Real-World Performance of the EasyPGX® Ready Epidermal Growth Factor Receptor Assay for Genomic Testing of Non-Small Cell Lung Cancer Samples
Abstract
1. Introduction
2. Materials and Methods
2.1. Technical Verification
2.2. Performance Evaluation Cohort
2.3. Specimen Preparation and Pathological Analysis
2.4. Nucleic Acid Extraction
2.5. Next Generation Sequencing
2.6. EasyPGX® Ready EGFR Assay
2.7. Turnaround Time
2.8. Statistical Analysis
3. Results
3.1. Verification of the EasyPGX® Ready EGFR Assay
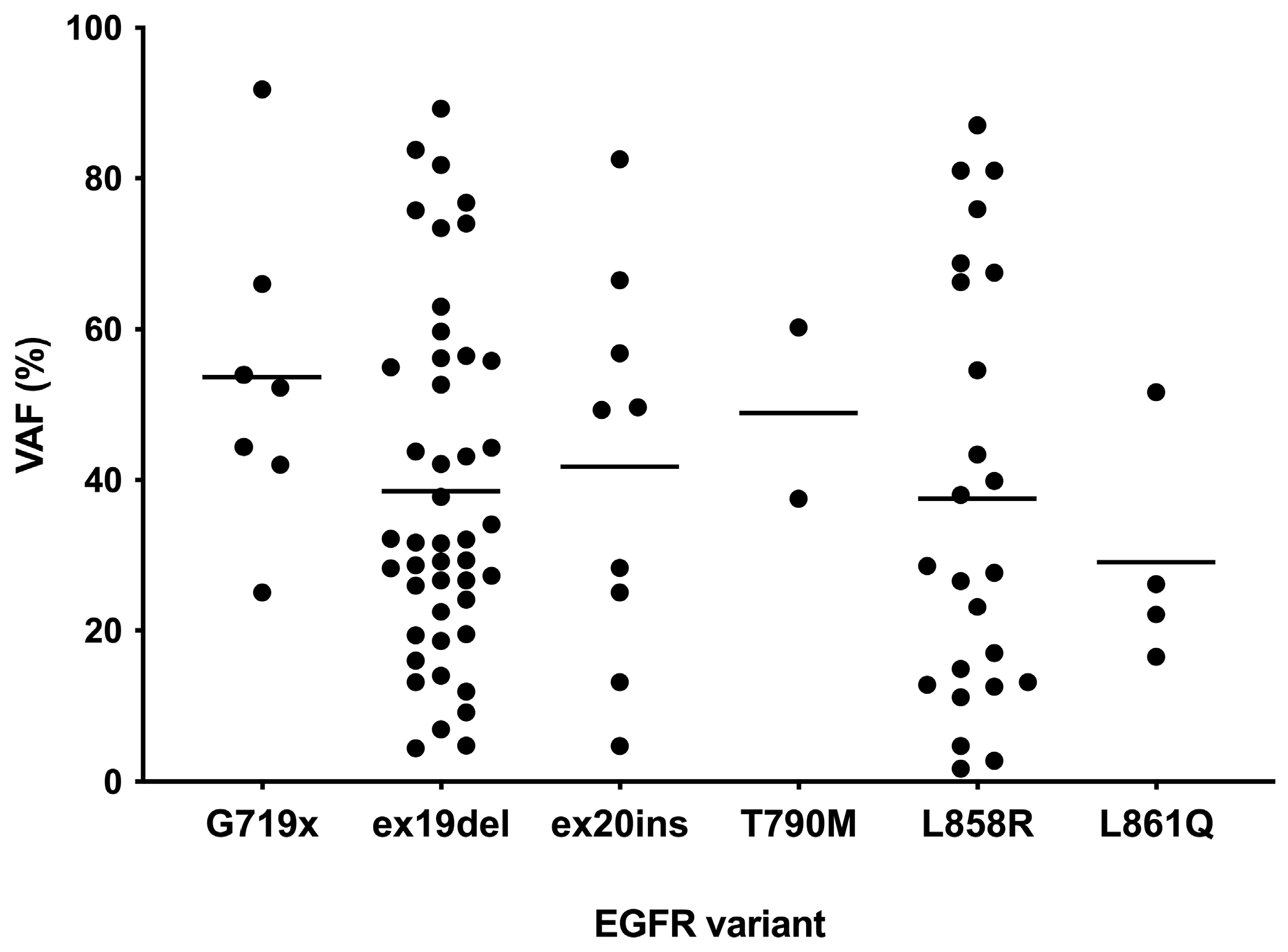
3.2. EGFR Variant Testing by NGS
3.3. EGFR Variant Testing by the EasyPGX® Ready EGFR Assay
3.4. Concordance Analysis
3.5. Discordant Samples
3.6. Failure Rate
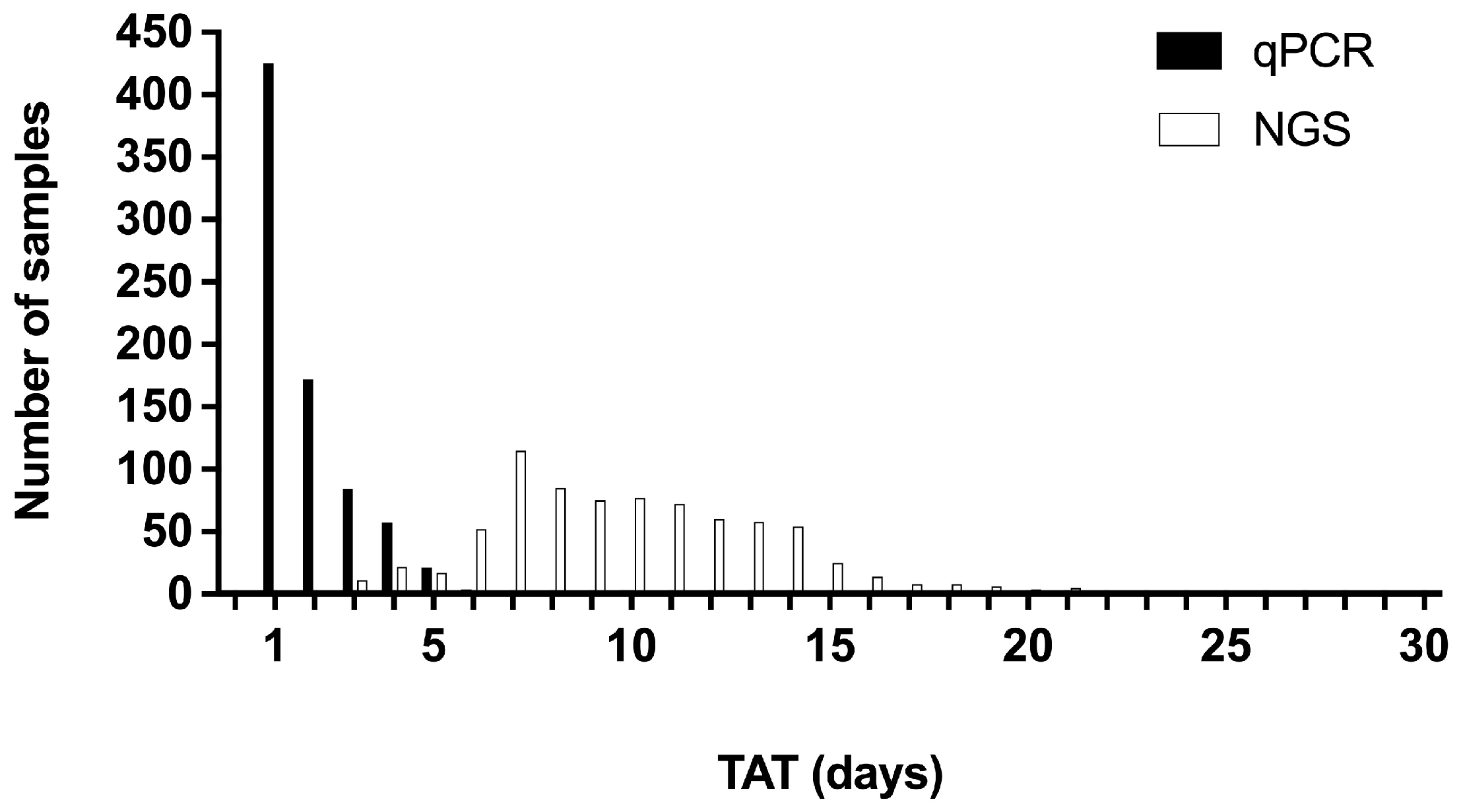
3.7. Turnaround Times
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic acid |
| EGFR | Epidermal growth factor receptor |
| FFPE | Formalin-fixed paraffin-embedded |
| FISH | Fluorescence in situ hybridization |
| H&E | Hematoxylin and eosin |
| IHC | Immunohistochemistry |
| LIMS | Laboratory information management system |
| NGS | Next generation sequencing |
| NSCLC | Non-small cell lung cancer |
| RT-qPCR | Real-time quantitative polymerase chain reaction |
| TAT | Turnaround time |
| TKI | Tyrosine kinase inhibitors |
| VAF | Variant allele frequency |
References
- Borgeaud, M.; Parikh, K.; Banna, G.L.; Kim, F.; Olivier, T.; Le, X.; Addeo, A. Unveiling the Landscape of Uncommon EGFR Mutations in NSCLC-A Systematic Review. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2024, 19, 973–983. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Midha, A.; Dearden, S.; McCormack, R. EGFR mutation incidence in non-small-cell lung cancer of adenocarcinoma histology: A systematic review and global map by ethnicity (mutMapII). Am. J. Cancer Res. 2015, 5, 2892–2911. [Google Scholar]
- Pretelli, G.; Spagnolo, C.C.; Ciappina, G.; Santarpia, M.; Pasello, G. Overview on Therapeutic Options in Uncommon EGFR Mutant Non-Small Cell Lung Cancer (NSCLC): New Lights for an Unmet Medical Need. Int. J. Mol. Sci. 2023, 24, 8878. [Google Scholar] [CrossRef] [PubMed]
- Passaro, A.; Leighl, N.; Blackhall, F.; Popat, S.; Kerr, K.; Ahn, M.J.; Arcila, M.E.; Arrieta, O.; Planchard, D.; de Marinis, F.; et al. ESMO expert consensus statements on the management of EGFR mutant non-small-cell lung cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2022, 33, 466–487. [Google Scholar] [CrossRef]
