A Prognostic Model for Senescence-Related LncRNA in a Novel Colon Adenocarcinoma Based on WGCNA and LASSO Regression
Abstract
1. Introduction
2. Materials and Methods
2.1. Public Database Acquisition and Processing
2.2. Screening of Senescence-Related lncRNAs
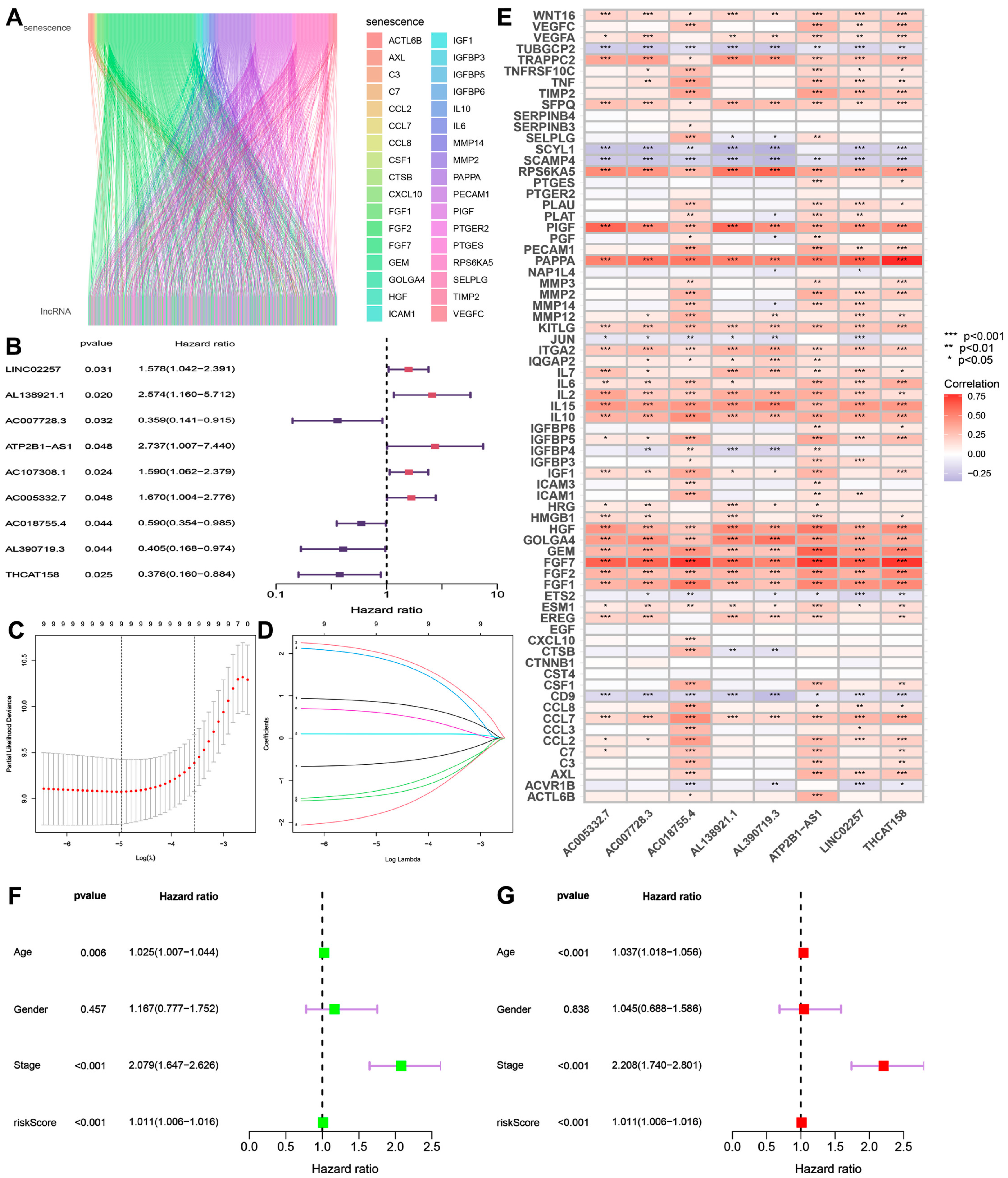
2.3. Construction of a Prognostic Model for Senescence-Related lncRNA
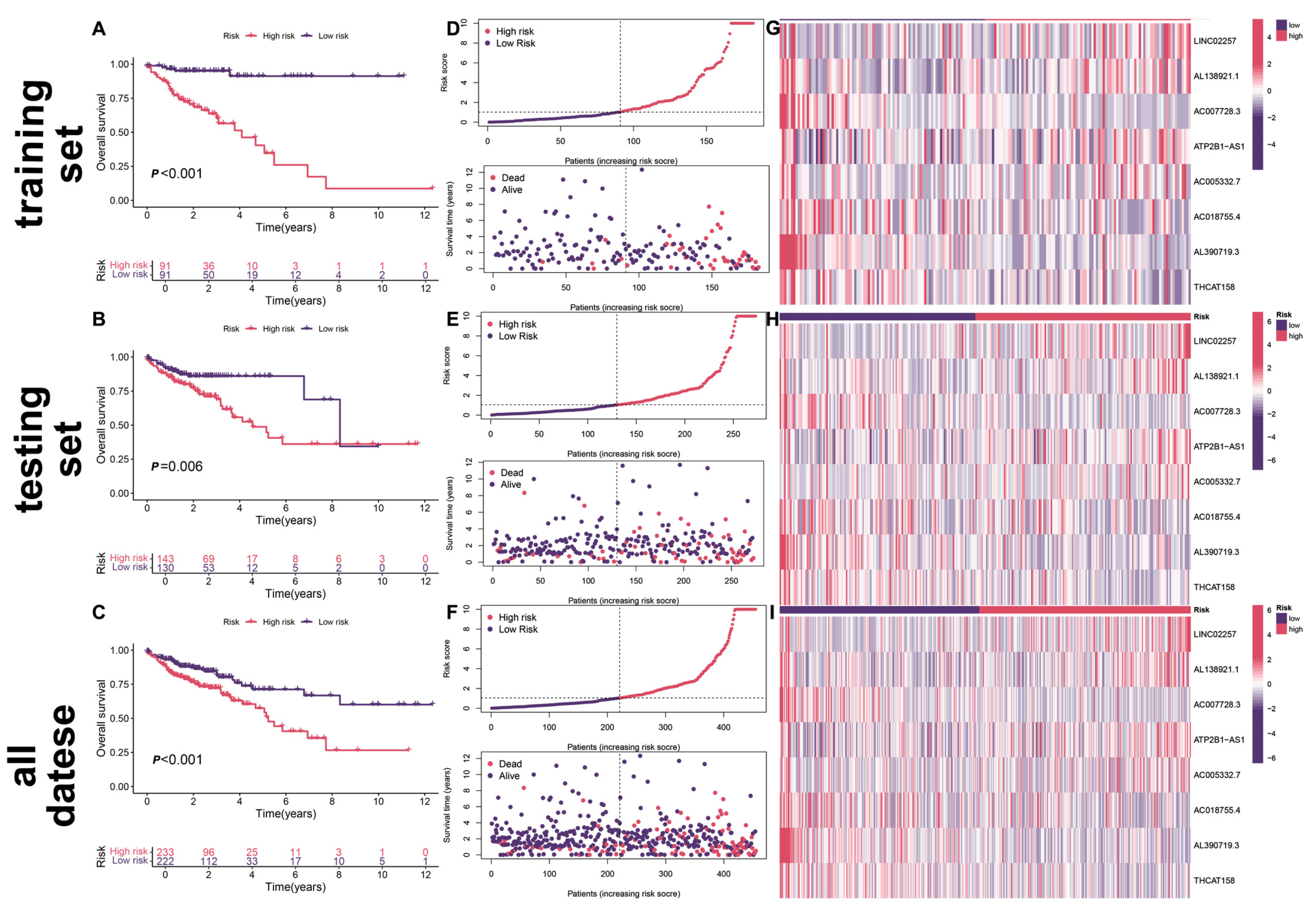
2.4. Validation of the Efficacy of Prognostic Models
2.5. Column Chart and Principal Component Analysis
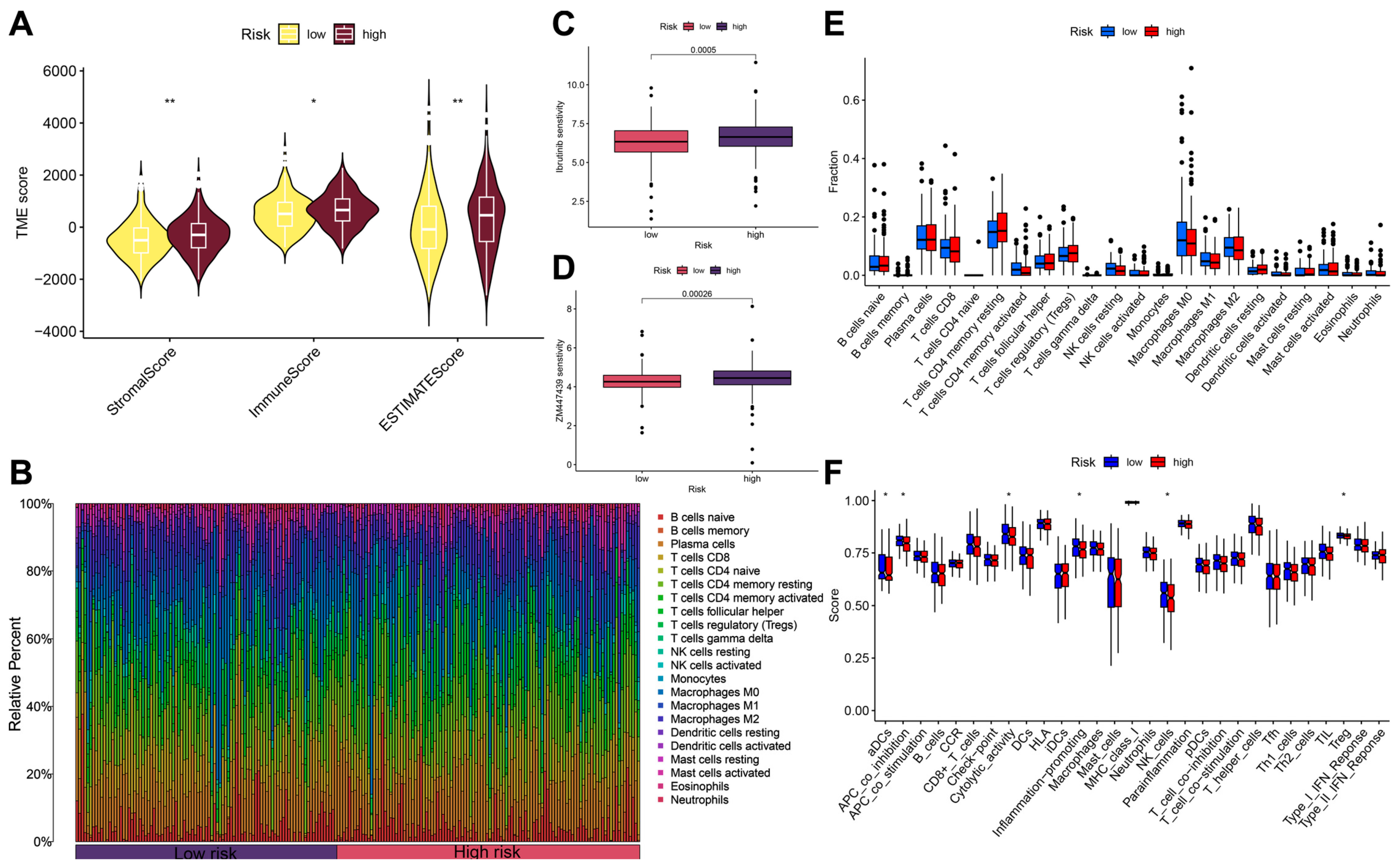
2.6. Evaluation of Tumor Microenvironment and Immune Cell Infiltration
2.7. Drug Sensitivity Analysis
2.8. Statistical Processing
3. Results
3.1. Construction of a Prognostic Model for Senescence-Related lncRNA Results
3.2. Independent Analysis of Prognostic Factors
3.3. Survival Analysis of Prognostic Models
3.4. Functional Enrichment Analysis
3.5. Immune Microenvironment Analysis and Drug Sensitivity Analysis
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Han, B.F.; Zheng, R.S.; Zeng, H.M.; Wang, S.M.; Sun, K.X.; Chen, R.; Li, L.; Wei, W.Q.; Jie, H. Cancer incidence and mortality in China, 2022. J. Natl. Cancer Cent. 2024, 4, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [PubMed]
- Wadhwa, V.; Patel, N.; Grover, D.; Ali, F.S.; Thosani, N. Interventional gastroenterology in oncology. CA Cancer J. Clin. 2023, 73, 286–319. