Therapeutic Target Discovery for Multiple Myeloma: Identifying Druggable Genes via Mendelian Randomization
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Antibodies and Reagents
2.3. Exposure Data
2.4. Instrumental Variable (IV) Selection
2.4.1. Statistical Significance (p < 5 × 10−8)
2.4.2. Linkage Disequilibrium (LD) Clumping (r2 < 0.1; Window Size = 10,000 kb)
2.4.3. Instrument Strength (F-Statistic > 10)
2.4.4. Software and Filtering Parameters
2.5. Outcome Data
2.6. Two-Sample Mendelian Randomization Analysis (Two-Sample MR)
2.7. Transcriptome-Wide Association Study (TWAS)
2.8. Colocalization
2.8.1. Single Causal Variant per Region
2.8.2. Effect Sizes Are Derived from GWAS and eQTL Summary Statistics
2.8.3. Rationale for PPH4 > 0.75 Threshold
2.8.4. Visualization of Colocalization Results
2.9. Mendelian Randomization Phenome-Wide Association Study (MR-PheWAS)
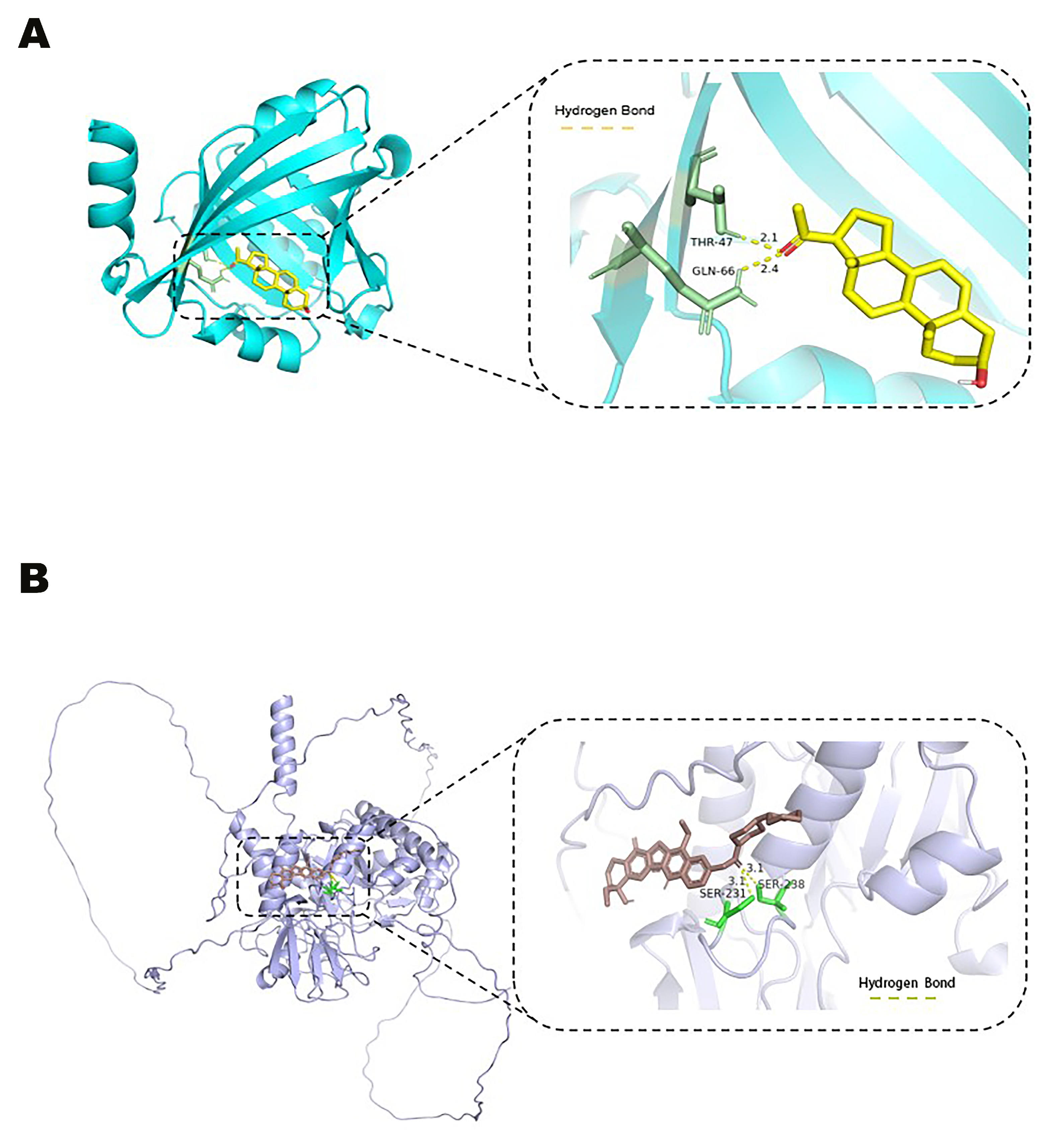
2.10. Molecular Docking
2.10.1. Retrieval and Preparation of Protein Structures
2.10.2. Selection and Preparation of Ligands
2.10.3. Docking Parameters and Scoring Functions
2.10.4. Visualization and Interaction Analysis
2.11. Cell Lines and Cell Culture
2.12. RT-qPCR
2.12.1. RNA Extraction and Quality Control
2.12.2. cDNA Synthesis
2.12.3. RT-qPCR Experimental Conditions
2.12.4. Normalization and Quantification Method
2.13. Western Blotting
2.13.1. Protein Extraction and Quantification
2.13.2. SDS-PAGE and Membrane Transfer
2.13.3. Blocking and Antibody Incubation
2.13.4. Quantification of Band Intensity
2.14. Cell Counting Kit-8 (CCK-8) Assays
2.14.1. Experimental Conditions
2.14.2. Drug Treatment and Experimental Setup
2.14.3. CCK-8 Assay
2.14.4. Data Analysis and Statistical Methods
2.15. Statistical Analysis
3. Results
3.1. Identification of Exposure Genes
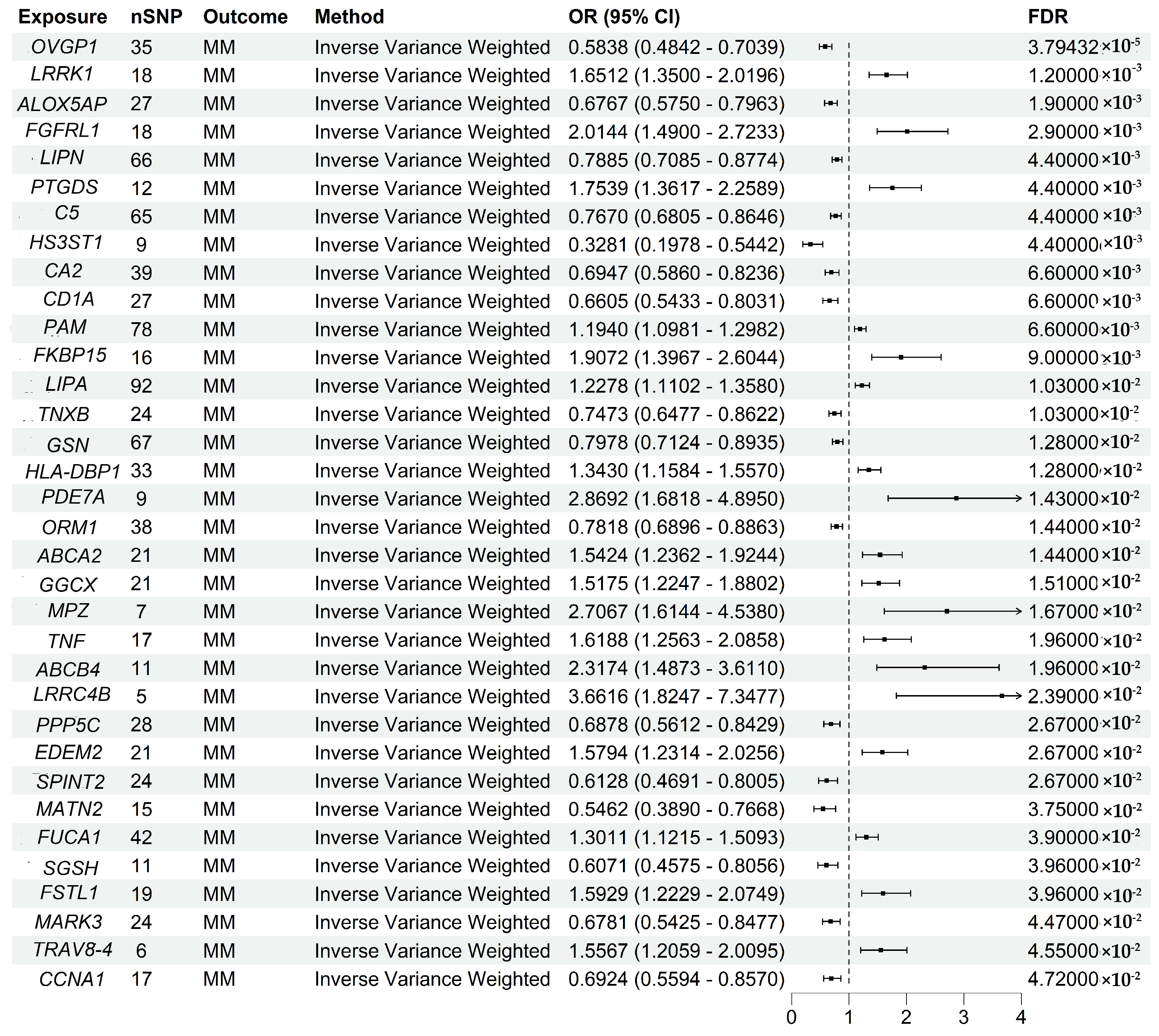
3.2. Two-Sample MR Analysis Validated Druggable Genes
3.3. Transcriptome-Wide Association Study (TWAS) Identified Susceptibility Genes for MM
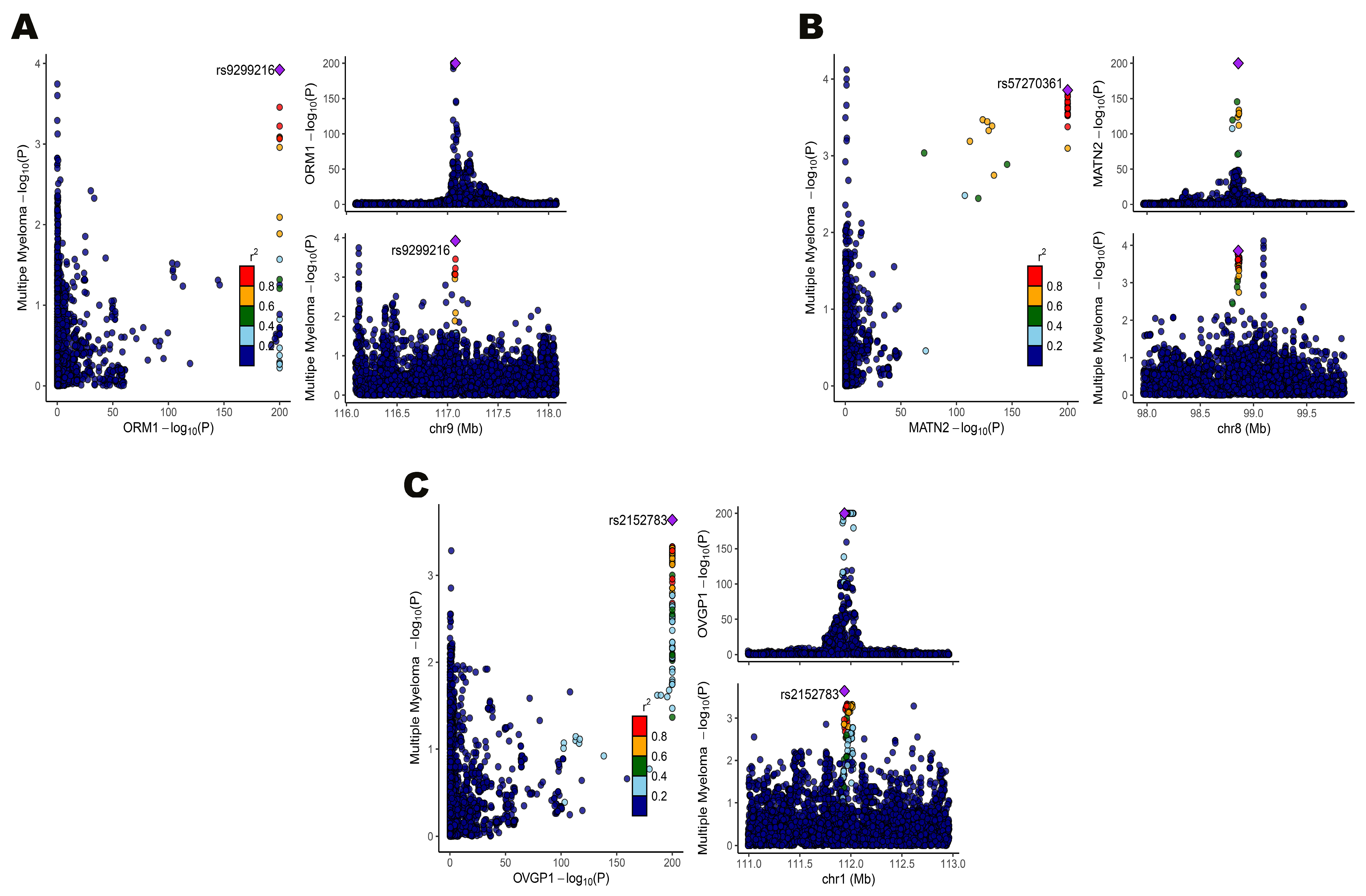
3.4. Colocalization Confirmed Shared Genetic Variants for Gene Expression and MM
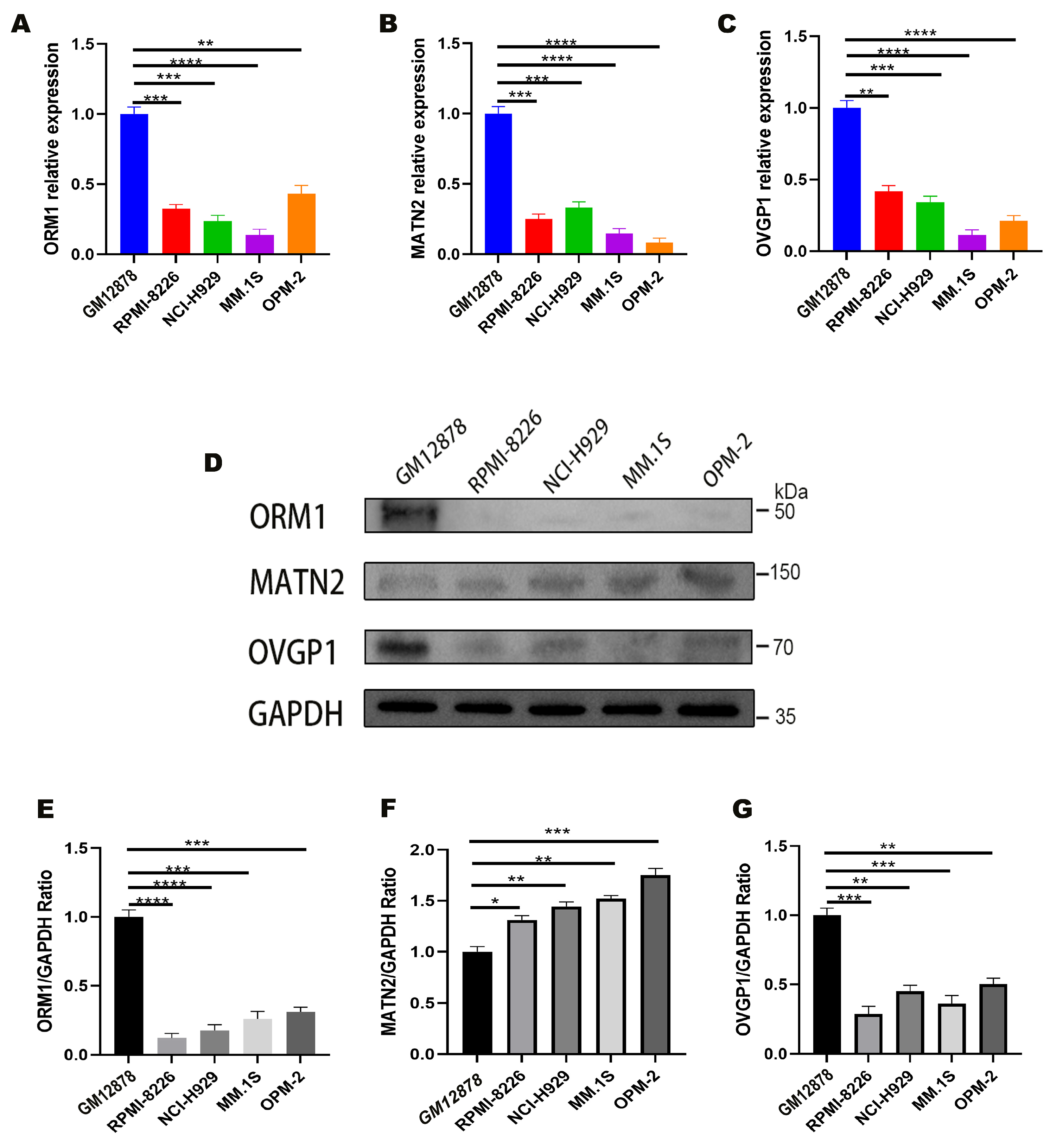
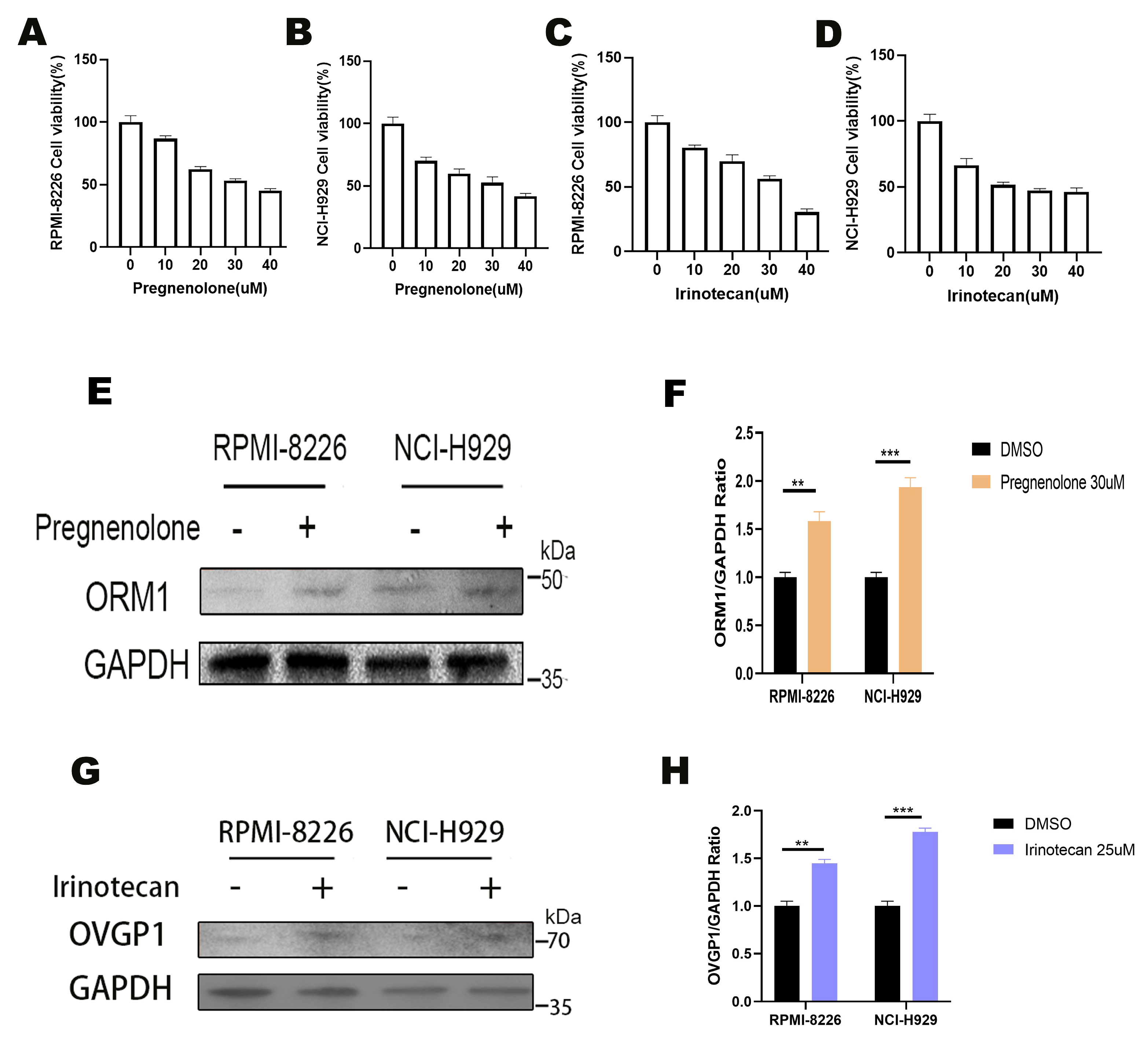
3.5. Experimental Validation of Target Genes
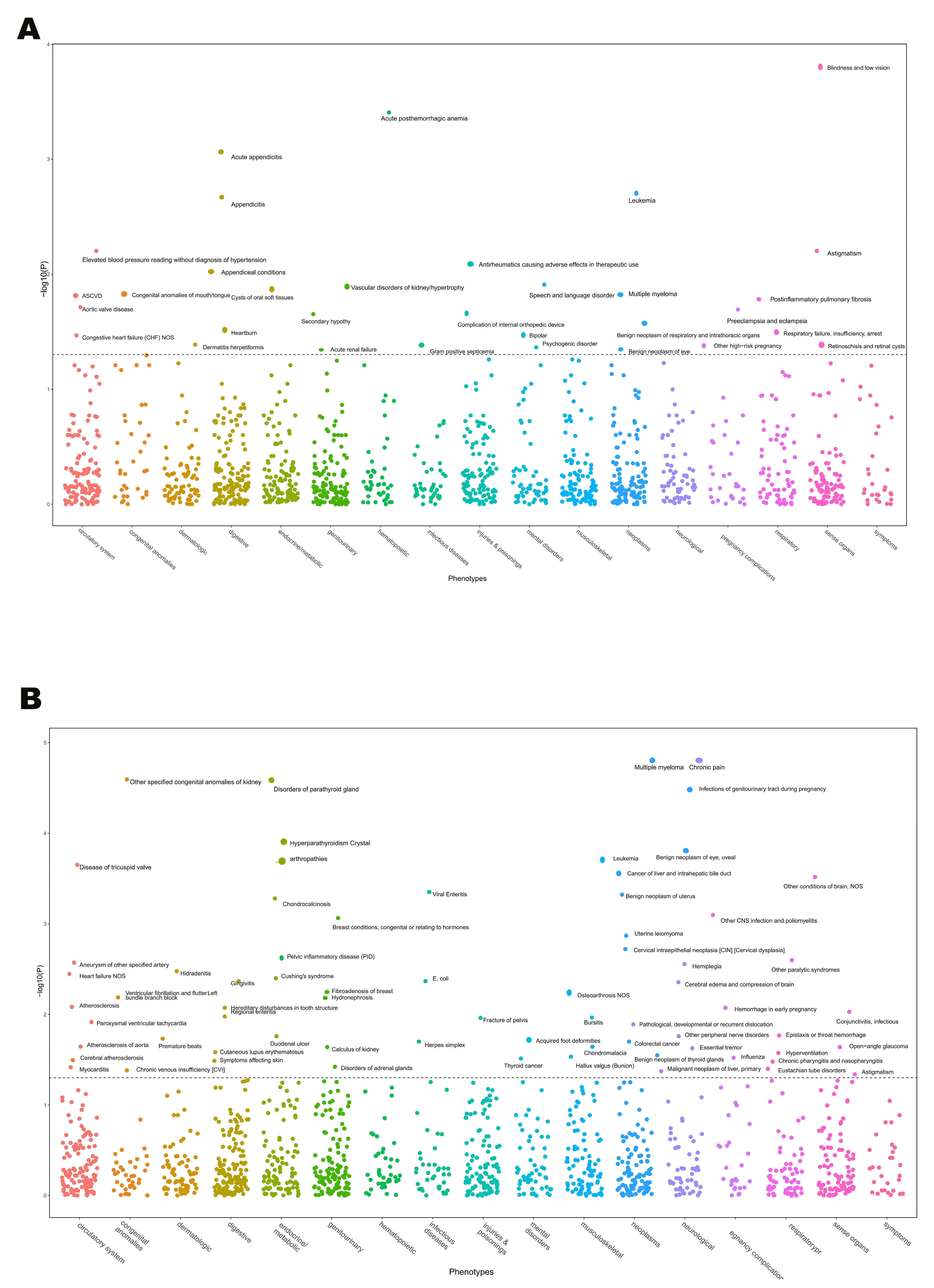
3.6. MR-PheWAS Explored Off-Target Effects of Target Genes
3.7. Pregnanolone and Irinotecan as Effective Agonists of ORM1 and OVGP1
3.8. Effects of Pregnanolone and Irinotecan on Multiple Myeloma Cell Viability
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ADCs | antibody–drug conjugates |
| BCL2 | B-Cell Lymphoma 2 |
| BiTEs | bispecific T cell engagers |
| CA2 | carbonic anhydrase 2 |
| CAR | chimeric antigen receptor |
| CCK-8 | Cell Counting Kit-8 |
| cis-eQTL | cis-acting Expression Quantitative Trait Loci |
| DMSO | dimethyl sulfoxide |
| DSigDB | Drug Signatures Database |
| DTMR | drug target Mendelian randomization |
| EBV | Epstein–Barr virus |
| ECL | Enhanced Chemiluminescence |
| ECM | extracellular matrix |
| ER | endoplasmic reticulum |
| FDR | false discovery rate |
| GPCRs | G-protein-coupled receptors |
| GWASs | genome-wide association studies |
| HDL-C | high-density lipoprotein cholesterol |
| HEIDI | Heterogeneity in Dependent Instruments |
| HRP | horseradish peroxidase |
| IL-1β | Interleukin 1 Beta |
| IL-6 | Interleukin 6 |
| IVs | instrumental variables |
| IVW | inverse variance weighted |
| LD | linkage disequilibrium |
| MAF | minor allele frequency |
| MATN2 | Matrilin-2 |
| MM | multiple myeloma |
| M-proteins | monoclonal immunoglobulins |
