A Retrospective Single-Center Analysis from Southern Italy on the Use of T2 Magnetic Resonance Assays as a Point-of-Care Method for Patients with Sepsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Methods: Study Design
2.2. Sample Collection and Processing
2.3. Statistical Analysis
3. Results
4. Discussion
Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, M.C.; Scherag, A.; Adhikari, N.K.J.; Hartog, C.; Tsaganos, T.; Schlattmann, P.; Angus, D.C.; Reinhart, K. Assessment of Global Incidence and Mortality of Hospital-treated Sepsis. Current Estimates and Limitations. Am. J. Respir. Crit. Care Med. 2016, 193, 259–272. [Google Scholar] [CrossRef]
- Wisplinghoff, H.; Bischoff, T.; Tallent, S.M.; Seifert, H.; Wenzel, R.P.; Edmond, M.B. Nosocomial Bloodstream Infections in US Hospitals: Analysis of 24,179 Cases from a Prospective Nationwide Surveillance Study. Clin. Infect. Dis. 2004, 39, 309–317. [Google Scholar] [CrossRef]
- Evans, L.; Rhodes, A.; Alhazzani, W.; Antonelli, M.; Coopersmith, C.M.; French, C.; Machado, F.R.; Mcintyre, L.; Ostermann, M.; Prescott, H.C.; et al. Surviving sepsis campaign: International guidelines for management of sepsis and septic shock 2021. Intensive Care Med. 2021, 47, 1181–1247. [Google Scholar] [CrossRef]
- Gotts, J.E.; Matthay, M.A. Sepsis: Pathophysiology and clinical management. BMJ 2016, 353, i1585. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, V.; Sarwar, S.; Walker, S.; Elligsen, M.; Palmay, L.; Daneman, N. Weighted-incidence syndromic combination antibiograms to guide empiric treatment of critical care infections: A retrospective cohort study. Crit. Care 2014, 18, R112. [Google Scholar] [CrossRef] [PubMed]
- Abe, T.; Ogura, H.; Kushimoto, S.; Shiraishi, A.; Sugiyama, T.; Deshpande, G.A.; Uchida, M.; Nagata, I.; Saitoh, D.; Fujishima, S.; et al. Variations in infection sites and mortality rates among patients in intensive care units with severe sepsis and septic shock in Japan. J. Intensive Care 2019, 7, 28. [Google Scholar] [CrossRef] [PubMed]
