Migrasome Marker Epidermal Growth Factor Domain-Specific O-GlcNAc Transferase: Pan-Cancer Angiogenesis Biomarker and the Potential Role of circ_0058189/miR-130a-3p/EOGT Axis in Hepatocellular Carcinoma Progression and Sorafenib Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Pan-Cancer Data Collection and Processing
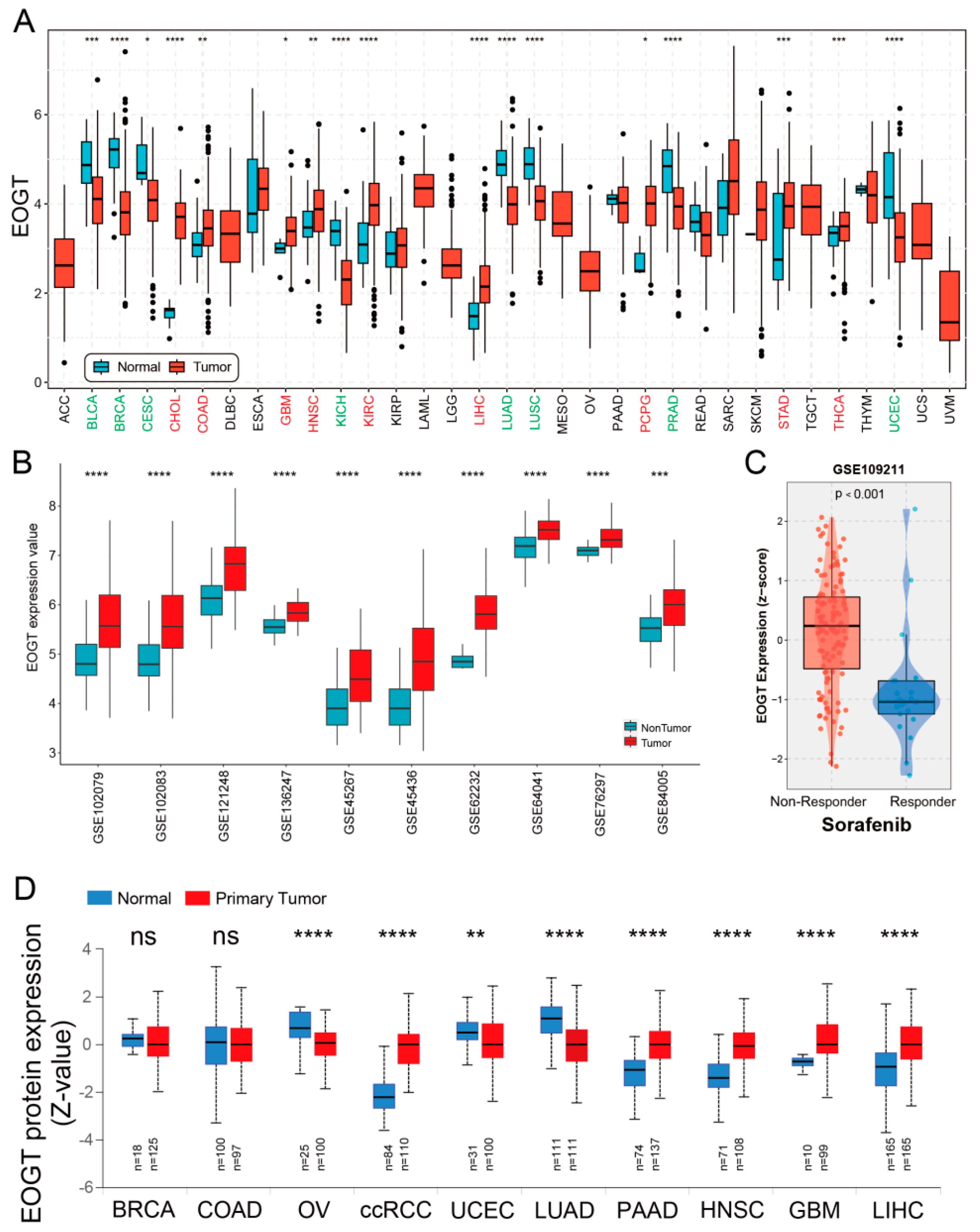
2.2. Pan-Cancer Analyses of Differential EOGT Expression
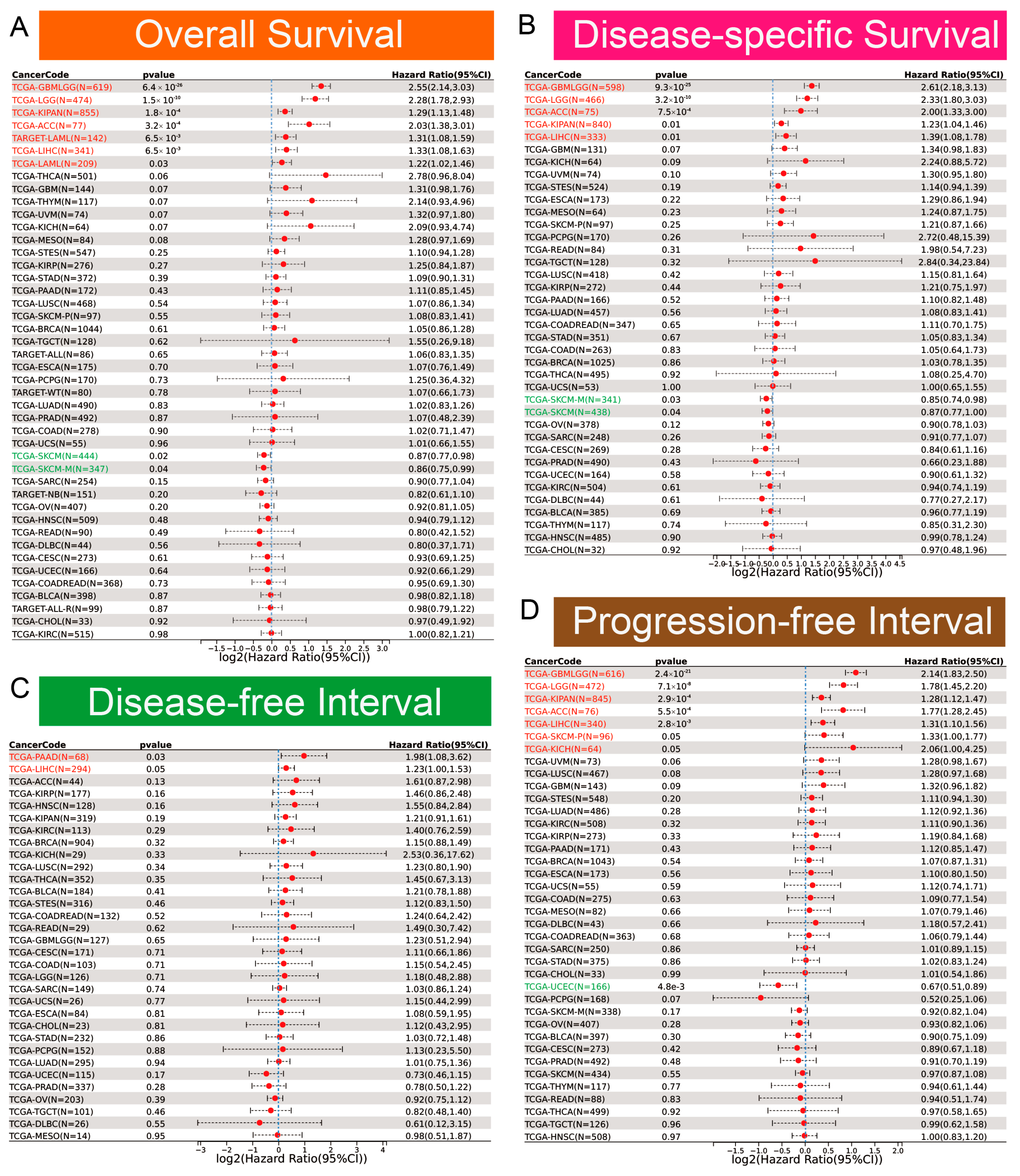
2.3. Diagnostic and Prognostic Evaluation
2.4. Genomic Alteration and Mutational Burden
2.5. DNA Repair, Stemness, and Epigenetic Analysis
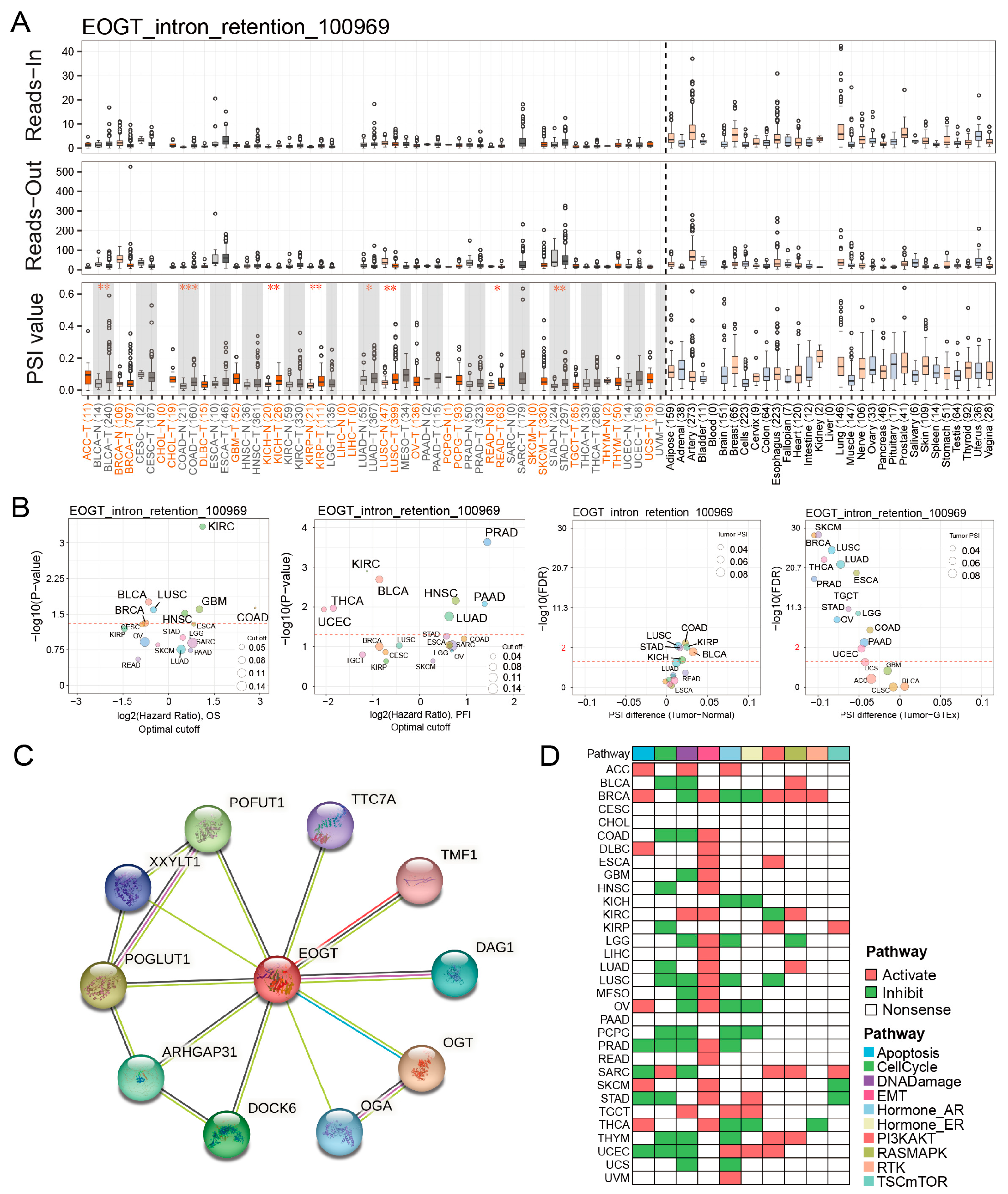
2.6. Alternative Splicing Analysis
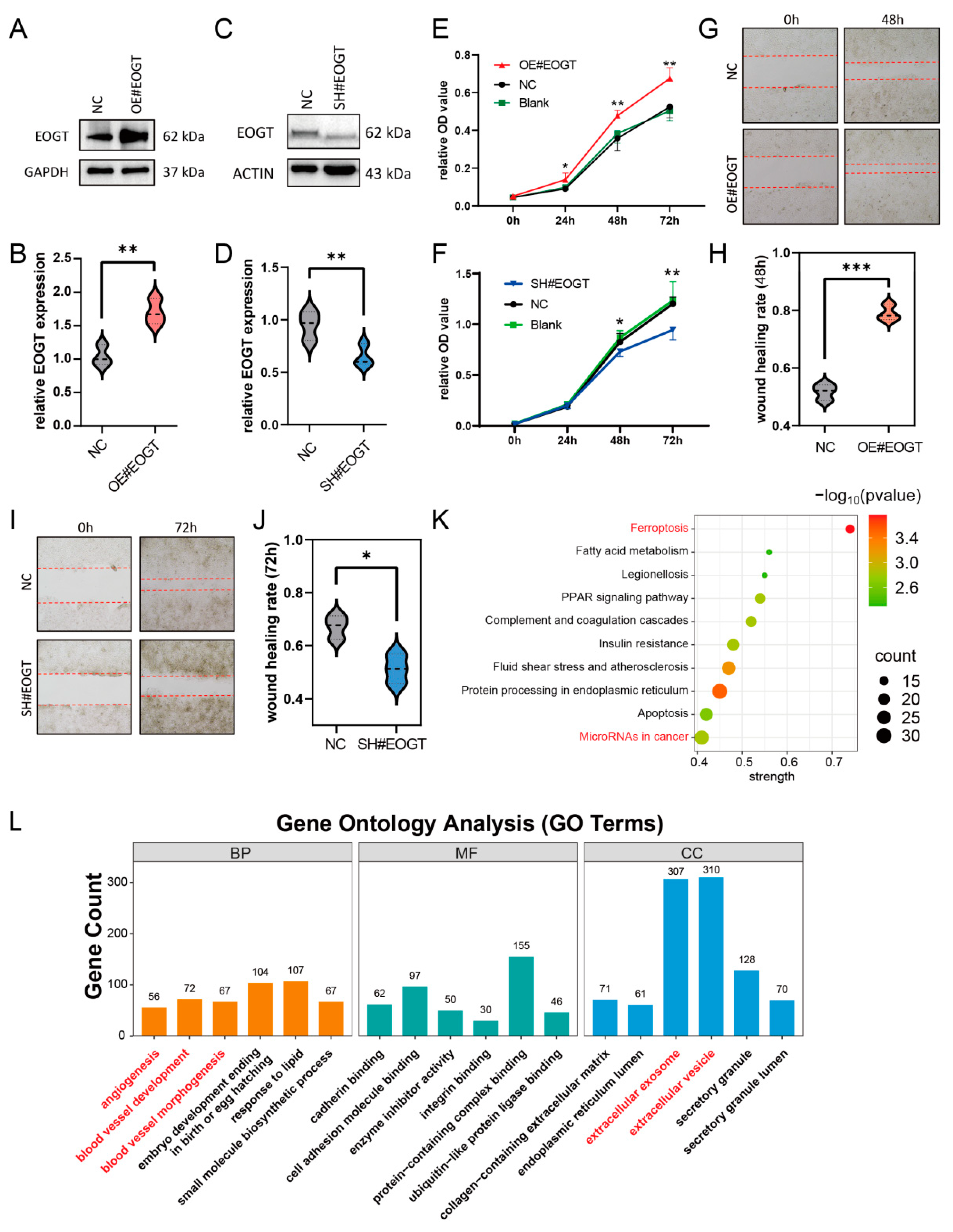
2.7. Protein Interaction and Functional Enrichment
2.8. Immune Response Analysis
2.9. Chemotherapeutic Drug Screening
2.10. Cell Line, Lentivirus Infection, and Transfection of miRNA Mimics
2.11. Western Blot Analysis
2.12. RNA-Seq and Bioinformatic Analysis
2.13. ceRNA Network Construction and Validation
2.14. Statistical Analysis
3. Results
3.1. Pan-Cancer Landscape of EOGT Expression and Clinical Relevance
3.2. Diagnostic and Prognostic Value of EOGT in Pan-Cancers
3.3. Genomic Alterations and Instability of EOGT
3.4. EOGT Expression and DNA Repair, Stemness, and Epigenetic Modifications
3.5. EOGT Alternative Splicing and Survival Outcomes
3.6. EOGT Promotes Cancer via EMT and DNA Damage Suppression
3.7. EOGT and Immune Cell Infiltration in Tumors
3.8. EOGT as a Biomarker for Tumor Angiogenesis
3.9. EOGT and Chemotherapy Sensitivity
3.10. EOGT Facilitates HCC Cell Proliferation and Migration
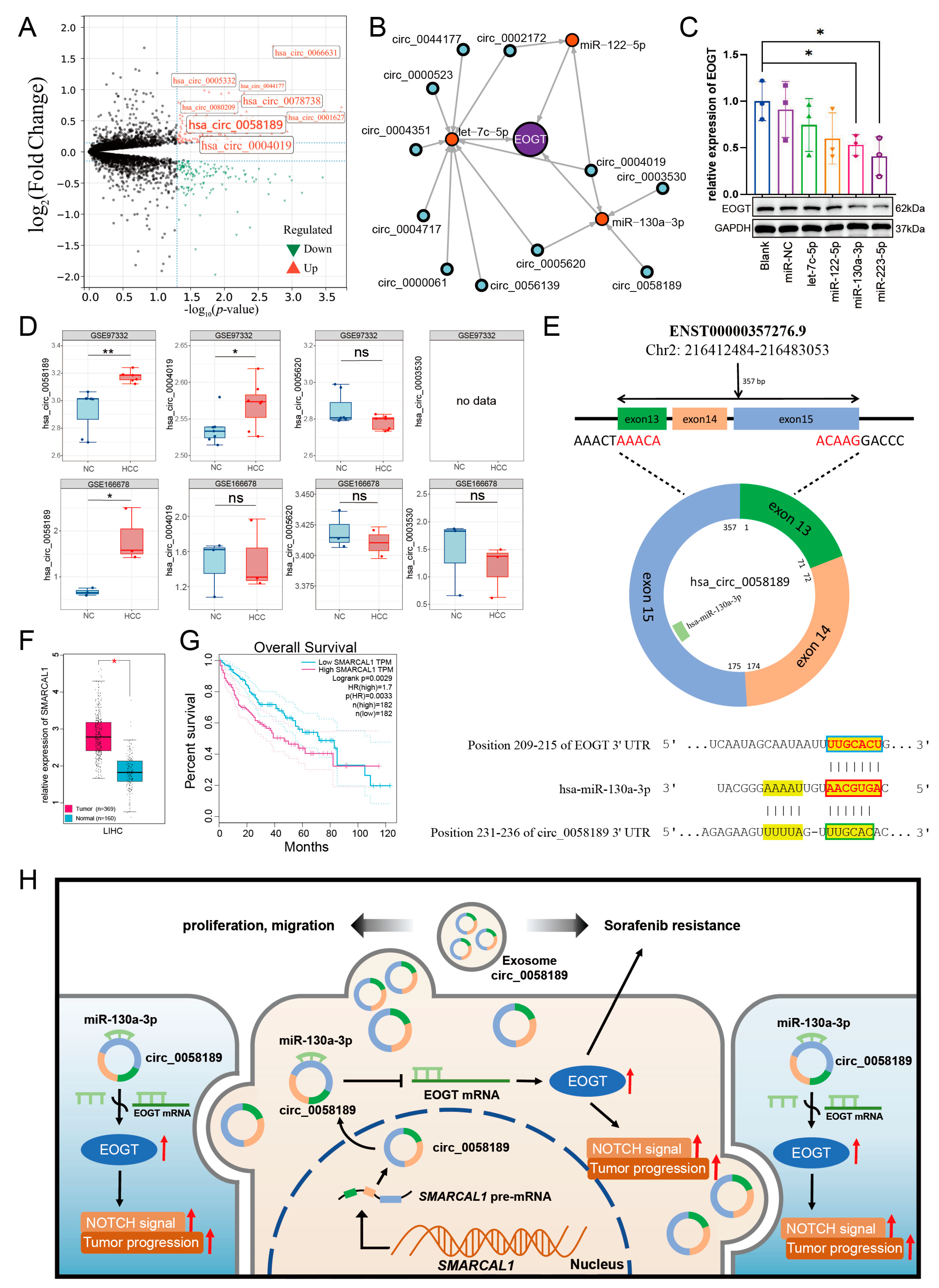
3.11. EOGT-Mediated ceRNA Network in HCC Progression and Sorafenib Resistance
3.12. Potential Critical Role of circ_0058189/miR-130a/EOGT Axis in HCC Progression and Sorafenib Resistance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- He, X.-F.; Hu, X.; Wen, G.-J.; Wang, Z.; Lin, W.-J. O-GlcNAcylation in cancer development and immunotherapy. Cancer Lett. 2023, 566, 216258. [Google Scholar] [CrossRef]