| MR | Mendelian randomization |
| MR-PheWAS | Mendelian randomization phenome-wide association |
| NAMPT | nicotinamide phosphoribosyl transferase |
| ORM1 | Orosomucoid 1 |
| OVGP1 | Oviductal Glycoprotein 1 |
| PBS | phosphate-buffered saline |
| RCT | randomized controlled trial |
| RT | Reverse transcription |
| SAIGE | Scalable and Accurate Implementation of Generalized Mixed Model |
| SDS-PAGE | sodium dodecyl sulfate–polyacrylamide gel electrophoresis |
| SNP | Single-Nucleotide Polymorphism |
| SOCS3 | suppressor of cytokine signaling 3 |
| TNF | Tumor Necrosis Factor |
| TWAS | transcriptome-wide association study |
| VZV | varicella zoster virus |
References
- Ludwig, H.; Novis Durie, S.; Meckl, A.; Hinke, A.; Durie, B. Multiple myeloma incidence and mortality around the globe; interrelations between health access and quality, economic resources, and patient empowerment. Oncologist 2020, 25, e1406–e1413. [Google Scholar] [CrossRef]
- Pertesi, M.; Went, M.; Hansson, M.; Hemminki, K.; Houlston, R.S.; Nilsson, B. Genetic predisposition for multiple myeloma. Leukemia 2020, 34, 697–708. [Google Scholar] [CrossRef]
- Ostendorf, L.; Burns, M.; Durek, P.; Heinz, G.A.; Heinrich, F.; Garantziotis, P.; Enghard, P.; Richter, U.; Biesen, R.; Schneider, U.; et al. Targeting CD38 with daratumumab in refractory systemic lupus erythematosus. N. Engl. J. Med. 2020, 383, 1149–1155. [Google Scholar] [CrossRef] [PubMed]
- Hvidtfeldt, U.A.; Chen, J.; Rodopoulou, S.; Strak, M.; de Hoogh, K.; Andersen, Z.J.; Bellander, T.; Brandt, J.; Forastiere, F.; Brynedal, B.; et al. Multiple myeloma risk in relation to long-term air pollution exposure—A pooled analysis of four European cohorts. Environ. Res. 2023, 239, 117230. [Google Scholar] [CrossRef]
- Went, M.; Sud, A.; Försti, A.; Halvarsson, B.-M.; Weinhold, N.; Kimber, S.; van Duin, M.; Thorleifsson, G.; Holroyd, A.; Johnson, D.C.; et al. Identification of multiple risk loci and regulatory mechanisms influencing susceptibility to multiple myeloma. Nat. Commun. 2018, 9, 3707. [Google Scholar] [CrossRef] [PubMed]
- White, B.S.; Lanc, I.; O’Neal, J.; Gupta, H.; Fulton, R.S.; Schmidt, H.; Fronick, C.; Belter, E.A.; Fiala, M.; King, J.; et al. A multiple myeloma-specific capture sequencing platform discovers novel translocations and frequent, risk-associated point mutations in IGLL5. Blood Cancer J. 2018, 8, 35. [Google Scholar] [CrossRef] [PubMed]
- Paner, A.; Patel, P.; Dhakal, B. The evolving role of translocation t(11;14) in the biology, prognosis, and management of multiple myeloma. Blood Rev. 2020, 41, 100643. [Google Scholar] [CrossRef]
- Fotiou, D.; Katodritou, E. From biology to clinical practice: The bone marrow microenvironment in multiple myeloma. J. Clin. Med. 2025, 14, 327. [Google Scholar] [CrossRef]
- García-Ortiz, A.; Rodríguez-García, Y.; Encinas, J.; Maroto-Martín, E.; Castellano, E.; Teixidó, J.; Martínez-López, J. The role of tumor microenvironment in multiple myeloma development and progression. Cancers 2021, 13, 217. [Google Scholar] [CrossRef]
- Alabanza, L.M.; Xiong, Y.; Vu, B.; Webster, B.; Wu, D.; Hu, P.; Zhu, Z.; Dropulic, B.; Dash, P.; Schneider, D. Armored BCMA CAR T cells eliminate multiple myeloma and are resistant to the suppressive effects of TGF-β. Front. Immunol. 2022, 13, 832645. [Google Scholar] [CrossRef]
- Swamydas, M.; Murphy, E.V.; Ignatz-Hoover, J.J.; Malek, E.; Driscoll, J.J. Deciphering mechanisms of immune escape to inform immunotherapeutic strategies in multiple myeloma. J. Hematol. Oncol. 2022, 15, 17. [Google Scholar] [CrossRef]