- Mosele, M.F.; Westphalen, C.B.; Stenzinger, A.; Barlesi, F.; Bayle, A.; Bièche, I.; Bonastre, J.; Castro, E.; Dienstmann, R.; Krämer, A.; et al. Recommendations for the use of next-generation sequencing (NGS) for patients with advanced cancer in 2024: A report from the ESMO Precision Medicine Working Group. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2024, 35, 588–606. [Google Scholar] [CrossRef]
- Wu, Y.L.; Tsuboi, M.; He, J.; John, T.; Grohe, C.; Majem, M.; Goldman, J.W.; Laktionov, K.; Kim, S.W.; Kato, T.; et al. Osimertinib in Resected EGFR-Mutated Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2020, 383, 1711–1723. [Google Scholar] [CrossRef]
- Pennell, N.A.; Neal, J.W.; Chaft, J.E.; Azzoli, C.G.; Jänne, P.A.; Govindan, R.; Evans, T.L.; Costa, D.B.; Wakelee, H.A.; Heist, R.S.; et al. SELECT: A Phase II Trial of Adjuvant Erlotinib in Patients with Resected Epidermal Growth Factor Receptor-Mutant Non-Small-Cell Lung Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2019, 37, 97–104. [Google Scholar] [CrossRef]
- Lv, C.; Fang, W.; Wu, N.; Jiao, W.; Xu, S.; Ma, H.; Wang, J.; Wang, R.; Ji, C.; Li, S.; et al. Osimertinib as neoadjuvant therapy in patients with EGFR-mutant resectable stage II-IIIB lung adenocarcinoma (NEOS): A multicenter, single-arm, open-label phase 2b trial. Lung Cancer 2023, 178, 151–156. [Google Scholar] [CrossRef]
- Blakely, C.M.; Urisman, A.; Gubens, M.A.; Mulvey, C.K.; Allen, G.M.; Shiboski, S.C.; Rotow, J.K.; Chakrabarti, T.; Kerr, D.L.; Aredo, J.V.; et al. Neoadjuvant Osimertinib for the Treatment of Stage I-IIIA Epidermal Growth Factor Receptor-Mutated Non-Small Cell Lung Cancer: A Phase II Multicenter Study. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2024, 42, 3105–3114. [Google Scholar] [CrossRef]
- Penault-Llorca, F.; Kerr, K.M.; Garrido, P.; Thunnissen, E.; Dequeker, E.; Normanno, N.; Patton, S.J.; Fairley, J.; Kapp, J.; de Ridder, D.; et al. Expert opinion on NSCLC small specimen biomarker testing–Part 1: Tissue collection and management. Virchows Arch. Int. J. Pathol. 2022, 481, 335–350. [Google Scholar] [CrossRef]
- Penault-Llorca, F.; Kerr, K.M.; Garrido, P.; Thunnissen, E.; Dequeker, E.; Normanno, N.; Patton, S.J.; Fairley, J.; Kapp, J.; de Ridder, D.; et al. Expert opinion on NSCLC small specimen biomarker testing–Part 2: Analysis, reporting, and quality assessment. Virchows Arch. Int. J. Pathol. 2022, 481, 351–366. [Google Scholar] [CrossRef] [PubMed]