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, S.; Liu, Y.; Zhang, C.; Li, H.; Lai, B. Metastatic patterns and survival outcomes in patients with stage IV colon cancer: A population-based analysis. Cancer Med. 2020, 9, 361–373. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lankhorst, L.; Bernards, R. Exploiting senescence for the treatment of cancer. Nat. Rev. Cancer 2022, 22, 340–355. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Shen, X. Long noncoding RNAs: Functions and mechanisms in colon cancer. Mol. Cancer 2020, 19, 167. [Google Scholar] [CrossRef] [PubMed]
- La, T.; Chen, S.; Zhao, X.H.; Zhou, S.; Xu, R.; Teng, L.; Zhang, Y.Y.; Ye, K.; Xu, L.; Guo, T.; et al. LncRNA LIMp27 Regulates the DNA Damage Response through p27 in p53-Defective Cancer Cells. Adv. Sci. 2023, 10, e2204599. [Google Scholar]
- Ungvari, Z.; Ungvari, A.; Bianchini, G.; Győrffy, B. Prognostic significance of a signature based on senescence-related genes in colorectal cancer. Geroscience 2024, 46, 4495–4504. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Zhang, Q.; Zhuang, Y.; Zhang, Y.; Li, L.; Pan, J.; Xu, L.; Ding, Y.; Wang, M.; Cong, Y.S. Reactivation of senescence-associated endogenous retroviruses by ATF3 drives interferon signaling in aging. Nat. Aging 2024, 4, 1794–1812. [Google Scholar] [CrossRef] [PubMed]
- Loison, I.; Pioger, A.; Paget, S.; Metatla, I.; OrgaRES Consortium; Vincent, A.; Abbadie, C.; Dehennaut, V. O-GlcNAcylation inhibition redirects the response of colon cancer cells to chemotherapy from senescence to apoptosis. Cell Death Dis. 2024, 15, 762. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Fu, F.; Nie, Y. Comprehensive genomics analysis of aging related gene signature to predict the prognosis and drug resistance of colon adenocarcinoma. Front. Pharmacol. 2023, 14, 1121634. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Chen, X.; Liu, X.; Jin, B.; Zhang, Y.; Wang, Y.; Xu, H.; Wan, X.; Zheng, Y.; Xu, L.; et al. LINC02257 regulates colorectal cancer liver metastases through JNK pathway. Heliyon 2024, 10, e30841. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Gao, X.; Sun, H.; Sun, L.; Hu, X. Expression characteristics of long non-coding RNA in colon adenocarcinoma and its potential value for judging the survival and prognosis of patients: Bioinformatics analysis based on The Cancer Genome Atlas database. J. Gastrointest. Oncol. 2022, 13, 1178–1187. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Chen, F.; Xu, L.; Zeng, J.; Wang, Y.; Zhang, S.; Ba, Y.; Zhang, H. Prognostic cellular senescence-related lncRNAs patterns to predict clinical outcome and immune response in colon cancer. Front. Immunol. 2024, 15, 1450135. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, D.; Li, Y.; Wu, Y.; Ma, H.; Jiang, X.; Fu, L.; Zhang, G.; Wang, H.; Liu, X.; et al. Constructing a novel signature and predicting the immune landscape of colon cancer using N6-methylandenosine-related lncRNAs. Front. Genet. 2023, 14, 906346. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liao, P.; Liu, X.; Yang, Z.; Wang, Y.; Zhong, W.; Wang, B. Construction and Validation of a Reliable Disulfidptosis-Related LncRNAs Signature of the Subtype, Prognostic, and Immune Landscape in Colon Cancer. Int. J. Mol. Sci. 2023, 24, 12915. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, C.; Jin, E. Regulation of overexpression lncRNA ATP2B1-AS1 on lung adenocarcinoma progression. J. Cardiothorac. Surg. 2024, 19, 88. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Sun, Y.; Zhao, X.; Li, J.; Fu, X.; Gong, T.; Zhang, X. Identification of m5C-related lncRNAs signature to predict prognosis and therapeutic responses in esophageal squamous cell carcinoma patients. Sci. Rep. 2023, 13, 14499. [Google Scholar] [CrossRef] [PubMed]


| Covariates | Typ | Total | Test | Train | p-Value |
|---|---|---|---|---|---|
| Age Age | ≤65 | 189 (41.54%) | 114 (41.76%) | 75 (41.21%) | 0.9845 |
| >65 | 266 (58.46%) | 159 (58.24%) | 107 (58.79%) | ||
| Gender Gender | Female | 216 (47.47%) | 131 (47.99%) | 85 (46.7%) | 0.8631 |
| Male | 239 (52.53%) | 142 (52.01%) | 97 (53.3%) | ||
| Stage | Stage I | 74 (16.26%) | 47 (17.22%) | 27 (14.84%) | 0.8117 |
| Stage II | 176 (38.68%) | 106 (38.83%) | 70 (38.46%) | ||
| Stage III | 128 (28.13%) | 73 (26.74%) | 55 (30.22%) | ||
| Stage IV | 65 (14.29%) | 40 (14.65%) | 25 (13.74%) | ||
| unknown | 11 (2.42%) | 6 (2.2%) | 5 (2.75%) | ||
| T | T1 | 11 (2.42%) | 8 (2.93%) | 3 (1.65%) | 0.791 |
| T2 | 77 (16.92%) | 44 (16.12%) | 33 (18.13%) | ||
| T3 | 310 (68.13%) | 187 (68.5%) | 123 (67.58%) | ||