- Lu, Q.; Yang, D.; Li, H.; Niu, T.; Tong, A. Multiple myeloma: Signaling pathways and targeted therapy. Mol. Biomed. 2024, 5, 25. [Google Scholar] [CrossRef]
- Wallington-Beddoe, C.T.; Sobieraj-Teague, M.; Kuss, B.J.; Pitson, S.M. Resistance to proteasome inhibitors and other targeted therapies in myeloma. Br. J. Haematol. 2018, 182, 11–28. [Google Scholar] [CrossRef]
- Moreau, P.; Attal, M.; Facon, T. Frontline therapy of multiple myeloma. Blood 2015, 125, 3076–3084. [Google Scholar] [CrossRef]
- Pinto, V.; Bergantim, R.; Caires, H.R.; Seca, H.; Guimarães, J.E.; Vasconcelos, M.H. Multiple myeloma: Available therapies and causes of drug resistance. Cancers 2020, 12, 407. [Google Scholar] [CrossRef]
- Alaterre, E.; Ovejero, S.; Herviou, L.; de Boussac, H.; Papadopoulos, G.; Kulis, M.; Boireau, S.; Robert, N.; Requirand, G.; Bruyer, A.; et al. Comprehensive characterization of the epigenetic landscape in multiple myeloma. Theranostics 2022, 12, 1715–1729. [Google Scholar] [CrossRef]
- Wu, Y.-X.; Yang, J.-H.; Saitsu, H. Bortezomib-resistance is associated with increased levels of proteasome subunits and apoptosis-avoidance. Oncotarget 2016, 7, 77622–77634. [Google Scholar] [CrossRef][Green Version]
- Pan, Y.; Zhang, Y.; Liu, W.; Huang, Y.; Shen, X.; Jing, R.; Pu, J.; Wang, X.; Ju, S.; Cong, H.; et al. LncRNA H19 overexpression induces bortezomib resistance in multiple myeloma by targeting MCL-1 via miR-29b-3p. Cell Death Dis. 2019, 10, 106. [Google Scholar] [CrossRef]
- Yue, T.; Sun, Y.; Dai, Y.; Jin, F. Mechanisms for resistance to BCMA-targeted immunotherapies in multiple myeloma. Blood Rev. 2025, 70, 101256. [Google Scholar] [CrossRef]
- Vaghari-Tabari, M.; Hassanpour, P.; Sadeghsoltani, F.; Malakoti, F.; Alemi, F.; Qujeq, D.; Asemi, Z.; Yousefi, B. CRISPR/Cas9 gene editing: A new approach for overcoming drug resistance in cancer. Cell. Mol. Biol. Lett. 2022, 27, 49. [Google Scholar] [CrossRef]
- Davies, N.M.; Holmes, M.V.; Smith, G.D. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef] [PubMed]
- Prats-Uribe, A.; Sayols-Baixeras, S.; Fernández-Sanlés, A.; Subirana, I.; Carreras-Torres, R.; Vilahur, G.; Civeira, F.; Marrugat, J.; Fitó, M.; Hernáez, Á.; et al. High-density lipoprotein characteristics and coronary artery disease: A Mendelian randomization study. Metabolism 2020, 112, 154351. [Google Scholar] [CrossRef]
- Georgiou, A.N.; Ntritsos, G.; Papadimitriou, N.; Dimou, N.; Evangelou, E. Cigarette smoking, coffee consumption, alcohol intake, and risk of Crohn’s disease and ulcerative colitis: A Mendelian randomization study. Inflamm. Bowel Dis. 2021, 27, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Yarmolinsky, J.; Bouras, E.; Constantinescu, A.; Burrows, K.; Bull, C.J.; Vincent, E.E.; Martin, R.M.; Dimopoulou, O.; Lewis, S.J.; Moreno, V.; et al. Genetically proxied glucose-lowering drug target perturbation and risk of cancer: A Mendelian randomisation analysis. Diabetologia 2023, 66, 1481–1500. [Google Scholar] [CrossRef] [PubMed]
- Yarmolinsky, J.; Díez-Obrero, V.; Richardson, T.G.; Pigeyre, M.; Sjaarda, J.; Paré, G.; Walker, V.M.; Vincent, E.E.; Tan, V.Y.; Obón-Santacana, M.; et al. Genetically proxied therapeutic inhibition of antihypertensive drug targets and risk of common cancers: A mendelian randomization analysis. PLoS Med. 2022, 19, e1003897. [Google Scholar] [CrossRef]
- Wang, Q.; Shi, Q.; Wang, Z.; Lu, J.; Hou, J. Integrating plasma proteomes with genome-wide association data for causal protein identification in multiple myeloma. BMC Med. 2023, 21, 377. [Google Scholar] [CrossRef]