- Zhang, X.; Qiao, Y.; Wu, Q.; Chen, Y.; Zou, S.; Liu, X.; Zhu, G.; Zhao, Y.; Chen, Y.; Yu, Y.; et al. The essential role of YAP O-GlcNAcylation in high-glucose-stimulated liver tumorigenesis. Nat. Commun. 2017, 8, 15280. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Qian, M.; Lu, L.; Chen, Y.; Zhang, X.; Wu, Q.; Liu, Y.; Bian, Z.; Yang, Y.; Guo, S.; et al. O-GlcNAcylation of YY1 stimulates tumorigenesis in colorectal cancer cells by targeting SLC22A15 and AANAT. Carcinogenesis 2019, 40, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Shang, M.; Yang, H.; Yang, R.; Chen, T.; Fu, Y.; Li, Y.; Fang, X.; Zhang, K.; Zhang, J.; Li, H.; et al. The folate cycle enzyme MTHFD2 induces cancer immune evasion through PD-L1 up-regulation. Nat. Commun. 2021, 12, 1940. [Google Scholar] [CrossRef] [PubMed]
- Hart, G.W. Nutrient regulation of signaling and transcription. J. Biol. Chem. 2019, 294, 2211–2231. [Google Scholar] [CrossRef]
- Lubas, W.A.; Frank, D.W.; Krause, M.; Hanover, J.A. O-Linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9316–9324. [Google Scholar] [CrossRef]
- Kreppel, L.K.; Blomberg, M.A.; Hart, G.W. Dynamic glycosylation of nuclear and cytosolic proteins. Cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 1997, 272, 9308–9315. [Google Scholar] [CrossRef]
- Sawaguchi, S.; Varshney, S.; Ogawa, M.; Sakaidani, Y.; Yagi, H.; Takeshita, K.; Murohara, T.; Kato, K.; Sundaram, S.; Stanley, P.; et al. O-GlcNAc on NOTCH1 EGF repeats regulates ligand-induced Notch signaling and vascular development in mammals. eLife 2017, 6, e24419. [Google Scholar] [CrossRef]
- Zhao, X.; Lei, Y.; Zheng, J.; Peng, J.; Li, Y.; Yu, L.; Chen, Y. Identification of markers for migrasome detection. Cell Discov. 2019, 5, 27. [Google Scholar] [CrossRef]
- Zhang, K.; Zhu, Z.; Jia, R.; Wang, N.A.; Shi, M.; Wang, Y.; Xiang, S.; Zhang, Q.; Xu, L. CD151-enriched migrasomes mediate hepatocellular carcinoma invasion by conditioning cancer cells and promoting angiogenesis. J. Exp. Clin. Cancer Res. CR 2024, 43, 160. [Google Scholar] [CrossRef]
- Yang, C.; Hu, J.-F.; Zhan, Q.; Wang, Z.-W.; Li, G.; Pan, J.-J.; Huang, L.; Liao, C.-Y.; Huang, Y.; Tian, Y.-F.; et al. SHCBP1 interacting with EOGT enhances O-GlcNAcylation of NOTCH1 and promotes the development of pancreatic cancer. Genomics 2021, 113, 827–842. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; He, L.; Gao, M.; Xiao, F.; Yang, J.; Wang, S.; Wei, H.; Zhang, F.; Wei, H. EOGT Correlated With Immune Infiltration: A Candidate Prognostic Biomarker for Hepatocellular Carcinoma. Front. Immunol. 