- Nieman, A.E.; Savelkoul, P.H.M.; Beishuizen, A.; Henrich, B.; Lamik, B.; MacKenzie, C.R.; Kindgen-Milles, D.; Helmers, A.; Diaz, C.; Sakka, S.G.; et al. A prospective multicenter evaluation of direct molecular detection of blood stream infection from a clinical perspective. BMC Infect. Dis. 2016, 16, 314. [Google Scholar] [CrossRef]
- Gupta, S.; Sakhuja, A.; Kumar, G.; McGrath, E.; Nanchal, R.S.; Kashani, K.B. Culture-Negative Severe Sepsis: Nationwide Trends and Outcomes. Chest 2016, 150, 1251–1259. [Google Scholar] [CrossRef] [PubMed]
- Giacobbe, D.R.; Giani, T.; Bassetti, M.; Marchese, A.; Viscoli, C.; Rossolini, G.M. Rapid microbiological tests for bloodstream infections due to multidrug resistant Gram-negative bacteria: Therapeutic implications. Clin. Microbiol. Infect. 2020, 26, 713–722. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Vena, A.; Brink, A. Challenges and research priorities to progress the impact of antimicrobial stewardship. Drugs Context 2019, 8, 212600. [Google Scholar] [CrossRef]
- Rhee, C.; Kadri, S.S.; Dekker, J.P.; Danner, R.L.; Chen, H.C.; Fram, D.; Zhang, F.; Wang, R.; Klompas, M.; CDC Prevention Epicenters Program. Prevalence of Antibiotic-Resistant Pathogens in Culture-Proven Sepsis and Outcomes Associated with Inadequate and Broad Spectrum Empiric Antibiotic Use. JAMA Netw. Open 2020, 3, e20289. [Google Scholar] [CrossRef] [PubMed]
- Paggi, R.; Cenci, E.; De Socio, G.V.; Belati, A.; Marini, D.; Gili, A.; Camilloni, B.; Mencacci, A. Accuracy and Impact on Patient Management of New Tools for Diagnosis of Sepsis: Experience with the T2 Magnetic Resonance Bacteria Panel. Pathogens 2021, 10, 1132. [Google Scholar] [CrossRef] [PubMed]
- Holmes, C.L.; Anderson, M.T.; Mobley, H.L.T.; Bachman, M.A. Pathogenesis of Gram-negative bacteremia. Clin. Microbiol. Rev. 2021, 34, e00234-20. [Google Scholar] [CrossRef]
- Diekema, D.J.; Hsueh, P.R.; Mendes, R.E.; Pfaller, M.A.; Rolston, K.V.; Sader, H.S.; Jones, R.N. The microbiology of bloodstream infection: 20-Year trends from the SENTRY Antimicrobial Surveillance Program. Antimicrob. Agents Chemother. 2019, 63, e00355-19. [Google Scholar] [CrossRef] [PubMed]