- VanderLaan, P.A.; Rangachari, D.; Costa, D.B. The rapidly evolving landscape of biomarker testing in non-small cell lung cancer. Cancer Cytopathol. 2021, 129, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Jing, C.; Mao, X.; Wang, Z.; Sun, K.; Ma, R.; Wu, J.; Cao, H. Next-generation sequencing-based detection of EGFR, KRAS, BRAF, NRAS, PIK3CA, Her-2 and TP53 mutations in patients with non-small cell lung cancer. Mol. Med. Rep. 2018, 18, 2191–2197. [Google Scholar] [CrossRef]
- Melosky, B.; Kambartel, K.; Häntschel, M.; Bennetts, M.; Nickens, D.J.; Brinkmann, J.; Kayser, A.; Moran, M.; Cappuzzo, F. Worldwide Prevalence of Epidermal Growth Factor Receptor Mutations in Non-Small Cell Lung Cancer: A Meta-Analysis. Mol. Diagn. Ther. 2022, 26, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Arcila, M.E.; Yang, S.R.; Momeni, A.; Mata, D.A.; Salazar, P.; Chan, R.; Elezovic, D.; Benayed, R.; Zehir, A.; Buonocore, D.J.; et al. Ultrarapid EGFR Mutation Screening Followed by Comprehensive Next-Generation Sequencing: A Feasible, Informative Approach for Lung Carcinoma Cytology Specimens With a High Success Rate. JTO Clin. Res. Rep. 2020, 1, 100077. [Google Scholar] [CrossRef]
- Offerman, S.; Prinsen, C.F.; Knol, A.; Methorst, N.; Kamphorst, J.; Niemantsverdriet, M. Short report: Performance evaluation of the IdyllaTM KRAS and EGFR mutation tests on paraffin-embedded cytological NSCLC samples. Diagn. Pathol. 2021, 16, 70. [Google Scholar] [CrossRef]
- Krogerus, L.; Kholová, I. Cell Block in Cytological Diagnostics: Review of Preparatory Techniques. Acta Cytol. 2018, 62, 237–243. [Google Scholar] [CrossRef]
- Hsieh, P.C.; Wu, Y.K.; Huang, C.Y.; Yang, M.C.; Kuo, C.Y.; Tzeng, I.S.; Lan, C.C. Comparison of T790M Acquisition After Treatment With First- and Second-Generation Tyrosine-Kinase Inhibitors: A Systematic Review and Network Meta-Analysis. Front. Oncol. 2022, 12, 869390. [Google Scholar] [CrossRef]
- Lee, E.; Jones, V.; Topkas, E.; Harraway, J. Reduced sensitivity for EGFR T790M mutations using the Idylla EGFR Mutation Test. J. Clin. Pathol. 2021, 74, 43–47. [Google Scholar] [CrossRef]
- Finall, A.; Davies, G.; Jones, T.; Emlyn, G.; Huey, P.; Mullard, A. Integration of rapid PCR testing as an adjunct to NGS in diagnostic pathology services within the UK: Evidence from a case series of non-squamous, non-small cell lung cancer (NSCLC) patients with follow-up. J. Clin. Pathol. 2023, 76, 391–399. [Google Scholar] [CrossRef] [PubMed]