2021, 12, 780509. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- Chandrashekar, D.S.; Karthikeyan, S.K.; Korla, P.K.; Patel, H.; Shovon, A.R.; Athar, M.; Netto, G.J.; Qin, Z.S.; Kumar, S.; Manne, U.; et al. UALCAN: An update to the integrated cancer data analysis platform. Neoplasia 2022, 25, 18–27. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Beroukhim, R.; Mermel, C.H.; Porter, D.; Wei, G.; Raychaudhuri, S.; Donovan, J.; Barretina, J.; Boehm, J.S.; Dobson, J.; Urashima, M.; et al. The landscape of somatic copy-number alteration across human cancers. Nature 2010, 463, 899–905. [Google Scholar] [CrossRef]
- Mayakonda, A.; Lin, D.-C.; Assenov, Y.; Plass, C.; Koeffler, H.P. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018, 28, 1747–1756. [Google Scholar] [CrossRef]
- Latham, A.; Srinivasan, P.; Kemel, Y.; Shia, J.; Bandlamudi, C.; Mandelker, D.; Middha, S.; Hechtman, J.; Zehir, A.; Dubard-Gault, M.; et al. Microsatellite Instability Is Associated with the Presence of Lynch Syndrome Pan-Cancer. J. Clin. Oncol. 2019, 37, 286–295. [Google Scholar] [CrossRef]
- Coleman, R.L.; Oza, A.M.; Lorusso, D.; Aghajanian, C.; Oaknin, A.; Dean, A.; Colombo, N.; Weberpals, J.I.; Clamp, A.; Scambia, G.; et al. ARIEL3 investigators, Rucaparib maintenance treatment for recurrent ovarian carcinoma after response to platinum therapy (ARIEL3): A randomised, double-blind, placebo-controlled, phase 3 trial. Lancet 2017, 390, 1949–1961. [Google Scholar] [CrossRef]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef] [PubMed]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kamińska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354.e15. [Google Scholar] [CrossRef] [PubMed]
- Lyko, F. The DNA methyltransferase family: A versatile toolkit for epigenetic regulation. Nat. Rev. Genet. 2018, 19, 81–92. [Google Scholar] [CrossRef]
- Shi, H.; Chai, P.; Jia, R.; Fan, X. Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol. Cancer 2020, 19, 78. [Google Scholar] [CrossRef]
- Zhang, Y.; Yao, X.; Zhou, H.; Wu, X.; Tian, J.; Zeng, J.; Yan, L.; Duan, C.; Liu, H.; Li, H.; et al. OncoSplicing: An updated database for clinically relevant alternative splicing in 33 human cancers. Nucleic Acids Res. 2022, 50, D1340–D1347. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Liu, C.-J.; Hu, F.-F.; Xie, G.-Y.; Miao, Y.-R.; Li, X.-W.; Zeng, Y.; Guo, A.-Y. GSCA: An integrated platform for gene set cancer analysis at genomic, pharmacogenomic and immunogenomic levels. Brief. Bioinf. 2023, 24, bbac558. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef]