- De Angelis, G.; Posteraro, B.; De Carolis, E.; Menchinelli, G.; Franceschi, F.; Tumbarello, M.; De Pascale, G.; Spanu, T.; Sanguinetti, M. T2Bacteria magnetic resonance assay for the rapid detection of ESKAPEc pathogens directly in whole blood. J. Antimicrob. Chemother. 2018, 73 (Suppl. S4), iv20–iv26. [Google Scholar] [CrossRef]
- Wang, M.C.; Lin, W.H.; Yan, J.J.; Fang, H.Y.; Kuo, T.H.; Tseng, C.C.; Wu, J.-J. Early identification of microorganisms in blood culture prior to the detection of a positive signal in the BACTEC FX system using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. J. Microbiol. Immunol. Infect. 2015, 48, 419–424. [Google Scholar] [CrossRef]
- Vrettou, C.S.; Douka, E.; Perivolioti, E.P.; Vassiliou, A.G.; Sarri, A.; Giannopoulou, V.; Trigkidis, K.K.; Jahaj, E.; Dimopoulou, I.; Kotanidou, A. Accuracy of T2 magnetic resonance assays as point-of-care methods in the intensive care unit. J. Hosp. Infect. 2023, 139, 240–248. [Google Scholar] [CrossRef] [PubMed]
- Clancy, C.J.; Pappas, P.G.; Vazquez, J.; Judson, M.A.; Kontoyiannis, D.P.; Thompson, G.R., III; Garey, K.W.; Reboli, A.; Greenberg, R.N.; Apewokin, S.; et al. Detecting Infections Rapidly and Easily for Candidemia Trial, Part 2 (DIRECT2): A Prospective, Multicenter Study of the T2Candida Panel. Clin. Infect. Dis. 2018, 66, 1678–1686. [Google Scholar] [CrossRef]
- Sanguinetti, M.; Posteraro, B.; Beigelman-Aubry, C.; Lamoth, F.; Dunet, V.; Slavin, M.; Richardson, M.D. Diagnosis and treatment of invasive fungal infections: Looking ahead. J. Antimicrob. Chemother. 2019, 74 (Suppl. S2), ii27–ii37. [Google Scholar] [CrossRef]
- Muñoz, P.; Vena, A.; Machado, M. T2Candida MR as a predictor of outcome in patients with suspected invasive candidiasis starting empirical antifungal treatment: A prospective pilot study. J. Antimicrob. Chemother. 2018, 73, iv6–iv12. [Google Scholar] [CrossRef] [PubMed]
- Mylonakis, E.; Clancy, C.J.; Ostrosky-Zeichner, L.; Garey, K.W.; Alangaden, G.J.; Vazquez, J.A.; Groeger, J.S.; Judson, M.A.; Vinagre, Y.M.; Heard, S.O.; et al. T2 magnetic resonance assay for the rapid diagnosis of candidemia in whole blood: A clinical trial. Clin. Infect. Dis. 2015, 60, 892–899. [Google Scholar] [CrossRef] [PubMed]
