Strategies for Anticancer Treatment in p53-Mutated Head and Neck Squamous Cell Carcinoma
Abstract
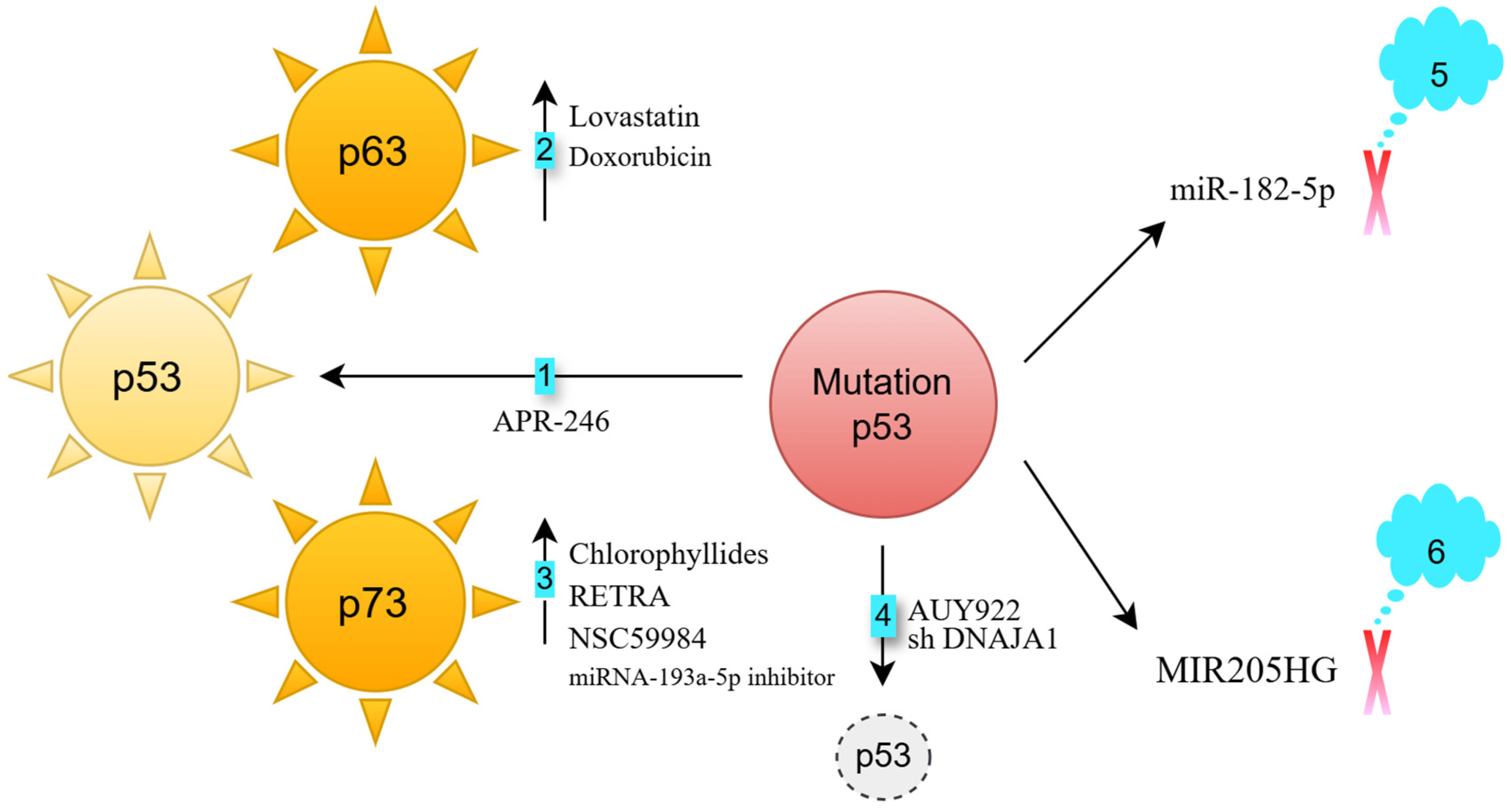
- Activation of p63: p63, a member of the p53 family, is rarely mutated in cancers [4]. Lovastatin activates p63, inducing cell death in FaDu HNSCC cells (p53 heterozygous R248L mutation) [5]. Doxorubicin enhances the binding of the c-Jun transcription factor to the TAp63 promoter, increasing p63 expression [6,7]. This response occurs in both HN30 cells (wild-type p53) and UMSCC10B cells (p53 heterozygous G245C mutation) [8]. Knocking down p63 partially reverses doxorubicin-induced apoptosis in UM-SCC10B cells, demonstrating that p63 is crucial for this apoptotic response [8].