- Ru, B.; Wong, C.N.; Tong, Y.; Zhong, J.Y.; Zhong, S.S.W.; Wu, W.C.; Chu, K.C.; Wong, C.Y.; Lau, C.Y.; Chen, I.; et al. TISIDB: An integrated repository portal for tumor-immune system interactions. Bioinformatics 2019, 35, 4200–4202. [Google Scholar] [CrossRef] [PubMed]
- Kalbasi, A.; Ribas, A. Tumour-intrinsic resistance to immune checkpoint blockade. Nat. Rev. Immunol. 2020, 20, 25–39. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Wong, C.J.; Yang, L.; Ouardaoui, N.; Li, D.; Zhang, W.; Gu, S.; Zhang, Y.; Liu, Y.; Wang, X.; et al. TISMO: Syngeneic mouse tumor database to model tumor immunity and immunotherapy response. Nucleic Acids Res. 2022, 50, D1391–D1397. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef]
- Becht, E.; Giraldo, N.A.; Lacroix, L.; Buttard, B.; Elarouci, N.; Petitprez, F.; Selves, J.; Laurent-Puig, P.; Sautès-Fridman, C.; Fridman, W.H.; et al. Erratum to: Estimating the population abundance of tissue-infiltrating immune and stromal cell populations using gene expression. Genome Biol. 2016, 17, 249. [Google Scholar] [CrossRef]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Li, T.; Fu, J.; Zeng, Z.; Cohen, D.; Li, J.; Chen, Q.; Li, B.; Liu, X.S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020, 48, W509–W514. [Google Scholar] [CrossRef]
- Yuan, H.; Yan, M.; Zhang, G.; Liu, W.; Deng, C.; Liao, G.; Xu, L.; Luo, T.; Yan, H.; Long, Z.; et al. CancerSEA: A cancer single-cell state atlas. Nucleic Acids Res. 2019, 47, D900–D908. [Google Scholar] [CrossRef]
- Yang, C.; Chen, M.; Wang, S.; Qian, R.; Huang, X.; Wang, J.; Liu, Z.; Qin, W.; Wang, C.; Hang, H.; et al. A survey of optimal strategy for signature-based drug repositioning and an application to liver cancer. eLife 2022, 11, e71880. [Google Scholar] [CrossRef]
- Shen, W.; Song, Z.; Zhong, X.; Huang, M.; Shen, D.; Gao, P.; Qian, X.; Wang, M.; He, X.; Wang, T.; et al. Sangerbox: A comprehensive, interaction-friendly clinical bioinformatics analysis platform. iMeta 2022, 1, e36. [Google Scholar] [CrossRef]