- Lin, Z.; Pan, W. A robust cis-Mendelian randomization method with application to drug target discovery. Nat. Commun. 2024, 15, 6072. [Google Scholar] [CrossRef]
- Schmidt, A.F.; Finan, C.; Gordillo-Marañón, M.; Asselbergs, F.W.; Freitag, D.F.; Patel, R.S.; Tyl, B.; Chopade, S.; Faraway, R.; Zwierzyna, M.; et al. Genetic drug target validation using Mendelian randomisation. Nat. Commun. 2020, 11, 3255. [Google Scholar] [CrossRef] [PubMed]
- Yeung, S.L.A.; Gill, D. Standardizing the reporting of Mendelian randomization studies. BMC Med. 2023, 21, 187. [Google Scholar] [CrossRef]
- Sun, Z.; Yun, Z.; Lin, J.; Sun, X.; Wang, Q.; Duan, J.; Li, C.; Zhang, X.; Xu, S.; Wang, Z.; et al. Comprehensive Mendelian randomization analysis of plasma proteomics to identify new therapeutic targets for the treatment of coronary heart disease and myocardial infarction. J. Transl. Med. 2024, 22, 404. [Google Scholar] [CrossRef]
- Liu, A.E.; Kang, H.M. Meta-imputation of transcriptome from genotypes across multiple datasets by leveraging publicly available summary-level data. PLoS Genet. 2022, 18, e1009571. [Google Scholar] [CrossRef]
- Evans, P.; Nagai, T.; Konkashbaev, A.; Zhou, D.; Knapik, E.W.; Gamazon, E.R. Transcriptome-Wide Association Studies (TWAS): Methodologies, Applications, and Challenges. Curr. Protoc. 2024, 4, e981. [Google Scholar] [CrossRef]
- Rasooly, D.; Peloso, G.M.; Giambartolomei, C. Bayesian genetic colocalization test of two traits using coloc. Curr. Protoc. 2022, 2, e627. [Google Scholar] [CrossRef]
- Agu, P.C.; Afiukwa, C.A.; Orji, O.U.; Ezeh, E.M.; Ofoke, I.H.; Ogbu, C.O.; Ugwuja, E.I.; Aja, P.M. Molecular docking as a tool for the discovery of molecular targets of nutraceuticals in diseases management. Sci. Rep. 2023, 13, 13398. [Google Scholar] [CrossRef]
- Gong, W.; Sun, P.; Li, X.; Wang, X.; Zhang, X.; Cui, H.; Yang, J. Investigating the molecular mechanisms of resveratrol in treating cardiometabolic multimorbidity: A network pharmacology and bioinformatics approach with molecular docking validation. Nutrients 2024, 16, 2488. [Google Scholar] [CrossRef]
- Finan, C.; Gaulton, A.; Kruger, F.A.; Lumbers, R.T.; Shah, T.; Engmann, J.; Galver, L.; Kelley, R.; Karlsson, A.; Santos, R.; et al. The druggable genome and support for target identification and validation in drug development. Sci. Transl. Med. 2017, 9, eaag1166. [Google Scholar] [CrossRef] [PubMed]
- Võsa, U.; Claringbould, A.; Westra, H.-J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-scale cis- and trans-eQTL analyses identify thousands of genetic loci and polygenic scores that regulate blood gene expression. Nat. Genet. 2021, 53, 1300–1310. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Peng, Z.; Lin, H.; Zhou, K.; Liang, L.; Cao, J.; Huang, Z.; Mei, J. Identifying potential drug targets for idiopathic pulmonary fibrosis: A Mendelian randomization study based on the druggable genes. Respir. Res. 2024, 25, 217. [Google Scholar] [CrossRef]
- Mathieu, S.; Briend, M.; Abner, E.; Couture, C.; Li, Z.; Bossé, Y.; Thériault, S.; Esko, T.; Arsenault, B.J.; Mathieu, P. Genetic association and Mendelian randomization for hypothyroidism highlight immune molecular mechanisms. iScience 2022, 25, 104992. [Google Scholar] [CrossRef]
- Yuan, S.; Chen, J.; Geng, J.; Zhao, S.S.; Yarmolinsky, J.; Arkema, E.V.; Abramowitz, S.; Levin, M.G.; Tsilidis, K.K.; Burgess, S.; et al. GWAS identifies genetic loci, lifestyle factors and circulating biomarkers that are risk factors for sarcoidosis. Nat. Commun. 2025, 16, 2481. [Google Scholar] [CrossRef]