- Activation of p73: p73, another p53 family member, also has a low mutation frequency in cancers [9]. Chlorophyllides activate p73, leading to apoptosis in Detroit 562 cells (p53 homozygous R175H mutation), as well as in TW01 and HONE-1 cells (both p53 heterozygous R280T mutation) [10]. RETRA and NSC59984 also activate p73, inducing cell death in Detroit 562 cells [11]. Additionally, miRNA-193a-5p inhibition can trigger p73 activation in JHU-029 HNSCC cells (p53 heterozygous G108Vfs*15 mutation) [12].
- Degradation of mutated p53: The chaperone proteins HSP40 and HSP90 stabilize mutated p53 [13,14]. DNAJA1, a member of the HSP40 family [15], supports mutant p53 stability. Its knockdown in CAL33 (p53 homozygous R175H mutation) and HN31 cells (p53 heterozygous C176F mutation) reduces p53 levels and impairs oncogenic traits such as cell migration and colony formation [14]. The HSP90 inhibitor AUY922 sensitizes FaDu (p53 heterozygous R248L mutation) and CAL-27 (p53 homozygous H193L mutation) cells to cisplatin [16].
- Blocking mutant p53-regulated oncogenic long non-coding RNAs: The long non-coding RNA MIR205HG is a downstream effector of mutant p53 [19]. Knocking down endogenous mutant p53 in CAL-27 cells reduces lncMIR205HG expression. Antisense targeting of MIR205HG impairs colony formation in CAL-27 and FaDu cells [19].
- Specific single-nucleotide polymorphism (SNP) in p53, p63, and p73 genes.
- 2.
- Aggregation type of p53 mutations
- 3.
- Homologous or heterologous p53 mutation
- 4.
- Two mutations in p53.
- 5.
- HPV-positive HNSCC
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 2013, 6, pl1. [Google Scholar] [CrossRef]
- Zhang, Q.; Bykov, V.J.N.; Wiman, K.G.; Zawacka-Pankau, J. APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis. 2018, 9, 439. [Google Scholar] [CrossRef] [PubMed]
- Hang, W.; Yin, Z.X.; Liu, G.; Zeng, Q.; Shen, X.F.; Sun, Q.H.; Li, D.D.; Jian, Y.P.; Zhang, Y.H.; Wang, Y.S.; et al. Piperlongumine and p53-reactivator APR-246 selectively induce cell death in HNSCC by targeting GSTP1. Oncogene 2018, 37, 3384–3398. [Google Scholar] [CrossRef] [PubMed]
- Hagiwara, K.; McMenamin, M.G.; Miura, K.; Harris, C.C. Mutational analysis of the p63/p73L/p51/p40/CUSP/KET gene in human cancer cell lines using intronic primers. Cancer Res. 1999, 59, 4165–4169. [Google Scholar]
- Yen, C.S.; Chen, J.C.; Chang, Y.F.; Hsu, Y.F.; Chiu, P.T.; Shiue, C.; Chuang, Y.F.; Ou, G.; Hsu, M.J. Lovastatin causes FaDu hypopharyngeal carcinoma cell death via AMPK-p63-survivin signaling cascade. Sci. Rep. 2016, 6, 25082. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.Y.; Pao, C.C.; Chen, J.K. Transcriptional activity of TAp63 promoter is regulated by c-jun. J. Cell Physiol. 2010, 225, 898–904. [Google Scholar] [CrossRef]
- Petitjean, A.; Cavard, C.; Shi, H.; Tribollet, V.; Hainaut, P.; Caron de Fromentel, C. The expression of TA and DeltaNp63 are regulated by different mechanisms in liver cells. Oncogene 2005, 24, 512–519. [Google Scholar] [CrossRef]
- Ongkeko, W.M.; An, Y.; Chu, T.S.; Aguilera, J.; Dang, C.L.; Wang-Rodriguez, J. Gleevec suppresses p63 expression in head and neck squamous cell carcinoma despite p63 activation by DNA-damaging agents. Laryngoscope 2006, 116, 1390–1396. [Google Scholar] [CrossRef]