- DSzklarczyk Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; Bork, P. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, L.; Weng, S.; Xu, H.; Xing, Z.; Ren, Y.; Ge, X.; Wang, L.; Guo, C.; Li, L.; et al. BEST: A web application for comprehensive biomarker exploration on large-scale data in solid tumors. J. Big Data 2023, 10, 165. [Google Scholar] [CrossRef]
- Zhang, Q.; Hu, W.; Xiong, L.; Wen, J.; Wei, T.; Yan, L.; Liu, Q.; Zhu, S.; Bai, Y.; Zeng, Y.; et al. IHGA: An interactive web server for large-scale and comprehensive discovery of genes of interest in hepatocellular carcinoma. Comput. Struct. Biotechnol. J. 2023, 21, 3987–3998. [Google Scholar] [CrossRef]
- Liu, Y.; Xun, Z.; Ma, K.; Liang, S.; Li, X.; Zhou, S.; Sun, L.; Liu, Y.; Du, Y.; Guo, X.; et al. Identification of a tumour immune barrier in the HCC microenvironment that determines the efficacy of immunotherapy. J. Hepatol. 2023, 78, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Amon, A. Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 2020, 21, 44–62. [Google Scholar] [CrossRef]
- Qu, X.; Li, H.; Braziel, R.M.; Passerini, V.; Rimsza, L.M.; Hsi, E.D.; Leonard, J.P.; Smith, S.M.; Kridel, R.; Press, O.; et al. Genomic alterations important for the prognosis in patients with follicular lymphoma treated in SWOG study S0016. Blood 2019, 133, 81–93. [Google Scholar] [CrossRef]
- Yu, F.; Zhang, Q.; Liu, H.; Liu, J.; Yang, S.; Luo, X.; Liu, W.; Zheng, H.; Liu, Q.; Cui, Y.; et al. Dynamic O-GlcNAcylation coordinates ferritinophagy and mitophagy to activate ferroptosis. Cell Discov. 2022, 8, 40. [Google Scholar] [CrossRef]
- Schröder, K.C.; Duman, D.; Tekin, M.; Schanze, D.; Sukalo, M.; Meester, J.; Wuyts, W.; Zenker, M. Adams-Oliver syndrome caused by mutations of the EOGT gene. Am. J. Med. Genet. A 2019, 179, 2246–2251. [Google Scholar] [CrossRef]
- Hao, X.; Li, Y.; Wang, J.; Ma, J.; Zhao, S.; Ye, X.; He, L.; Yang, J.; Gao, M.; Xiao, F.; et al. Deficient O-GlcNAc Glycosylation Impairs Regulatory T Cell Differentiation and Notch Signaling in Autoimmune Hepatitis. Front. Immunol. 2018, 9, 2089. [Google Scholar] [CrossRef]
- Deng, S.; Wu, Y.; Huang, S.; Yang, X. Novel insights into the roles of migrasome in cancer. Discov. Oncol. 2024, 15, 166. [Google Scholar] [CrossRef]