- Petiteau, C.; Robinet-Zimmermann, G.; Riot, A.; Dorbeau, M.; Richard, N.; Blanc-Fournier, C.; Bibeau, F.; Deshayes, S.; Bergot, E.; Gervais, R.; et al. Contribution of the IdyllaTM System to Improving the Therapeutic Care of Patients with NSCLC through Early Screening of EGFR Mutations. Curr. Oncol. Tor. Ont. 2021, 28, 4432–4445. [Google Scholar] [CrossRef]
- Sharma, S.; Satapathy, A.; Aggarwal, A.; Dewan, A.; Jain, E.; Katara, R.; Kumar, V.; Pal, R.; Pandey, S.; Naidu, M.M.; et al. Comparison of epidermal growth factor receptor mutation detection turnaround times and concordance among real-time polymerase chain reaction, high-throughput next-generation sequencing and the Biocartis IdyllaTM platforms in non-small cell lung carcinomas. Pathol. Res. Pract. 2021, 220, 153394. [Google Scholar] [CrossRef] [PubMed]
- Banyi, N.; Alex, D.; Hughesman, C.; McNeil, K.; NIonescu, D.; Ma, C.; Yip, S.; Melosky, B. Improving Time-to-Treatment for Advanced Non-Small Cell Lung Cancer Patients through Faster Single Gene EGFR Testing Using the IdyllaTM EGFR Testing Platform. Curr. Oncol. Tor. Ont. 2022, 29, 7900–7911. [Google Scholar] [CrossRef]
- Hofman, P.; Calabrese, F.; Kern, I.; Adam, J.; Alarcão, A.; Alborelli, I.; Anton, N.T.; Arndt, A.; Avdalyan, A.; Barberis, M.; et al. Real-world EGFR testing practices for non-small-cell lung cancer by thoracic pathology laboratories across Europe. ESMO Open 2023, 8, 101628. [Google Scholar] [CrossRef] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Aisner, D.L.; Arcila, M.E.; Beasley, M.B.; Bernicker, E.H.; Colasacco, C.; Dacic, S.; Hirsch, F.R.; Kerr, K.; et al. Updated Molecular Testing Guideline for the Selection of Lung Cancer Patients for Treatment with Targeted Tyrosine Kinase Inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. Arch. Pathol. Lab. Med. 2018, 142, 321–346. [Google Scholar] [CrossRef]
- Friedlaender, A.; Perol, M.; Banna, G.L.; Parikh, K.; Addeo, A. Oncogenic alterations in advanced NSCLC: A molecular super-highway. Biomark. Res. 2024, 12, 24. [Google Scholar] [CrossRef]
- Shen, C.I.; Chiang, C.L.; Shiao, T.H.; Luo, Y.H.; Chao, H.S.; Huang, H.C.; Chiu, C.H. Real-world evidence of the intrinsic limitations of PCR-based EGFR mutation assay in non-small cell lung cancer. Sci. Rep. 2022, 12, 13566. [Google Scholar] [CrossRef]