- Han, S.; Semba, S.; Abe, T.; Makino, N.; Furukawa, T.; Fukushige, S.; Takahashi, H.; Sakurada, A.; Sato, M.; Shiiba, K.; et al. Infrequent somatic mutations of the p73 gene in various human cancers. Eur. J. Surg. Oncol. 1999, 25, 194–198. [Google Scholar] [CrossRef]
- Cai, B.H.; Wang, Y.T.; Chen, C.C.; Yeh, F.Y.; Lin, Y.R.; Lin, Y.C.; Wu, T.Y.; Wu, K.Y.; Lien, C.F.; Shih, Y.C.; et al. Chlorophyllides repress gain-of-function p53 mutated HNSCC cell proliferation via activation of p73 and repression of p53 aggregation in vitro and in vivo. Biochim. Biophys. Acta Mol. Basis Dis. 2025, 1871, 167662. [Google Scholar] [CrossRef]
- Cai, B.H.; Bai, Z.Y.; Lien, C.F.; Yu, S.J.; Lu, R.Y.; Wu, M.H.; Wu, W.C.; Chen, C.C.; Hsu, Y.C. NAMPT Inhibitor and P73 Activator Represses P53 R175H Mutated HNSCC Cell Proliferation in a Synergistic Manner. Biomolecules 2022, 12, 438. [Google Scholar] [CrossRef]
- Ory, B.; Ramsey, M.R.; Wilson, C.; Vadysirisack, D.D.; Forster, N.; Rocco, J.W.; Rothenberg, S.M.; Ellisen, L.W. A microRNA-dependent program controls p53-independent survival and chemosensitivity in human and murine squamous cell carcinoma. J. Clin. Invest. 2011, 121, 809–820. [Google Scholar] [CrossRef] [PubMed]
- Kaida, A.; Iwakuma, T. Regulation of p53 and Cancer Signaling by Heat Shock Protein 40/J-Domain Protein Family Members. Int. J. Mol. Sci. 2021, 22, 13527. [Google Scholar] [CrossRef]
- Alexandrova, E.M.; Yallowitz, A.R.; Li, D.; Xu, S.; Schulz, R.; Proia, D.A.; Lozano, G.; Dobbelstein, M.; Moll, U.M. Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 2015, 523, 352–356. [Google Scholar] [CrossRef] [PubMed]
- de Jesus, J.R.; Linhares, L.A.; Aragão, A.Z.B.; Arruda, M.A.Z.; Ramos, C.H.I. The stability and function of human cochaperone Hsp40/DNAJA1 are affected by zinc removal and partially restored by copper. Biochimie 2023, 213, 123–129. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, M.; Barker, H.E.; Khan, A.A.; Pedersen, M.; Dillon, M.; Mansfield, D.C.; Patel, R.; Kyula, J.N.; Bhide, S.A.; Newbold, K.L.; et al. HSP90 inhibition sensitizes head and neck cancer to platin-based chemoradiotherapy by modulation of the DNA damage response resulting in chromosomal fragmentation. BMC Cancer 2017, 17, 86. [Google Scholar] [CrossRef]
- Madrigal, T.; Ortega-Bernal, D.; Herrera, L.A.; González-De la Rosa, C.H.; Domínguez-Gómez, G.; Aréchaga-Ocampo, E.; Díaz-Chávez, J. Mutant p53 Gain-of-Function Induces Migration and Invasion through Overexpression of miR-182-5p in Cancer Cells. Cells 2023, 12, 2506. [Google Scholar] [CrossRef]
- Wang, L.; Jiang, H.; Li, W.; Jia, C.; Zhang, H.; Sun, Y.; Chen, X.; Song, X. Overexpression of TP53 mutation-associated microRNA-182 promotes tumor cell proliferation and migration in head and neck squamous cell carcinoma. Arch. Oral Biol. 2017, 73, 105–112. [Google Scholar] [CrossRef]
- Di Agostino, S.; Valenti, F.; Sacconi, A.; Fontemaggi, G.; Pallocca, M.; Pulito, C.; Ganci, F.; Muti, P.; Strano, S.; Blandino, G. Long Non-coding MIR205HG Depletes Hsa-miR-590-3p Leading to Unrestrained Proliferation in Head and Neck Squamous Cell Carcinoma. Theranostics 2018, 8, 1850–1868. [Google Scholar] [CrossRef]