- Wanna-Udom, S.; Terashima, M.; Lyu, H.; Ishimura, A.; Takino, T.; Sakari, M.; Tsukahara, T.; Suzuki, T. The m6A methyltransferase METTL3 contributes to transforming growth factor-beta-induced epithelial-mesenchymal transition of lung cancer cells through the regulation of JUNB. Biochem. Biophys. Res. Commun. 2020, 524, 150–155. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhu, Z.; Sheng, H.; Sun, J.; Cao, C. TRIM21 suppresses invasion of hepatocellular carcinoma cells by promoting β-catenin ubiquitylation and degradation. J. South. Med. Univ. 2022, 42, 55–62. [Google Scholar] [CrossRef]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.; Fan, T.; Xiao, C.; Tian, H.; Zheng, Y.; Li, C.; He, J. TGF-β signaling in health, disease, and therapeutics. Signal Transduct. Target. Ther. 2024, 9, 61. [Google Scholar] [CrossRef]
- Wang, X.; Eichhorn, P.J.A.; Thiery, J.P. TGF-β, EMT, and resistance to anti-cancer treatment. Semin. Cancer Biol. 2023, 97, 1–11. [Google Scholar] [CrossRef]
- Watabe, T.; Takahashi, K.; Pietras, K.; Yoshimatsu, Y. Roles of TGF-β signals in tumor microenvironment via regulation of the formation and plasticity of vascular system. Semin. Cancer Biol. 2023, 92, 130–138. [Google Scholar] [CrossRef]
- Moreau, J.M.; Velegraki, M.; Bolyard, C.; Rosenblum, M.D.; Li, Z. Transforming growth factor-β1 in regulatory T cell biology. Sci. Immunol. 2022, 7, eabi4613. [Google Scholar] [CrossRef]
- Sakano, Y.; Noda, T.; Kobayashi, S.; Sasaki, K.; Iwagami, Y.; Yamada, D.; Tomimaru, Y.; Akita, H.; Gotoh, K.; Takahashi, H.; et al. Tumor endothelial cell-induced CD8+ T-cell exhaustion via GPNMB in hepatocellular carcinoma. Cancer Sci. 2022, 113, 1625–1638. [Google Scholar] [CrossRef]
- Ladd, A.D.; Duarte, S.; Sahin, I.; Zarrinpar, A. Mechanisms of drug resistance in HCC. Hepatology 2024, 79, 926–940. [Google Scholar] [CrossRef]
- Conn, V.M.; Chinnaiyan, A.M.; Conn, S.J. Circular RNA in cancer. Nat. Rev. Cancer 2024, 24, 597–613. [Google Scholar] [CrossRef]
- Guo, X.; Gao, C.; Yang, D.-H.; Li, S. Exosomal circular RNAs: A chief culprit in cancer chemotherapy resistance. Drug Resist. Updates Rev. Comment. Antimicrob. Anticancer Chemother. 2023, 67, 100937. [Google Scholar] [CrossRef]
- Yuan, P.; Song, J.; Wang, F.; Chen, B. Exosome-transmitted circ_002136 promotes hepatocellular carcinoma progression by miR-19a-3p/RAB1A pathway. BMC Cancer 2022, 22, 1284. [Google Scholar] [CrossRef]
