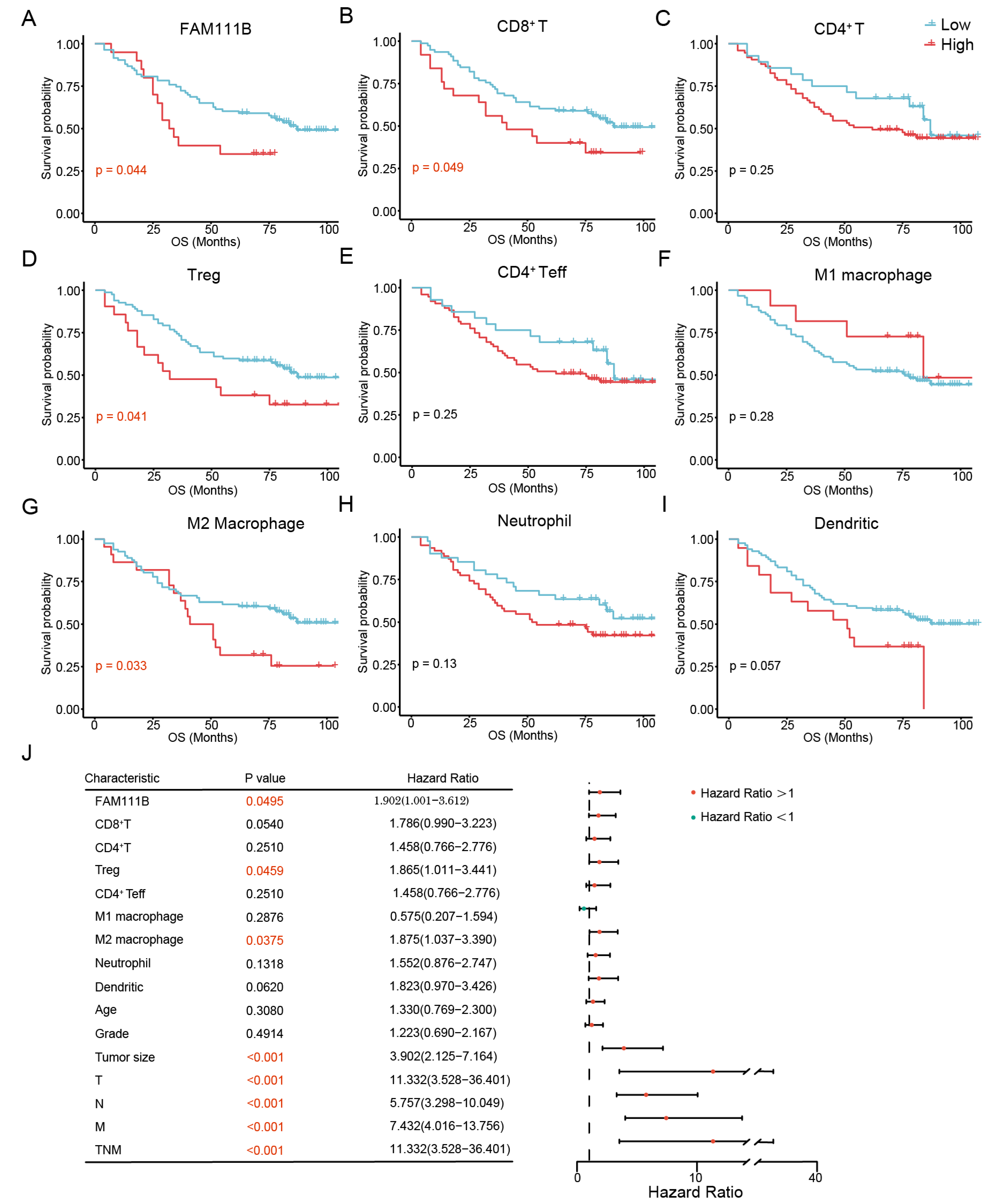
3.6. FAM111B and Immune Cell Subtypes Are Associated with the Prognosis of OC
3.7. FAM111B Expression Is Associated with Immune Cell Distributions, Immune Checkpoint Expression, and Immune Scores in OC Patients from the TCGA Dataset
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| OC | Ovarian cancer |
| TIME | Tumor immune microenvironment |
| Tregs | Regulatory T cells |
| FAM111B | Family with sequence similarity 111 member B |
| PD-L1 | Programmed death-ligand 1 |
| IHC | Immunohistochemistry |
| mIF | Multiplex immunofluorescence |
| TCGA | The cancer genome atlas |
| FIGO | International federation of gynecology and obstetrics |
| OS | Overall survival |
| PANCK | Pan Cytokeratin |
| mm2 | Square millimeter |
| CTLA-4 | Cytotoxic T-lymphocyte antigen 4 |
| IDO1 | Indoleamine 2,3-dioxygenase 1 |
| ROC | Receiver operating characteristic |
| AUC | Area under the curve |
| CD4+ Teff cells | CD4+ effector T cells |
| DCs | Dendritic cells |
| ATM | Ataxia telangiectasia mutated |
| NF-κB | Nuclear factor-kappa B |
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Konstantinopoulos, P.A.; Matulonis, U.A. Clinical and translational advances in ovarian cancer therapy. Nat. Cancer 2023, 4, 1239–1257. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, D.K.; Alvarez, R.D.; Bakkum-Gamez, J.N.; Barroilhet, L.; Behbakht, K.; Berchuck, A.; Chen, L.M.; Cristea, M.; DeRosa, M.; Eisenhauer, E.L.; et al. Ovarian Cancer, Version 2.2020, NCCN Clinical Practice Guidelines in Oncology. J. Natl. Compr. Cancer Netw. 2021, 19, 191–226. [Google Scholar] [CrossRef] [PubMed]
- Kurnit, K.C.; Fleming, G.F.; Lengyel, E. Updates and New Options in Advanced Epithelial Ovarian Cancer Treatment. Obstet. Gynecol. 2021, 137, 108–121. [Google Scholar] [CrossRef]
- Cai, L.; Li, Y.; Tan, J.; Xu, L.; Li, Y. Targeting LAG-3, TIM-3, and TIGIT for cancer immunotherapy. J. Hematol. Oncol. 2023, 16, 101. [Google Scholar] [CrossRef]
- Ghisoni, E.; Morotti, M.; Sarivalasis, A.; Grimm, A.J.; Kandalaft, L.; Laniti, D.D.; Coukos, G. Immunotherapy for ovarian cancer: Towards a tailored immunophenotype-based approach. Nat. Rev. Clin. Oncol. 2024, 21, 801–817. [Google Scholar] [CrossRef]