- Ørsted, D.D.; Bojesen, S.E.; Tybjaerg-Hansen, A.; Nordestgaard, B.G. Tumor suppressor p53 Arg72Pro polymorphism and longevity, cancer survival, and risk of cancer in the general population. J. Exp. Med. 2007, 204, 1295–1301. [Google Scholar] [CrossRef]
- Marin, M.C.; Jost, C.A.; Brooks, L.A.; Irwin, M.S.; O’Nions, J.; Tidy, J.A.; James, N.; McGregor, J.M.; Harwood, C.A.; Yulug, I.G.; et al. A common polymorphism acts as an intragenic modifier of mutant p53 behaviour. Nat. Genet. 2000, 25, 47–54. [Google Scholar] [CrossRef]
- Bergamaschi, D.; Gasco, M.; Hiller, L.; Sullivan, A.; Syed, N.; Trigiante, G.; Yulug, I.; Merlano, M.; Numico, G.; Comino, A.; et al. p53 polymorphism influences response in cancer chemotherapy via modulation of p73-dependent apoptosis. Cancer Cell 2003, 3, 387–402. [Google Scholar] [CrossRef] [PubMed]
- Hientz, K.; Mohr, A.; Bhakta-Guha, D.; Efferth, T. The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 2017, 8, 8921–8946. [Google Scholar] [CrossRef]
- Zhang, N.; Huo, Q.; Wang, X.; Chen, X.; Long, L.; Guan, X.; Jiang, L.; Ma, T.; Hu, W.; Yang, Q. A genetic variant in p63 (rs17506395) is associated with breast cancer susceptibility and prognosis. Gene 2014, 535, 170–176. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Du, M.; Ma, L.; Chu, H.; Lv, Q.; Ye, D.; Guo, J.; Gu, C.; Xia, G.; Zhu, Y.; et al. A functional variant in TP63 at 3q28 associated with bladder cancer risk by creating an miR-140-5p binding site. Int. J. Cancer 2016, 139, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Yang, Y.; Ma, S. A functional Variant (Rs35592567) in TP63 at 3q28 is Associated with Gastric Cancer Risk via Modifying its Regulation by MicroRNA-140. Cell Physiol. Biochem. 2018, 47, 235–244. [Google Scholar] [CrossRef]
- Yu, X.J.; Fang, F.; Xie, J. Relationship between TP73 polymorphism (G4C14-A4T14) and cancer risk: A meta-analysis based on literatures. Gene 2011, 484, 42–46. [Google Scholar] [CrossRef]
- Kaghad, M.; Bonnet, H.; Yang, A.; Creancier, L.; Biscan, J.C.; Valent, A.; Minty, A.; Chalon, P.; Lelias, J.M.; Dumont, X.; et al. Monoallelically expressed gene related to p53 at 1p36, a region frequently deleted in neuroblastoma and other human cancers. Cell 1997, 90, 809–819. [Google Scholar] [CrossRef]
- Li, F.; Sturgis, E.M.; Zafereo, M.E.; Liu, Z.; Wang, L.E.; Wei, Q.; Li, G. p73 G4C14-to-A4T14 polymorphism and risk of second primary malignancy after index squamous cell carcinoma of head and neck. Int. J. Cancer 2009, 125, 2660–2665. [Google Scholar] [CrossRef]
- Li, J.; Guo, M.; Chen, L.; Chen, Z.; Fu, Y.; Chen, Y. Amyloid aggregates induced by the p53-R280T mutation lead to loss of p53 function in nasopharyngeal carcinoma. Cell Death Dis. 2024, 15, 35. [Google Scholar] [CrossRef]
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, S.; Shi, Y.P.; Jones, S.N.; Vogel, H.; Bradley, A.; Pinkel, D.; Donehower, L.A. Retention of wild-type p53 in tumors from p53 heterozygous mice: Reduction of p53 dosage can promote cancer formation. EMBO J. 1998, 17, 4657–4667. [Google Scholar] [CrossRef]