- Chiarini, A.; Palmeri, A.; Amato, T.; Immordino, R.; Distefano, S.; Giammanco, A. Detection of bacterial and yeast species with the Bactec 9120 automated system with routine use of aerobic, anaerobic, and fungal media. J Clin Microbiol. 2008, 46, 4029–4033. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Chen, J.R.; Lee, S.Y.; Yang, B.H.; Lu, J.J. Rapid identification and susceptibility testing using the VITEK 2 system using culture fluids from positive BacT/ALERT blood cultures. J. Microbiol. Immunol. Infect. 2008, 41, 259–264. [Google Scholar] [PubMed]
- Mylonakis, E.; Zacharioudakis, I.M.; Clancy, C.J.; Nguyen, M.H.; Pappas, P.G. Efficacy of T2 Magnetic Resonance Assay in Monitoring Candidemia after Initiation of Antifungal Therapy: The Serial Therapeutic and Antifungal Monitoring Protocol (STAMP) Trial. J. Clin. Microbiol. 2018, 56, e01756-17. [Google Scholar] [CrossRef] [PubMed] [PubMed Central]
- Pfaller, M.A.; Wolk, D.M.; Lowery, T.J. T2MR and T2Candida: Novel technology for the rapid diagnosis of candidemia and invasive candidiasis. Future Microbiol. 2015, 11, 103–117. [Google Scholar] [CrossRef]
- European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 15.0. 2025. Available online: https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/v_15.0_Breakpoint_Tables.pdf (accessed on 15 April 2025).
- Voigt, C.; Silbert, S.; Widen, R.H.; Marturano, J.E.; Lowery, T.J.; Ashcraft, D.; Pankey, G. The T2Bacteria Assay is a sensitive and rapid detector of bacteremia that can be initiated in the emergency department and has potential to favorably influence subsequent therapy. J. Emerg. Med. 2020, 58, 785–796. [Google Scholar] [CrossRef]
- Farrell, J.J.; Hujer, A.M.; Sampath, R.; Bonomo, R.A. Salvage microbiology: Opportunities and challenges in the detection of bacterial pathogens following initiation of antimicrobial treatment. Expert Rev. Mol. Diagn. 2015, 15, 349–360. [Google Scholar] [CrossRef]
- Lodes, U.; Bohmeier, B.; Lippert, H.; König, B.; Meyer, F. PCR-based rapid sepsis diagnosis effectively guides clinical treatment in patients with new onset of SIRS. Langenbecks Arch. Surg. 2012, 397, 447–455. [Google Scholar] [CrossRef]