- Blanc-Durand, F.; Pautier, P.; Michels, J.; Leary, A. Targeting the immune microenvironment in ovarian cancer therapy-mission impossible? ESMO Open 2024, 9, 102936. [Google Scholar] [CrossRef]
- Wang, Z.B.; Zhang, X.; Fang, C.; Liu, X.T.; Liao, Q.J.; Wu, N.; Wang, J. Immunotherapy and the ovarian cancer microenvironment: Exploring potential strategies for enhanced treatment efficacy. Immunology 2024, 173, 14–32. [Google Scholar] [CrossRef]
- Morand, S.; Devanaboyina, M.; Staats, H.; Stanbery, L.; Nemunaitis, J. Ovarian Cancer Immunotherapy and Personalized Medicine. Int. J. Mol. Sci. 2021, 22, 6532. [Google Scholar] [CrossRef]
- Mollaoglu, G.; Tepper, A.; Falcomatà, C.; Potak, H.T.; Pia, L.; Amabile, A.; Mateus-Tique, J.; Rabinovich, N.; Park, M.D.; LaMarche, N.M.; et al. Ovarian cancer-derived IL-4 promotes immunotherapy resistance. Cell 2024, 187, 7492–7510.e22. [Google Scholar] [CrossRef]
- Wu, H.; Liang, C. Pan-Cancer Analysis of the Tumorigenic Effect and Prognostic Diagnostic Value of FAM111B in Human Carcinomas. Int. J. Gen. Med. 2023, 16, 1845–1865. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Gu, Y.; Ni, H.; Quan, Q.; Guo, L. Silencing of FAM111B inhibits tumor growth and promotes apoptosis by decreasing AKT activity in ovarian cancer. Exp. Biol. Med. (Maywood) 2023, 248, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Yu, G.; Si, C.; Chao, T.; Xiong, H.; Zhang, L. High FAM111B expression predicts aggressive clinicopathologic features and poor prognosis in ovarian cancer. Transl. Oncol. 2023, 32, 101659. [Google Scholar] [CrossRef] [PubMed]
- Lao, Y.; Li, T.; Xie, X.; Chen, K.; Li, M.; Huang, L. MiR-195-3p is a Novel Prognostic Biomarker Associated with Immune Infiltrates of Lung Adenocarcinoma. Int. J. Gen. Med. 2022, 15, 191–203. [Google Scholar] [CrossRef]
- Li, W.; Feng, S.S.; Wu, H.; Deng, J.; Zhou, W.Y.; Jia, M.X.; Shi, Y.; Ma, L.; Zeng, X.X.; Zuberi, Z.; et al. Comprehensive Analysis of CDK1-Associated ceRNA Network Revealing the Key Pathways LINC00460/LINC00525-Hsa-Mir-338-FAM111/ZWINT as Prognostic Biomarkers in Lung Adenocarcinoma Combined with Experiments. Cells 2022, 11, 1220. [Google Scholar] [CrossRef]