- Zhang, P.-F.; Gao, C.; Huang, X.-Y.; Lu, J.-C.; Guo, X.-J.; Shi, G.-M.; Cai, J.-B.; Ke, A.-W. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol. Cancer 2020, 19, 110. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Wang, R.; Qin, S.; Liu, J.; Su, F.; Yang, Y.; Zhao, F.; Wang, Z.; Wu, Q. MiR-130a-3p regulates cell migration and invasion via inhibition of Smad4 in gemcitabine resistant hepatoma cells. J. Exp. Clin. Cancer Res. CR 2016, 35, 19. [Google Scholar] [CrossRef]
- Zhu, J.; Luo, Y.; Zhao, Y.; Kong, Y.; Zheng, H.; Li, Y.; Gao, B.; Ai, L.; Huang, H.; Huang, J.; et al. circEHBP1 promotes lymphangiogenesis and lymphatic metastasis of bladder cancer via miR-130a-3p/TGFβR1/VEGF-D signaling. Mol. Ther. 2021, 29, 1838–1852. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Fu, Q.; Jing, C.; Zhang, X.; Qin, T.; Pan, Y. LncRNA HOTAIR knockdown inhibits glycolysis by regulating miR-130a-3p/HIF1A in hepatocellular carcinoma under hypoxia. Biomed. Pharmacother. 2020, 125, 109703. [Google Scholar] [CrossRef]
- Song, G.-L.; Xiao, M.; Wan, X.-Y.; Deng, J.; Ling, J.-D.; Tian, Y.-G.; Li, M.; Yin, J.; Zheng, R.-Y.; Tang, Y.; et al. MiR-130a-3p suppresses colorectal cancer growth by targeting Wnt Family Member 1 (WNT1). Bioengineered 2021, 12, 8407–8418. [Google Scholar] [CrossRef]
- Poodineh, J.; Sirati-Sabet, M.; Rajabibazl, M.; Mohammadi-Yeganeh, S. MiR-130a-3p blocks wnt signaling cascade in the triple-negative breast cancer by targeting the key players at multiple points. Heliyon 2020, 6, e05434. [Google Scholar] [CrossRef]
- Wang, G.; Popovic, B.; Tao, J.; Jiang, A. Overexpression of COX7RP promotes tumor growth and metastasis by inducing ROS production in hepatocellular carcinoma cells. Am. J. Cancer Res. 2020, 10, 1366–1383. [Google Scholar]



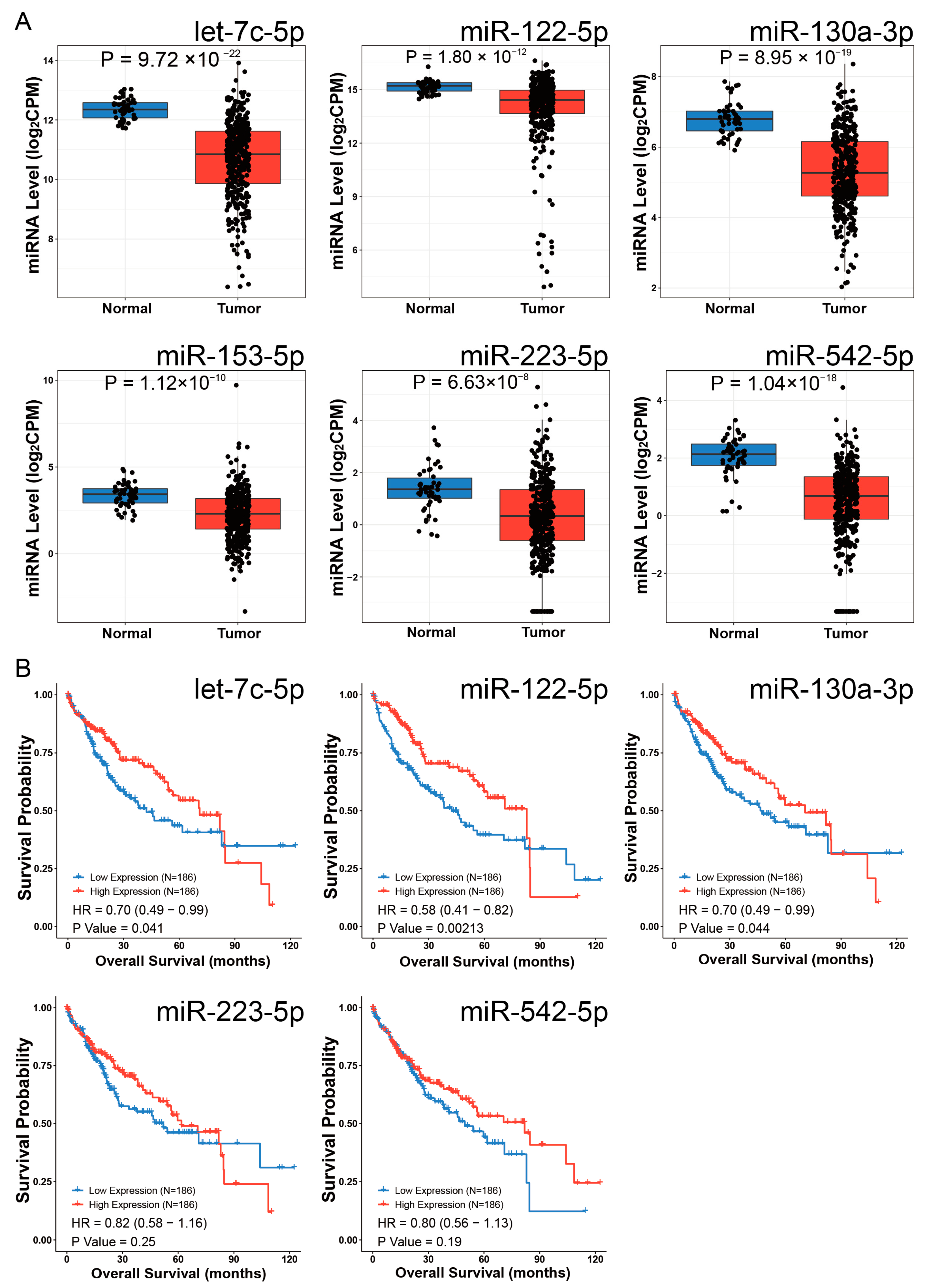
| miRNA ID | Source ID | Design | logFC | Experiment ID |
| hsa-let-7c-5p | GSE115016 | cancer vs. normal | −0.8 | EXP00501 |
| SRP049590 | cancer vs. normal | −0.69 | EXP00719 | |
| hsa-miR-122-5p | GSE147889 | cancer vs. normal | −0.69 | EXP00576 |
| hsa-miR-130a-3p | GSE147889 | cancer vs. normal | −1.23 | EXP00576 |
| E_MTAB_4170 | cancer vs. normal | −1.3 | EXP00627 | |
| SRP049590 | cancer vs. normal | −0.98 | EXP00719 | |