| T4 | 56 (12.31%) | 33 (12.09%) | 23 (12.64%) | ||
| unknown | 1 (0.22%) | 1 (0.37%) | 0 (0%) | ||
| M | M0 | 333 (73.19%) | 199 (72.89%) | 134 (73.63%) | 0.8971 |
| M1 | 65 (14.29%) | 40 (14.65%) | 25 (13.74%) | ||
| unknown | 57 (12.53%) | 34 (12.45%) | 23 (12.64%) | ||
| N | N0 | 267 (58.68%) | 165 (60.44%) | 102 (56.04%) | 0.3945 |
| N1 | 105 (23.08%) | 57 (20.88%) | 48 (26.37%) | ||
| N2 | 83 (18.24%) | 51 (18.68%) | 32 (17.58%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Y.; Min, G.; Wang, H.; Jiang, L. A Prognostic Model for Senescence-Related LncRNA in a Novel Colon Adenocarcinoma Based on WGCNA and LASSO Regression. Biomedicines 2025, 13, 1088. https://doi.org/10.3390/biomedicines13051088
Huang Y, Min G, Wang H, Jiang L. A Prognostic Model for Senescence-Related LncRNA in a Novel Colon Adenocarcinoma Based on WGCNA and LASSO Regression. Biomedicines. 2025; 13(5):1088. https://doi.org/10.3390/biomedicines13051088
Chicago/Turabian StyleHuang, Yichu, Guangtao Min, Hongpeng Wang, and Lei Jiang. 2025. "A Prognostic Model for Senescence-Related LncRNA in a Novel Colon Adenocarcinoma Based on WGCNA and LASSO Regression" Biomedicines 13, no. 5: 1088. https://doi.org/10.3390/biomedicines13051088
APA StyleHuang, Y., Min, G., Wang, H., & Jiang, L. (2025). A Prognostic Model for Senescence-Related LncRNA in a Novel Colon Adenocarcinoma Based on WGCNA and LASSO Regression. Biomedicines, 13(5), 1088. https://doi.org/10.3390/biomedicines13051088