- Bankhead, P.; Loughrey, M.B.; Fernández, J.A.; Dombrowski, Y.; McArt, D.G.; Dunne, P.D.; McQuaid, S.; Gray, R.T.; Murray, L.J.; Coleman, H.G.; et al. QuPath: Open source software for digital pathology image analysis. Sci. Rep. 2017, 7, 16878. [Google Scholar] [CrossRef]
- Fan, J.Q.; Wang, M.F.; Chen, H.L.; Shang, D.; Das, J.K.; Song, J. Current advances and outlooks in immunotherapy for pancreatic ductal adenocarcinoma. Mol. Cancer 2020, 19, 32. [Google Scholar] [CrossRef]
- Xiao, Z.; Todd, L.; Huang, L.; Noguera-Ortega, E.; Lu, Z.; Huang, L.; Kopp, M.; Li, Y.; Pattada, N.; Zhong, W.; et al. Desmoplastic stroma restricts T cell extravasation and mediates immune exclusion and immunosuppression in solid tumors. Nat. Commun. 2023, 14, 5110. [Google Scholar] [CrossRef]
- Naicker, D.; Rhoda, C.; Sunda, F.; Arowolo, A. Unravelling the Intricate Roles of FAM111A and FAM111B: From Protease-Mediated Cellular Processes to Disease Implications. Int. J. Mol. Sci. 2024, 25, 2845. [Google Scholar] [CrossRef]
- Gupta, S.; Silveira, D.A.; Hashimoto, R.F. A Boolean model of the oncogene role of FAM111B in lung adenocarcinoma. Comput. Biol. Chem. 2023, 106, 107926. [Google Scholar] [CrossRef]
- Sun, H.; Liu, K.; Huang, J.; Sun, Q.; Shao, C.; Luo, J.; Xu, L.; Shen, Y.; Ren, B. FAM111B, a direct target of p53, promotes the malignant process of lung adenocarcinoma. OncoTargets Ther. 2019, 12, 2829–2842. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Xue, C.; Kang, X.; Jia, X.; Wang, L.; Younis, M.H.; Liu, D.; Huo, N.; Han, Y.; Chen, Z.; et al. DNMT3B-mediated FAM111B methylation promotes papillary thyroid tumor glycolysis, growth and metastasis. Int. J. Biol. Sci. 2022, 18, 4372–4387. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; He, H.Y.; Fan, Z.H.; Li, C.M.; Gong, Y.; Wang, X.J.; Xiong, H.J.; Xie, C.M.; Bie, P. Silencing of FAM111B inhibited proliferation, migration and invasion of hepatoma cells through activating p53 pathway. Dig. Liver Dis. 2023, 55, 1679–1689. [Google Scholar] [CrossRef] [PubMed]