- Reers, Y.; Idelevich, E.A.; Pätkau, H.; Sauerland, M.C.; Tafelski, S.; Nachtigall, I.; Berdel, W.E.; Peters, G.; Silling, G.; Becker, K.; et al. Multiplex PCR assay underreports true bloodstream infections with coagulase-negative staphylococci in hematological patients with febrile neutropenia. Diagn. Microbiol. Infect. Dis. 2016, 85, 413–415. [Google Scholar] [CrossRef]
- Al Hennawi, H.E.T.; Mahdi, E.M.; Memish, Z.A. Native valve Staphylococcus capitis infective endocarditis: A mini review. Infection 2020, 48, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Vasconcellos, D.; Weng, B.; Wu, P.; Thompson, G.; Sutjita, M. Staphylococcus hominis infective endocarditis presenting with embolic splenic and renal infarcts and spinal discitis. Case Rep. Infect. Dis. 2022, 2022, 7183049. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.H.; Clancy, C.J.; Pasculle, A.W.; Pappas, P.G.; Alangaden, G.; Pankey, G.A.; Schmitt, B.H.; Rasool, A.; Weinstein, M.P.; Widen, R.; et al. Performance of the T2Bacteria panel for diagnosing bloodstream infections. Ann. Intern. Med. 2019, 170, 845–852. [Google Scholar] [CrossRef]
- Di Lodovico, S.; Fasciana, T.; Di Giulio, M.; Cellini, L.; Giammanco, A.; Rossolini, G.M.; Antonelli, A. Spread of multidrug-resistant microorganisms. Antibiotics 2022, 11, 832. [Google Scholar] [CrossRef] [PubMed]
- McCarty, T.; Leal, S.M.; Lee, R.A.; White, C.; Pappas, P. False negative rate of T2Candida assay in blood culture positive candidemia. Open Forum Infect. Dis. 2019, 6 (Suppl. S2), S143–S144. [Google Scholar] [CrossRef]
- Bogan, C.; Marchaim, D. The role of antimicrobial stewardship in curbing carbapenem resistance. Future Microbiol. 2013, 8, 979–991. [Google Scholar] [CrossRef]
| Parameters | Sample |
|---|---|
| No. of patients | 81 |
| Age at test | |
| Mean ± SD | 64.2 ± 21.6 |
| Median (IQR) | 71.5 (53.0, 80.0) |
| Gender | |
| Male | 63.0% (51) |
| Female | 37.0% (30) |
| SOFA score | |
| Mean ± SD | 4.9 ± 2.5 |
| Median (IQR) | 4.0 (3.0, 6.0) |
| Comorbidity † | 54.3% (44) |
| Chronic liver disease | 11.4% (5) |
| Chronic renal failure | 13.6% (6) |
| Diabetes | 15.9% (7) |
| Hematological disease | 2.3% (1) |
| History of cardiovascular disease | 31.8% (14) |