- Garfield, V.; Salzmann, A.; Burgess, S.; Chaturvedi, N. A guide for selection of genetic instruments in Mendelian randomization studies of type 2 diabetes and HbA1c: Toward an integrated approach. Diabetes 2023, 72, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Bycroft, C.; Freeman, C.; Petkova, D.; Band, G.; Elliott, L.T.; Sharp, K.; Motyer, A.; Vukcevic, D.; Delaneau, O.; O’Connell, J.; et al. The UK Biobank resource with deep phenotyping and genomic data. Nature 2018, 562, 203–209. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Nielsen, J.B.; Fritsche, L.G.; Dey, R.; Gabrielsen, M.E.; Wolford, B.N.; LeFaive, J.; VandeHaar, P.; Gagliano, S.A.; Gifford, A.; et al. Efficiently controlling for case–control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018, 50, 1335–1341. [Google Scholar] [CrossRef]
- Burgess, S.; Foley, C.N.; Allara, E.; Staley, J.R.; Howson, J.M.M. A robust and efficient method for Mendelian randomization with hundreds of genetic variants. Nat. Commun. 2020, 11, 376. [Google Scholar] [CrossRef]
- Chen, S.; Sun, S.; Cai, M.; Zhou, Z.; Ma, Y.; Zhou, Z.; Wang, F.; Liu, J.; Song, W.; Liu, Y.; et al. A metabolome-wide Mendelian randomization study prioritizes causal circulating metabolites for reproductive disorders including primary ovarian insufficiency, polycystic ovary syndrome, and abnormal spermatozoa. J. Ovarian Res. 2024, 17, 166. [Google Scholar] [CrossRef]
- Mai, J.; Lu, M.; Gao, Q.; Zeng, J.; Xiao, J. Transcriptome-wide association studies: Recent advances in methods, applications and available databases. Commun. Biol. 2023, 6, 899. [Google Scholar] [CrossRef]
- Zuber, V.; Grinberg, N.F.; Gill, D.; Manipur, I.; Slob, E.A.; Patel, A.; Wallace, C.; Burgess, S. Combining evidence from Mendelian randomization and colocalization: Review and comparison of approaches. Am. J. Hum. Genet. 2022, 109, 767–782. [Google Scholar] [CrossRef]
- Li, Y.; Song, X.; Huang, Y.; Zhou, S.; Zhong, L. Genetic associations of plasma metabolites with immune cells in hyperthyroidism revealed by Mendelian randomization and GWAS-sc-eQTLs xQTLbiolinks analysis. Sci. Rep. 2025, 15, 1377. [Google Scholar] [CrossRef]
- Wallace, C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020, 16, e1008720. [Google Scholar] [CrossRef]
- Wang, Z.; Li, S.; Cai, G.; Gao, Y.; Yang, H.; Li, Y.; Liang, J.; Zhang, S.; Hu, J.; Zheng, J. Mendelian randomization analysis identifies druggable genes and drugs repurposing for chronic obstructive pulmonary disease. Front. Cell. Infect. Microbiol. 2024, 14, 1386506. [Google Scholar] [CrossRef]
- Yuan, S.; Sun, J.; Lu, Y.; Xu, F.; Li, D.; Jiang, F.; Wan, Z.; Li, X.; Qin, L.-Q.; Larsson, S.C. Health effects of milk consumption: Phenome-wide Mendelian randomization study. BMC Med. 2022, 20, 55. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Lophatananon, A.; Mekli, K.; Muir, K.R.; Guo, H. Exploring the causal role of the immune response to varicella-zoster virus on multiple traits: A phenome-wide Mendelian randomization study. BMC Med. 2023, 21, 143. [Google Scholar] [CrossRef]
- Gibson, M.J.; Lawlor, D.A.; Millard, L.A.C. Identifying the potential causal role of insomnia symptoms on 11,409 health-related outcomes: A phenome-wide Mendelian randomization analysis in UK Biobank. BMC Med. 2023, 21, 128. [Google Scholar] [CrossRef] [PubMed]
- Valdés-Tresanco, M.S.; Valdés-Tresanco, M.E.; Valiente, P.A.; Moreno, E. AMDock: A versatile graphical tool for assisting molecular docking with Autodock Vina and Autodock4. Biol. Direct. 2020, 15, 12. [Google Scholar] [CrossRef]