- Havel, J.J.; Chowell, D.; Chan, T.A. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat. Rev. Cancer 2019, 19, 133–150. [Google Scholar] [CrossRef]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef]
- So, L.; Obata-Ninomiya, K.; Hu, A.; Muir, V.S.; Takamori, A.; Song, J.; Buckner, J.H.; Savan, R.; Ziegler, S.F. Regulatory T cells suppress CD4+ effector T cell activation by controlling protein synthesis. J. Exp. Med. 2023, 220, e20221676. [Google Scholar] [CrossRef]
- Marangoni, F.; Zhakyp, A.; Corsini, M.; Geels, S.N.; Carrizosa, E.; Thelen, M.; Mani, V.; Prüßmann, J.N.; Warner, R.D.; Ozga, A.J.; et al. Expansion of tumor-associated Treg cells upon disruption of a CTLA-4-dependent feedback loop. Cell 2021, 184, 3998–4015.e19. [Google Scholar] [CrossRef]
- Wang, H.; Wang, X.; Zhang, X.; Xu, W. The promising role of tumor-associated macrophages in the treatment of cancer. Drug Resist. Updat. 2024, 73, 101041. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, X.; Hao, H. Macrophage phenotype-switching in cancer. Eur. J. Pharmacol. 2022, 931, 175229. [Google Scholar] [CrossRef]
- Wei, F.; Li, W.; Zhou, T.; Yuan, X.; Zhang, L. Unveiling FAM111B: A Pan-Cancer Biomarker for DNA Repair and Immune Infiltration. Int. J. Mol. Sci. 2025, 26, 3151. [Google Scholar] [CrossRef]
- Zhao, Y.; Simon, M.; Seluanov, A.; Gorbunova, V. DNA damage and repair in age-related inflammation. Nat. Rev. Immunol. 2023, 23, 75–89. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Xiong, Y.; Sha, Z.; Xue, W.; Xu, B.; Tan, S.; Guo, D.; Lin, F.; Wang, L.; Ji, J.; et al. SEPTIN2 suppresses an IFN-γ-independent, proinflammatory macrophage activation pathway. Nat. Commun. 2023, 14, 7441. [Google Scholar] [CrossRef] [PubMed]
- Heras-Murillo, I.; Adán-Barrientos, I.; Galán, M.; Wculek, S.K.; Sancho, D. Dendritic cells as orchestrators of anticancer immunity and immunotherapy. Nat. Rev. Clin. Oncol. 2024, 21, 257–277. [Google Scholar] [CrossRef] [PubMed]