| Hypertension | 15.9% (7) |
| Solid malignancy | 13.6% (6) |
| Hospital setting | |
| Emergency room | 24.7% (20) |
| Infectious diseases | 17.3% (14) |
| Intensive care | 17.3% (14) |
| Medicine | 24.7% (20) |
| Surgical | 3.7% (3) |
| Other | 12.3% (10) |
| Leukocytes (103/µL): Mean ± SD | 13.4 ± 12.2 |
| % patients with normal values | 39.5% (32) |
| % patients with abnormal values | 60.5% (49) |
| % patients with abnormal values | |
| Leukocytosis (>11 × 103/µL) | 79.6% (39) |
| Leukopenia (<4.3 × 103/µL) | 20.4% (10) |
| Platelets (103/µL): Mean ± SD | 226.4 ± 182.0 |
| % patients with normal values | 53.1% (43) |
| % patients with abnormal values | 46.9% (38) |
| PCR (mg/L): Mean ± SD | 149.38 ± 117.6 |
| % patients with normal values | 8.6% (7) |
| % patients with abnormal values | 91.4% (74) |
| PCT (ng/mL): Mean ± SD | 17.01 ± 32.88 |
| % patients with normal values | 3.7% (3) |
| % patients with abnormal values | 91.4% (78) |
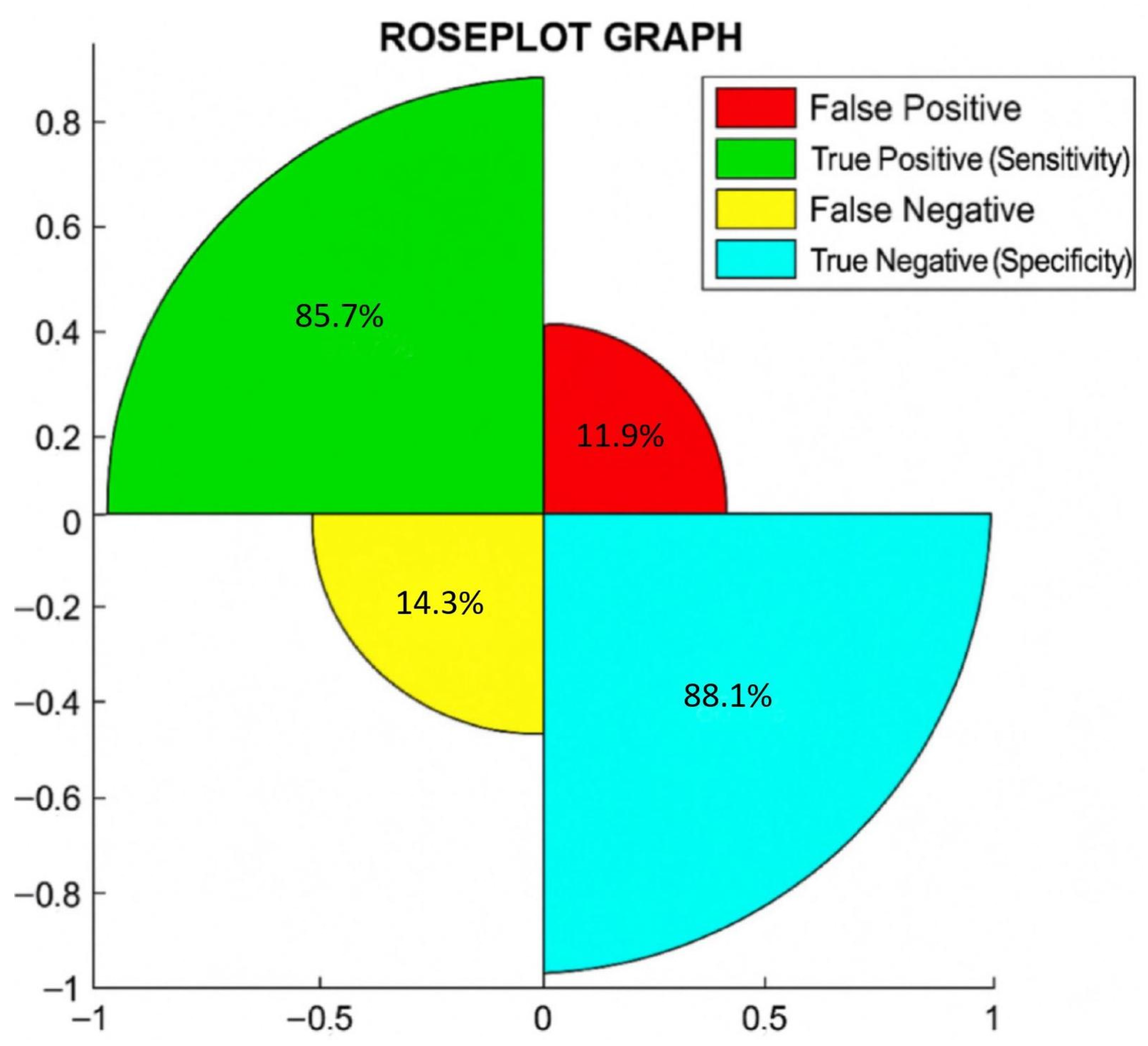
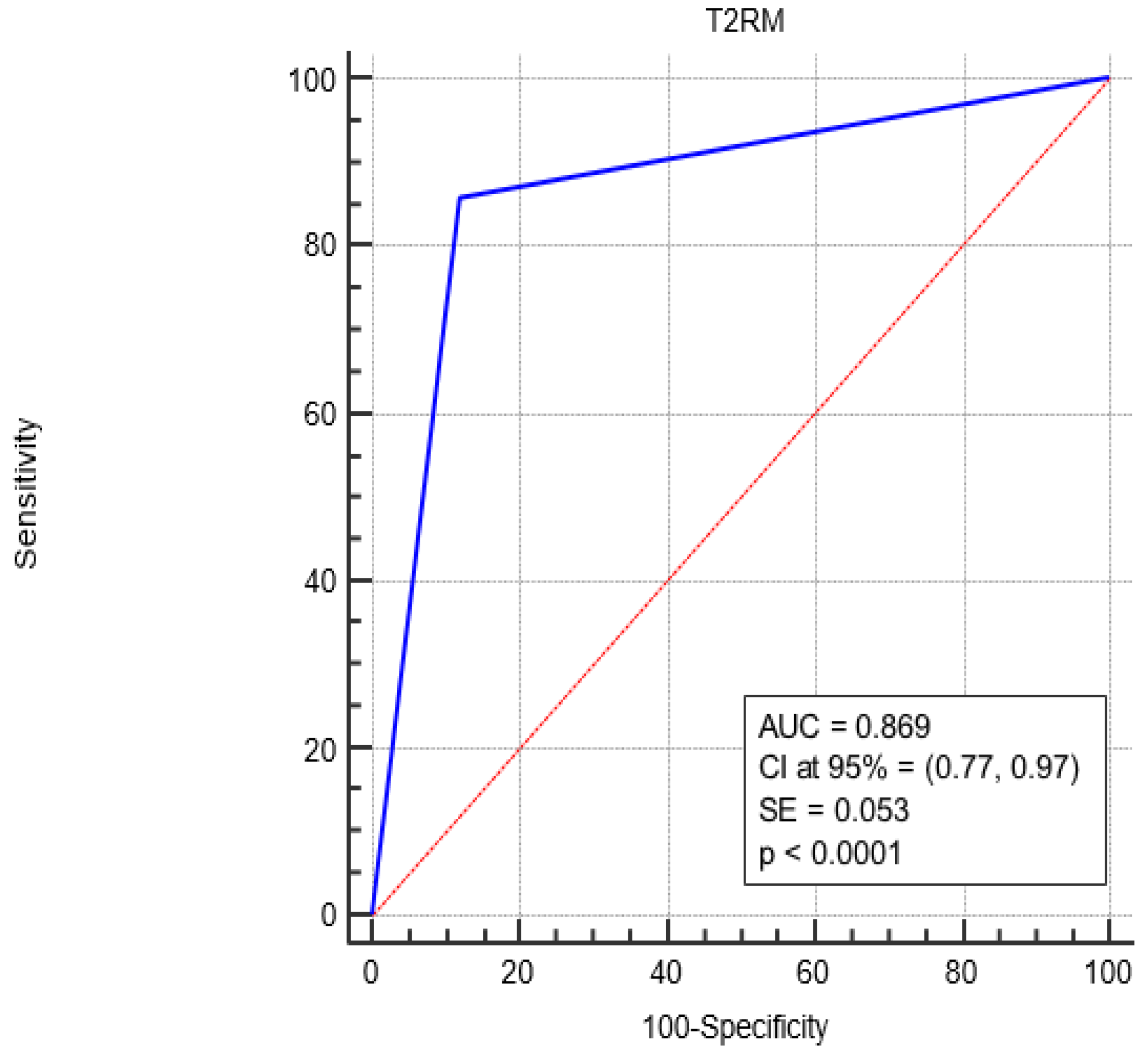
| Diagnostic | Sensitivity (CI at 95%) | Specificity (CI at 95%) | Accuracy (CI at 95%) |
|---|---|---|---|
| T2MR | 85.7% (75.8%, 92.4%) | 88.1% (78.5%, 94.2%) | 87.2% (78.0%, 93.9%) |
| BC | Both Dignostics | T2MR |
|---|---|---|
| Staphylococcus spp. (2) | S. aureus (5) | S. aureus (2) |
| E. coli (1) | K. pneumoniae (3) | K. pneumoniae (1) |
| E. coli (3) | E. coli (1) | |
| A. baumannii (1) | ||
| E. faecium + P. aeruginosa + C. krusei/C. glabrata (1) | ||
| K. pneumoniae + E. faecium (1) | ||
| C. albicans/C. tropicalis + C. krusei/C. glabrata (1) | ||
| C. albicans/C. tropicalis (1) |
| Parameters | BC | T2RM | BC vs. T2RM p-Value (Test) |
|---|---|---|---|
| No. of Patients | 81 | 81 | |
| Blood culture | Cohen’s Kappa unweighted = 0.63 (substantial agreement) Standard error = 0.11 CI at 95% = (0.42, 0.84) p < 0.0001 * | ||
| positive | (14) | (20) | |
| negative | (67) | (61) | |
| Blood culture: characterization # | |||
| Bacteria | Cohen’s Kappa unweighted = 0.63 (substantial agreement) Standard error = 0.066 CI at 95% = (0.50, 0.76) p < 0.0001 * | ||
| S. epidermidis | 7.1% (1/14) | 0.0% (0) | |