- Liu, Y.; Bodmer, W.F. Analysis of P53 mutations and their expression in 56 colorectal cancer cell lines. Proc. Natl. Acad. Sci. USA 2006, 103, 976–981. [Google Scholar] [CrossRef]
- Meng, L.; Lin, L.; Zhang, H.; Nassiri, M.; Morales, A.R.; Nadji, M. Multiple mutations of the p53 gene in human mammary carcinoma. Mutat. Res. 1999, 435, 263–269. [Google Scholar] [CrossRef] [PubMed]
- Burd, E.M. Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 2003, 16, 1–17. [Google Scholar] [CrossRef]
- Fakhry, C.; Gillison, M.L. Clinical implications of human papillomavirus in head and neck cancers. J. Clin. Oncol. 2006, 24, 2606–2611. [Google Scholar] [CrossRef]
- Lechner, M.; Liu, J.; Masterson, L.; Fenton, T.R. HPV-associated oropharyngeal cancer: Epidemiology, molecular biology and clinical management. Nat. Rev. Clin. Oncol. 2022, 19, 306–327. [Google Scholar] [CrossRef] [PubMed]
- Munger, K.; White, E.A. What are the essential determinants of human papillomavirus carcinogenesis? mBio 2024, 15, e0046224. [Google Scholar] [CrossRef]
- Crook, T.; Tidy, J.A.; Vousden, K.H. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell 1991, 67, 547–556. [Google Scholar] [CrossRef]
- Ruttkay-Nedecky, B.; Jimenez Jimenez, A.M.; Nejdl, L.; Chudobova, D.; Gumulec, J.; Masarik, M.; Adam, V.; Kizek, R. Relevance of infection with human papillomavirus: The role of the p53 tumor suppressor protein and E6/E7 zinc finger proteins (Review). Int. J. Oncol. 2013, 43, 1754–1762. [Google Scholar] [CrossRef]
- Qian, G.; Wang, D.; Magliocca, K.R.; Hu, Z.; Nannapaneni, S.; Kim, S.; Chen, Z.; Sun, S.Y.; Shin, D.M.; Saba, N.F.; et al. Human papillomavirus oncoprotein E6 upregulates c-Met through p53 downregulation. Eur. J. Cancer 2016, 65, 21–32. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Li, F.; Ji, M.; Lin, L.; Hu, C. Evaluating the prognostic significance of p53 and TP53 mutations in HPV-negative hypopharyngeal carcinoma patients: A 5-year follow-up retrospective study. BMC Cancer 2023, 23, 324. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Liu, Z.; Myers, J.N. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J. Cell Biochem. 2016, 117, 2682–2692. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, B.-H.; Chen, C.-C.; Sung, Y.-T.; Shih, Y.-C.; Lien, C.-F. Strategies for Anticancer Treatment in p53-Mutated Head and Neck Squamous Cell Carcinoma. Biomedicines 2025, 13, 1165. https://doi.org/10.3390/biomedicines13051165
Cai B-H, Chen C-C, Sung Y-T, Shih Y-C, Lien C-F. Strategies for Anticancer Treatment in p53-Mutated Head and Neck Squamous Cell Carcinoma. Biomedicines. 2025; 13(5):1165. https://doi.org/10.3390/biomedicines13051165
Chicago/Turabian StyleCai, Bi-He, Chia-Chi Chen, Yu-Te Sung, Yu-Chen Shih, and Ching-Feng Lien. 2025. "Strategies for Anticancer Treatment in p53-Mutated Head and Neck Squamous Cell Carcinoma" Biomedicines 13, no. 5: 1165. https://doi.org/10.3390/biomedicines13051165
APA StyleCai, B.-H., Chen, C.-C., Sung, Y.-T., Shih, Y.-C., & Lien, C.-F. (2025). Strategies for Anticancer Treatment in p53-Mutated Head and Neck Squamous Cell Carcinoma. Biomedicines, 13(5), 1165. https://doi.org/10.3390/biomedicines13051165