- Eberhardt, J.; Santos-Martins, D.; Tillack, A.F.; Forli, S. AutoDock Vina 1.2.0: New docking methods, expanded force field, and Python bindings. J. Chem. Inf. Model. 2021, 61, 3891–3898. [Google Scholar] [CrossRef]
- Nguyen, N.T.; Nguyen, T.H.; Pham, T.N.H.; Huy, N.T.; Bay, M.V.; Pham, M.Q.; Nam, P.C.; Vu, V.V.; Ngo, S.T. AutoDock Vina adopts more accurate binding poses but AutoDock4 forms better binding affinity. J. Chem. Inf. Model. 2020, 60, 204–211. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef]
- Taylor, S.C.; Rosselli-Murai, L.K.; Crobeddu, B.; Plante, I. A critical path to producing high quality, reproducible data from quantitative western blot experiments. Sci. Rep. 2022, 12, 17599. [Google Scholar] [CrossRef]
- Gamazon, E.R.; GTEx Consortium; Wheeler, H.E.; Shah, K.P.; Mozaffari, S.V.; Aquino-Michaels, K.; Carroll, R.J.; Eyler, A.E.; Denny, J.C.; Nicolae, D.L.; et al. A gene-based association method for mapping traits using reference transcriptome data. Nat. Genet. 2015, 47, 1091–1098. [Google Scholar] [CrossRef]
- Huang, M.; Wu, Y.; Li, Y.; Chen, X.; Feng, J.; Li, Z.; Li, J.; Chen, J.; Lu, Y.; Feng, Y. Circadian clock-related genome-wide Mendelian randomization identifies putatively genes for ulcerative colitis and its comorbidity. BMC Genom. 2024, 25, 130. [Google Scholar] [CrossRef]
- Lu, Y.; Lu, D.; Zhang, H.; Li, H.; Yu, B.; Zhang, Y.; Hu, H.; Sheng, H. Causality between ankylosing spondylitis and osteoarthritis in European ancestry: A bidirectional Mendelian randomization study. Front. Immunol. 2024, 15, 1297454. [Google Scholar] [CrossRef]
- Xu, J.; Si, S.; Han, Y.; Zeng, L.; Zhao, J. Genetic insight into dissecting the immunophenotypes and inflammatory profiles in the pathogenesis of Sjogren syndrome. J. Transl. Med. 2025, 23, 56. [Google Scholar] [CrossRef]
- Ocio, E.M.; Motlló, C.; Rodríguez-Otero, P.; Martínez-López, J.; Cejalvo, M.J.; Martín-Sánchez, J.; Bladé, J.; García-Malo, M.D.; Dourdil, M.V.; García-Mateo, A.; et al. Filanesib in combination with pomalidomide and dexamethasone in refractory MM patients: Safety and efficacy, and association with alpha 1-acid glycoprotein (AAG) levels. Phase Ib/II Pomdefil clinical trial conducted by the Spanish MM group. Br. J. Haematol. 2021, 192, 522–530. [Google Scholar] [CrossRef]
- Gao, Y.; Liu, X.; Li, T.; Wei, L.; Yang, A.; Lu, Y.; Zhang, J.; Li, L.; Wang, S.; Yin, F. Cross-validation of genes potentially associated with overall survival and drug resistance in ovarian cancer. Oncol. Rep. 2017, 37, 3084–3092. [Google Scholar] [CrossRef]
- Gabbasov, R.; Xiao, F.; Howe, C.G.; Bickel, L.E.; O’Brien, S.W.; Benrubi, D.; Do, T.-V.; Zhou, Y.; Nicolas, E.; Cai, K.Q.; et al. NEDD9 promotes oncogenic signaling, a stem/mesenchymal gene signature, and aggressive ovarian cancer growth in mice. Oncogene 2018, 37, 4854–4870. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.-J.; Zhu, Y.-P.; Sun, J.; Ding, X.-Y.; Wang, X.; Zhang, Q.-Z. PTGES2 and RNASET2 identified as novel potential biomarkers and therapeutic targets for basal cell carcinoma: Insights from proteome-wide Mendelian randomization, colocalization, and MR-PheWAS analyses. Front. Pharmacol. 2024, 15, 1418560. [Google Scholar] [CrossRef]
- Schverer, M.; Lanfumey, L.; Baulieu, E.-E.; Froger, N.; Villey, I. Neurosteroids: Non-genomic pathways in neuroplasticity and involvement in neurological diseases. Pharmacol. Ther. 2018, 191, 190–206. [Google Scholar] [CrossRef]