- Mayoux, M.; Roller, A.; Pulko, V.; Sammicheli, S.; Chen, S.; Sum, E.; Jost, C.; Fransen, M.F.; Buser, R.B.; Kowanetz, M.; et al. Dendritic cells dictate responses to PD-L1 blockade cancer immunotherapy. Sci. Transl. Med. 2020, 12, eaav7431. [Google Scholar] [CrossRef]
- Yin, H.; Gao, S.; Chen, Q.; Liu, S.; Shoucair, S.; Ji, Y.; Lou, W.; Yu, J.; Wu, W.; Pu, N. Tumor-associated N1 and N2 neutrophils predict prognosis in patients with resected pancreatic ductal adenocarcinoma: A preliminary study. MedComm (2020) 2022, 3, e183. [Google Scholar] [CrossRef]
- Wang, J.X.; Choi, S.Y.C.; Niu, X.; Kang, N.; Xue, H.; Killam, J.; Wang, Y. Lactic Acid and an Acidic Tumor Microenvironment suppress Anticancer Immunity. Int. J. Mol. Sci. 2020, 21, 8363. [Google Scholar] [CrossRef]
- Meagher, N.S.; Hamilton, P.; Milne, K.; Thornton, S.; Harris, B.; Weir, A.; Alsop, J.; Bisinoto, C.; Brenton, J.D.; Brooks-Wilson, A.; et al. Profiling the immune landscape in mucinous ovarian carcinoma. Gynecol. Oncol. 2023, 168, 23–31. [Google Scholar] [CrossRef]
- Pankowska, K.A.; Będkowska, G.E.; Chociej-Stypułkowska, J.; Rusak, M.; Dąbrowska, M.; Osada, J. Crosstalk of Immune Cells and Platelets in an Ovarian Cancer Microenvironment and Their Prognostic Significance. Int. J. Mol. Sci. 2023, 24, 9279. [Google Scholar] [CrossRef]
- Hensler, M.; Kasikova, L.; Fiser, K.; Rakova, J.; Skapa, P.; Laco, J.; Lanickova, T.; Pecen, L.; Truxova, I.; Vosahlikova, S.; et al. M2-like macrophages dictate clinically relevant immunosuppression in metastatic ovarian cancer. J. Immunother. Cancer. 2020, 8, e000979. [Google Scholar] [CrossRef]
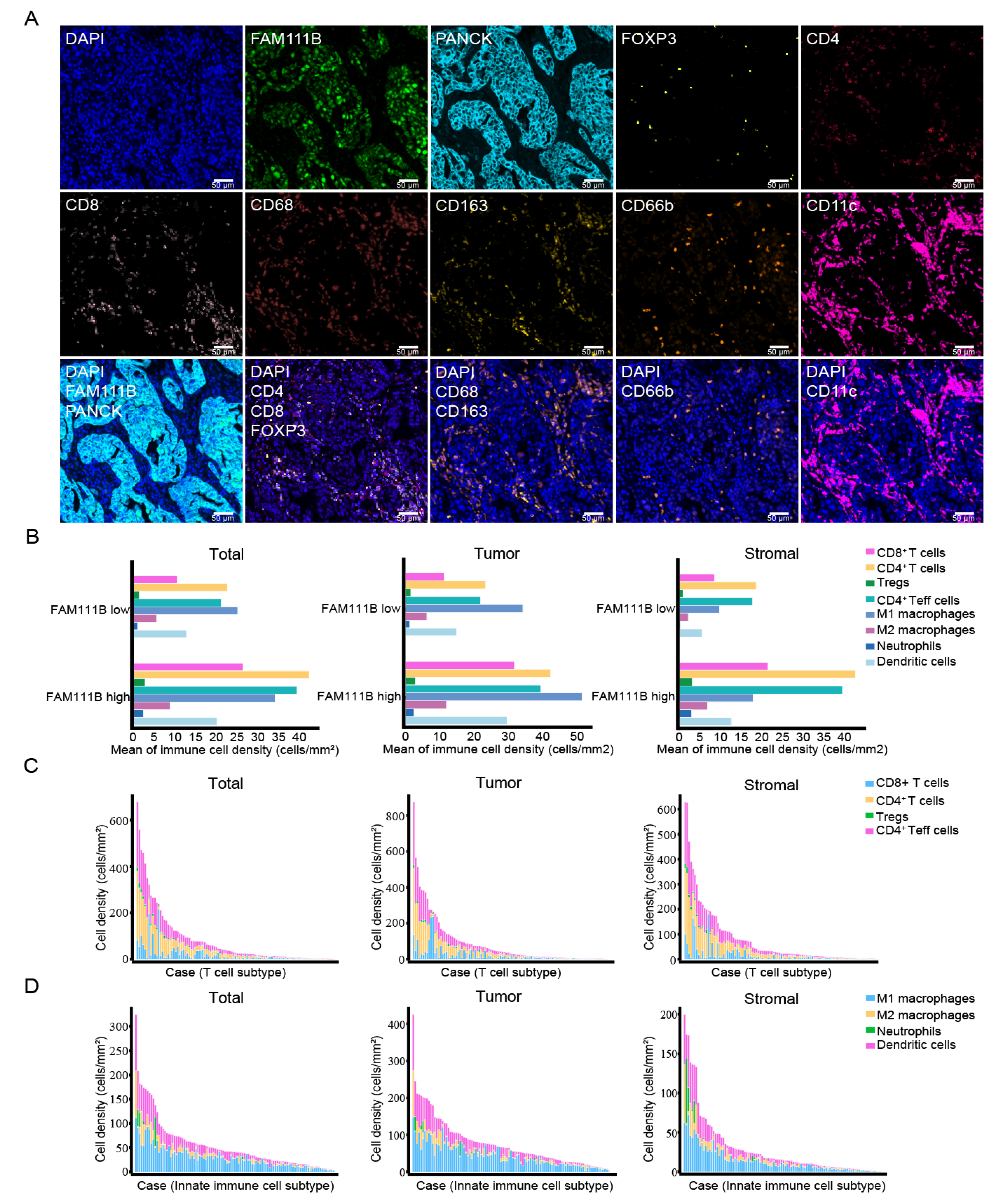

| Cell Type | Total | Tumor | Stromal | ||||||
|---|---|---|---|---|---|---|---|---|---|
| (Cells/mm2) (Mean) | FAM111B Low | FAM111B High | p Value | FAM111B Low | FAM111B High | p Value | FAM111B Low | FAM111B High | p Value |
| CD8+ T cells | 0.1596–108.5405 (10.6339) | 0.1825–216.0015 (26.4687) | 0.0035 | 0–66.5236 (11.1890) | 0.3183–236.5694 (31.6968) | 0.0118 | 0–133.0868 (8.5752) | 0–180.3608 (21.4257) | 0.0176 |
| CD4+ T cells | 0.0847–252.4395 (22.6482) | 0.0976–297.8265 (42.2519) | 0.0040 | 0–178.7593 (23.3493) | 0–368.7631 (42.2549) | 0.0086 | 0–112.1557 (18.6738) | 0–283.3256 (42.6883) | 0.3296 |