- Pisapia, P.; Russo, A.; De Luca, C.; Pepe, F.; Drago, F.; Rolfo, C.; Troncone, G.; Malapelle, U. The relevance of the reference range for EGFR testing in non-small cell lung cancer patients. Lung Cancer 2024, 198, 108002. [Google Scholar] [CrossRef]
- Duke, E.S.; Stapleford, L.; Drezner, N.; Amatya, A.K.; Mishra-Kalyani, P.S.; Shen, Y.L.; Maxfield, K.; Zirkelbach, J.F.; Bi, Y.; Liu, J.; et al. FDA Approval Summary: Mobocertinib for Metastatic Non-Small Cell Lung Cancer with EGFR Exon 20 Insertion Mutations. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2023, 29, 508–512. [Google Scholar] [CrossRef]
- Chon, K.; Larkins, E.; Chatterjee, S.; Mishra-Kalyani, P.S.; Aungst, S.; Wearne, E.; Subramaniam, S.; Li, Y.; Liu, J.; Sun, J.; et al. FDA Approval Summary: Amivantamab for the Treatment of patients with non-small cell lung cancer with EGFR exon 20 insertion mutations. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2023, 29, 3262–3266. [Google Scholar] [CrossRef]
- Jin, R.; Peng, L.; Shou, J.; Wang, J.; Jin, Y.; Liang, F.; Zhao, J.; Wu, M.; Li, Q.; Zhang, B.; et al. EGFR-Mutated Squamous Cell Lung Cancer and Its Association With Outcomes. Front. Oncol. 2021, 11, 680804. [Google Scholar] [CrossRef]
| Characteristic | Number (%) |
|---|---|
| Age (in years) Mean ± standard deviation Range | 69.6 ± 10.1 28–98 |
| Gender Male Female | 421 (52.4) 383 (47.6) |
| Histological type NSCLC, non-squamous | 804 (100) |
| Sample type FFPE, biopsy FFPE, resection Cytology, cell block Cytology, cytological specimens | 487 (60.6) 193 (24.0) 106 (13.2) 18 (2.2) |
| EGFR mutation status (NGS-based) Positive Negative No valid result | 89 (11.1) 713 (88.6) 2 (0.3) |
| EGFR variants (NGS-based) Exon 19 deletion p.L858R Exon 20 insertion p.G719X p.L861Q p.T790M a Total | 45 (49.4) 24 (26.4) 9 (9.9) 7 (7.7) 4 (4.4) 2 (2.2) 91 (100) |
| EasyPGX® ready EGFR assay | NGS | |||||||||
| Ex19del | p.L858R | Ex20ins | p.G719X | p.L861Q | p.T790M | Negative | Not Conclusive | Total | ||
| Ex19del | 44 | 44 | ||||||||
| p.L858R | 24 b | 24 | ||||||||
| Ex20ins | 5 | 2 | 7 | |||||||
| p.G719X | 7 | 1 | 8 | |||||||
| p.L861Q | 4 | 4 | ||||||||
| p.T790M | 2 b | 2 | ||||||||
| Negative | 4 | 672 | 676 | |||||||
| Not conclusive | 1 | 38 | 2 | 41 | ||||||
| Total | 45 | 24 | 9 | 7 | 4 | 2 | 713 | 2 | 806 b | |