| S. capitis | 7.1% (1/14) | 0.0% (0) | |
| S. aureus | 35.7% (5/14) | 35.0% (7/20) | |
| K. pneumoniae | 21.4% (3/14) | 25.0% (5/20) | |
| E. faecium | 0.0% (0) | 10.0% (2/20) | |
| A. baumannii | 0.0% (0) | 5.0% (1/20) | |
| E. coli | 28.6% (4/14) | 20.0% (4/20) | |
| P. aeruginosa | 0.0% (0) | 5.0% (1/20) | |
| Yeast | |||
| C. krusei/glabrata | 0.0% (0) | 10.0% (2) | |
| Candida albicans/tropicalis | 0.0% (0) | 10.0% (2) | |
| Time (hours) | |||
| Mean ± SD | 125.6 ± 6.7 | 5.8 ± 2.1 | p < 0.0001 * (W) |
| Median (IQR) | 122.0 (113.1, 135.2) | 5.2 (4.5, 6.5) | |
| Multiple infection | 0.0% (0/14) | 20.0% (4/20) | 0.0153 * (Mc) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bona, M.M.; Carelli, V.M.; Serra, N.; Amico, S.; Bartolini, R.; Giammanco, A.; Di Carlo, P.; Fasciana, T.; Andriolo, M. A Retrospective Single-Center Analysis from Southern Italy on the Use of T2 Magnetic Resonance Assays as a Point-of-Care Method for Patients with Sepsis. Biomedicines 2025, 13, 999. https://doi.org/10.3390/biomedicines13040999
Bona MM, Carelli VM, Serra N, Amico S, Bartolini R, Giammanco A, Di Carlo P, Fasciana T, Andriolo M. A Retrospective Single-Center Analysis from Southern Italy on the Use of T2 Magnetic Resonance Assays as a Point-of-Care Method for Patients with Sepsis. Biomedicines. 2025; 13(4):999. https://doi.org/10.3390/biomedicines13040999
Chicago/Turabian StyleBona, Mariarita Margherita, Vincenza Maria Carelli, Nicola Serra, Salvatore Amico, Roberta Bartolini, Anna Giammanco, Paola Di Carlo, Teresa Fasciana, and Maria Andriolo. 2025. "A Retrospective Single-Center Analysis from Southern Italy on the Use of T2 Magnetic Resonance Assays as a Point-of-Care Method for Patients with Sepsis" Biomedicines 13, no. 4: 999. https://doi.org/10.3390/biomedicines13040999
APA StyleBona, M. M., Carelli, V. M., Serra, N., Amico, S., Bartolini, R., Giammanco, A., Di Carlo, P., Fasciana, T., & Andriolo, M. (2025). A Retrospective Single-Center Analysis from Southern Italy on the Use of T2 Magnetic Resonance Assays as a Point-of-Care Method for Patients with Sepsis. Biomedicines, 13(4), 999. https://doi.org/10.3390/biomedicines13040999