- Bailly, C. Irinotecan: 25 years of cancer treatment. Pharmacol. Res. 2019, 148, 104398. [Google Scholar] [CrossRef]
- Cheng, X.; Liu, D.; Xing, R.; Song, H.; Tian, X.; Yan, C.; Han, Y. Orosomucoid 1 attenuates doxorubicin-induced oxidative stress and apoptosis in cardiomyocytes via Nrf2 signaling. Biomed Res. Int. 2020, 2020, 5923572. [Google Scholar] [CrossRef]
- Yin, D.-M.; Yuan, D.; Sun, R.-J.; Xu, H.-Z.; Hun, S.-Y.; Sui, X.-H.; Shan, N.-N. Identification of ORM1, vWF, SPARC, and PPBP as immune-related proteins involved in immune thrombocytopenia by quantitative LC–MS/MS. Clin. Proteom. 2023, 20, 24. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Wang, F.; Zhou, Z.; Teng, J.; Su, Y.; Chi, H.; Wang, Z.; Hu, Q.; Jia, J.; Liu, T.; et al. Urinary proteomics identifying novel biomarkers for the diagnosis of adult-onset Still’s disease. Front. Immunol. 2020, 11, 2112. [Google Scholar] [CrossRef]
- Ramsay, T.G.; Stoll, M.J.; Blomberg, L.A.; Caperna, T.J. Regulation of cytokine gene expression by orosomucoid in neonatal swine adipose tissue. J. Anim. Sci. Biotechnol. 2016, 7, 25. [Google Scholar] [CrossRef][Green Version]
- Qiong, L.; Yin, J. Orosomucoid 1 promotes epirubicin resistance in breast cancer by upregulating the expression of matrix metalloproteinases 2 and 9. Bioengineered 2021, 12, 8822–8832. [Google Scholar] [CrossRef]
- He, J.; Zheng, Z.; Liu, T.; Ao, Y.; Yang, Y.; Hu, H. Salivary orosomucoid 1 as a biomarker of hepatitis B associated hepatocellular carcinoma. Sci. Rep. 2022, 12, 15347. [Google Scholar] [CrossRef]
- Yue, L.; Xu, X.; Dai, S.; Xu, F.; Zhao, W.; Gu, J.; Dai, X.; Qian, X. Orosomucoid 1 promotes colorectal cancer progression and liver metastasis by affecting PI3K/AKT pathway and inducing macrophage M2 polarization. Sci. Rep. 2023, 13, 14092. [Google Scholar] [CrossRef]
- Meng, H.; Jiang, X.; Huang, H.; Shen, N.; Guo, C.; Yu, C.; Yin, G.; Wang, Y. A MUCINs expression signature impacts overall survival in patients with clear cell renal cell carcinoma. Cancer Med. 2021, 10, 5823–5838. [Google Scholar] [CrossRef]
- Flesken-Nikitin, A.; Ralston, C.Q.; Fu, D.-J.; De Micheli, A.J.; Phuong, D.J.; Harlan, B.A.; Ashe, C.S.; Armstrong, A.P.; McKellar, D.W.; Ghuwalewala, S.; et al. Preciliated tubal epithelial cells are prone to initiation of high-grade serous ovarian carcinoma. Nat. Commun. 2024, 15, 8641. [Google Scholar] [CrossRef]
- García-Vázquez, F.A.; Garrappa, G.; Luongo, C.; Hamze, J.G.; Caballero, M.; Marco-Jiménez, F.; Antón, J.S.V.; Molina-Cuberos, G.J.; Jiménez-Movilla, M. Magnetic-assisted control of eggs and embryos via zona pellucida-linked nanoparticles. Adv. Sci. 2024, 11, e2306901. [Google Scholar] [CrossRef]
- Laheri, S.; Ashary, N.; Bhatt, P.; Modi, D. Oviductal glycoprotein 1 (OVGP1) is expressed by endometrial epithelium that regulates receptivity and trophoblast adhesion. J. Assist. Reprod. Genet. 2018, 35, 1419–1429. [Google Scholar] [CrossRef]
- Cosentino, G.; Romero-Cordoba, S.; Plantamura, I.; Cataldo, A.; Iorio, M.V. miR-9-mediated inhibition of EFEMP1 contributes to the acquisition of pro-tumoral properties in normal fibroblasts. Cells 2020, 9, 2143. [Google Scholar] [CrossRef] [PubMed]
- Thomas, A.; Pommier, Y. Targeting Topoisomerase I in the era of precision medicine. Clin. Cancer Res. 2019, 25, 6581–6589. [Google Scholar] [CrossRef]
- Chen, C.-I.; Chen, Y.-C.; Kao, Y.-K.; Chen, C.-H.; Yang, P.-W.; Chen, P.-C.; Song, L.-C.; Tsai, K.L. Taurine protects irinotecan-induced muscle dysfunction by modulating oxidative stress and endoplasmic reticulum stress in human skeletal muscle cells. Anticancer Res. 2024, 44, 5371–5378. [Google Scholar] [CrossRef]