| Concordance rate (%) | 97.8 | 100.0 | 55.6 | 100.0 | 100.0 | 100.0 | 94.3 | 100.0 | ||
| Discordant Sample | qPCR | NGS | DNA Input Category | Sample Type |
|---|---|---|---|---|
| p.S768_D770dup (exon 20) | negative | positive | high | Cytology, cell block |
| p.S768_D770dup (exon 20) | negative | positive | intermediate | FFPE resection |
| p.N771delinsGF (exon 20) | negative | positive | intermediate | FFPE biopsy |
| p.E746_A750del (exon 19) | negative | positive | low | FFPE biopsy |
| Exon 20 insertion | positive | negative | low | FFPE biopsy |
| Exon 20 insertion | positive | negative | intermediate | FFPE biopsy |
| p.G719X (exon 18) | positive | negative | intermediate | FFPE biopsy |
| DNA Input Category | Number (%) | Failure Rate (%) |
|---|---|---|
| High (>15 ng) | 276 (34.3) | 0 |
| Intermediate (5–15 ng) | 273 (34.0) | 0.4 |
| Low (<5 ng) | 255 (31.7) | 15.7 |
| Overall | 804 (100) | 5.1 |
| Sample Type | Number (%) | Failure Rate (%) |
|---|---|---|
| FFPE biopsy | 487 (60.6) | 5.7 |
| FFPE resection | 193 (24.0) | 0.5 |
| Cytology, cell block | 106 (13.2) | 8.4 |
| Cytology, cytological specimen | 18 (2.2) | 16.7 |
| Overall | 804 (100) | 5.1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmid, M.B.; Demmer, I.; Floriani, S.; Born, D.; Jochum, W. Real-World Performance of the EasyPGX® Ready Epidermal Growth Factor Receptor Assay for Genomic Testing of Non-Small Cell Lung Cancer Samples. Biomedicines 2025, 13, 814. https://doi.org/10.3390/biomedicines13040814
Schmid MB, Demmer I, Floriani S, Born D, Jochum W. Real-World Performance of the EasyPGX® Ready Epidermal Growth Factor Receptor Assay for Genomic Testing of Non-Small Cell Lung Cancer Samples. Biomedicines. 2025; 13(4):814. https://doi.org/10.3390/biomedicines13040814
Chicago/Turabian StyleSchmid, Michael Bento, Izadora Demmer, Sandra Floriani, Diana Born, and Wolfram Jochum. 2025. "Real-World Performance of the EasyPGX® Ready Epidermal Growth Factor Receptor Assay for Genomic Testing of Non-Small Cell Lung Cancer Samples" Biomedicines 13, no. 4: 814. https://doi.org/10.3390/biomedicines13040814
APA StyleSchmid, M. B., Demmer, I., Floriani, S., Born, D., & Jochum, W. (2025). Real-World Performance of the EasyPGX® Ready Epidermal Growth Factor Receptor Assay for Genomic Testing of Non-Small Cell Lung Cancer Samples. Biomedicines, 13(4), 814. https://doi.org/10.3390/biomedicines13040814