| Tregs | 0–21.5237 (1.5045) | 0–17.2489 (2.8593) | 0.0141 | 0–12.6141 (1.5222) | 0–22.1824 (2.8326) | 0.0274 | 0–10.6848 (0.9342) | 0–25.3217 (3.1866) | 0.3521 |
| CD4+Teff cells | 0.0847–230.9158 (21.1436) | 0.0976–280.5775 (39.3925) | 0.0047 | 0–166.1453 (21.8271) | 0–346.5806 (39.4222) | 0.0087 | 0–109.7016 (17.7395) | 0–258.0039 (39.5017) | 0.3103 |
| M1 macrophages | 1.8225–94.1098 (25.1451) | 4.6628–111.4464 (34.1032) | 0.0042 | 2.2875–112.5114 (34.2119) | 7.3032–147.1362 (51.3765) | 0.0004 | 0–43.4280 (9.7239) | 0.1643–72.6733 (17.8694) | 0.0059 |
| M2 macrophages | 0–69.2255 (5.6479) | 0–96.9348 (8.8280) | 0.1868 | 0–62.4555 (6.2439) | 0–127.3834 (11.9483) | 0.0536 | 0–13.9997 (2.2146) | 0–73.6824 (6.6828) | 0.0017 |
| Neutrophils | 0–34.3665 (1.0833) | 0–60.8612 (2.4294) | 0.0509 | 0–38.7365 (1.2878) | 0–40.4351 (2.3880) | 0.0375 | 0–7.5640 (0.2748) | 0–82.2875 (3.0029) | 0.0997 |
| Dendritic cells | 0.1041–80.3256 (12.8441) | 0.0356–115.0961 (20.1329) | 0.0084 | 0–95.2228 (14.8661) | 0.0661–149.5789 (29.5424) | < 0.0001 | 0–40.4262 (5.5252) | 0–65.9148 (12.5496) | 0.0028 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, W.; Wei, F.; Zhou, T.; Feng, L.; Zhang, L. FAM111B Overexpression and Immune Cell Infiltration: Implications for Ovarian Cancer Immunotherapy. Biomedicines 2025, 13, 1295. https://doi.org/10.3390/biomedicines13061295
Li W, Wei F, Zhou T, Feng L, Zhang L. FAM111B Overexpression and Immune Cell Infiltration: Implications for Ovarian Cancer Immunotherapy. Biomedicines. 2025; 13(6):1295. https://doi.org/10.3390/biomedicines13061295
Chicago/Turabian StyleLi, Wanying, Fang Wei, Ting Zhou, Lijuan Feng, and Lihong Zhang. 2025. "FAM111B Overexpression and Immune Cell Infiltration: Implications for Ovarian Cancer Immunotherapy" Biomedicines 13, no. 6: 1295. https://doi.org/10.3390/biomedicines13061295
APA StyleLi, W., Wei, F., Zhou, T., Feng, L., & Zhang, L. (2025). FAM111B Overexpression and Immune Cell Infiltration: Implications for Ovarian Cancer Immunotherapy. Biomedicines, 13(6), 1295. https://doi.org/10.3390/biomedicines13061295