| hsa-miR-223-5p | SRP049590 | cancer vs. normal | −1.1 | EXP00719 |
| hsa-miR-542-5p | GSE21362 | cancer vs. normal | −1.2 | EXP00117 |
| GSE40744 | cancer vs. normal | −0.81 | EXP00213 | |
| GSE36915 | cancer vs. normal | −0.68 | EXP00221 | |
| GSE147889 | cancer vs. normal | −0.81 | EXP00576 |
| miRNA | circRNA |
| hsa-let-7c-5p | hsa_circ_0044177 |
| hsa_circ_0005620 | |
| hsa_circ_0002172 | |
| hsa_circ_0000523 | |
| hsa_circ_0004351 | |
| hsa_circ_0004717 | |
| hsa_circ_0000061 | |
| hsa_circ_0056139 | |
| hsa_circ_0004019 | |
| hsa-miR-122-5p | hsa_circ_0002172 |
| hsa_circ_0004019 | |
| hsa-miR-130a-3p | hsa_circ_0005620 |
| hsa_circ_0058189 | |
| hsa_circ_0004019 | |
| hsa_circ_0003530 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, Z.; Luo, J.; An, W.; Wei, H.; Li, M.; He, L.; Xiao, F.; Wei, H. Migrasome Marker Epidermal Growth Factor Domain-Specific O-GlcNAc Transferase: Pan-Cancer Angiogenesis Biomarker and the Potential Role of circ_0058189/miR-130a-3p/EOGT Axis in Hepatocellular Carcinoma Progression and Sorafenib Resistance. Biomedicines 2025, 13, 773. https://doi.org/10.3390/biomedicines13040773
Yu Z, Luo J, An W, Wei H, Li M, He L, Xiao F, Wei H. Migrasome Marker Epidermal Growth Factor Domain-Specific O-GlcNAc Transferase: Pan-Cancer Angiogenesis Biomarker and the Potential Role of circ_0058189/miR-130a-3p/EOGT Axis in Hepatocellular Carcinoma Progression and Sorafenib Resistance. Biomedicines. 2025; 13(4):773. https://doi.org/10.3390/biomedicines13040773
Chicago/Turabian StyleYu, Zhe, Jing Luo, Wen An, Herui Wei, Mengqi Li, Lingling He, Fan Xiao, and Hongshan Wei. 2025. "Migrasome Marker Epidermal Growth Factor Domain-Specific O-GlcNAc Transferase: Pan-Cancer Angiogenesis Biomarker and the Potential Role of circ_0058189/miR-130a-3p/EOGT Axis in Hepatocellular Carcinoma Progression and Sorafenib Resistance" Biomedicines 13, no. 4: 773. https://doi.org/10.3390/biomedicines13040773
APA StyleYu, Z., Luo, J., An, W., Wei, H., Li, M., He, L., Xiao, F., & Wei, H. (2025). Migrasome Marker Epidermal Growth Factor Domain-Specific O-GlcNAc Transferase: Pan-Cancer Angiogenesis Biomarker and the Potential Role of circ_0058189/miR-130a-3p/EOGT Axis in Hepatocellular Carcinoma Progression and Sorafenib Resistance. Biomedicines, 13(4), 773. https://doi.org/10.3390/biomedicines13040773