- Giannotta, C.; Autino, F.; Massaia, M. The immune suppressive tumor microenvironment in multiple myeloma: The contribution of myeloid-derived suppressor cells. Front. Immunol. 2023, 13, 1102471. [Google Scholar] [CrossRef] [PubMed]
- Maiso, P.; Mogollón, P.; Ocio, E.M.; Garayoa, M. Bone marrow mesenchymal stromal cells in multiple myeloma: Their role as active contributors to myeloma progression. Cancers 2021, 13, 2542. [Google Scholar] [CrossRef]
- Agrawal, B.; Gupta, N.; Konowalchuk, J.D. MUC1 mucin: A putative regulatory (checkpoint) molecule of T cells. Front. Immunol. 2018, 9, 2391. [Google Scholar] [CrossRef]
- Zhao, L.; Cheng, S.; Fan, L.; Zhang, B.; Xu, S. TIM-3: An update on immunotherapy. Int. Immunopharmacol. 2021, 99, 107933. [Google Scholar] [CrossRef]
- Osorio, D.; Yu, X.; Yu, P.; Serpedin, E.; Cai, J.J. Single-cell RNA sequencing of a European and an African lymphoblastoid cell line. Sci. Data 2019, 6, 112. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Zhang, J.; Wu, X.; Du, W.; Zhu, Y.; Liu, X.; Liu, Z.; Meng, B.; Guo, J.; Yang, Q.; et al. Blocking glycine utilization inhibits multiple myeloma progression by disrupting glutathione balance. Nat. Commun. 2022, 13, 4007. [Google Scholar] [CrossRef]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Yarmolinsky, J.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Higgins, J.P.T.; Timpson, N.J.; Dimou, N.; et al. Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomization (STROBE-MR) Statement. JAMA 2021, 326, 1614–1621. [Google Scholar]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Davies, N.M.; Swanson, S.A.; VanderWeele, T.J.; Timpson, N.J.; Higgins, J.P.T.; Dimou, N.; Langenberg, C.; et al. Strengthening the Reporting of Observational Studies in Epidemiology using Mendelian Randomisation (STROBE-MR): Explanation and Elaboration. BMJ 2021, 375, n2233. [Google Scholar]

| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| ORM1 | ACCTACATGCTTGCTTTTGACG | CCCCCAAGTCTCTGTCCTGA |
| MATN2 | GTGTCAACACCCATGACTATGC | CATCAGGACCAATGTCCAAG |
| OVGP1 | AGCGAAGAAGCACTGGATTGA | ATTCACAGCAGATGACAGCCA |
| GAPDH | GAAGGTGAAGGTCGGAGTC | GAAGATGGTGATGGGATTTC |
| ID | CHR | SNPID | pos | Z | FDR |
|---|---|---|---|---|---|
| OVGP1 | 1 | rs1264878 | 111947430 | −3.442623 | 0.013824 |
| ORM1 | 9 | rs7851482 | 117078286 | −2.95 | 0.03416 |
| ALOX5AP | 13 | rs6490461 | 30976845 | −2.8571 | 0.03416 |
| PTGDS | 9 | rs2811786 | 139683224 | 2.75 | 0.03594 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, S.; Fan, F.; Li, Q.; Zuo, L.; Xu, A.; Sun, C. Therapeutic Target Discovery for Multiple Myeloma: Identifying Druggable Genes via Mendelian Randomization. Biomedicines 2025, 13, 885. https://doi.org/10.3390/biomedicines13040885
Jiang S, Fan F, Li Q, Zuo L, Xu A, Sun C. Therapeutic Target Discovery for Multiple Myeloma: Identifying Druggable Genes via Mendelian Randomization. Biomedicines. 2025; 13(4):885. https://doi.org/10.3390/biomedicines13040885
Chicago/Turabian StyleJiang, Shijun, Fengjuan Fan, Qun Li, Liping Zuo, Aoshuang Xu, and Chunyan Sun. 2025. "Therapeutic Target Discovery for Multiple Myeloma: Identifying Druggable Genes via Mendelian Randomization" Biomedicines 13, no. 4: 885. https://doi.org/10.3390/biomedicines13040885
APA StyleJiang, S., Fan, F., Li, Q., Zuo, L., Xu, A., & Sun, C. (2025). Therapeutic Target Discovery for Multiple Myeloma: Identifying Druggable Genes via Mendelian Randomization. Biomedicines, 13(4), 885. https://doi.org/10.3390/biomedicines13040885






