Dasatinib Inhibits Basal B Breast Cancer Through ETS1-Mediated Extracellular Matrix Remodeling
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Viability Assay
2.3. Incucyte 96-Well Scratch Wound-Healing Assays
2.4. Transwell Assay
2.5. Colony Formation Assay
2.6. Plasmids and Lentivirus Infection
2.7. Western Blot
2.8. Immunofluorescence Staining
2.9. Quantitative Real-Time PCR
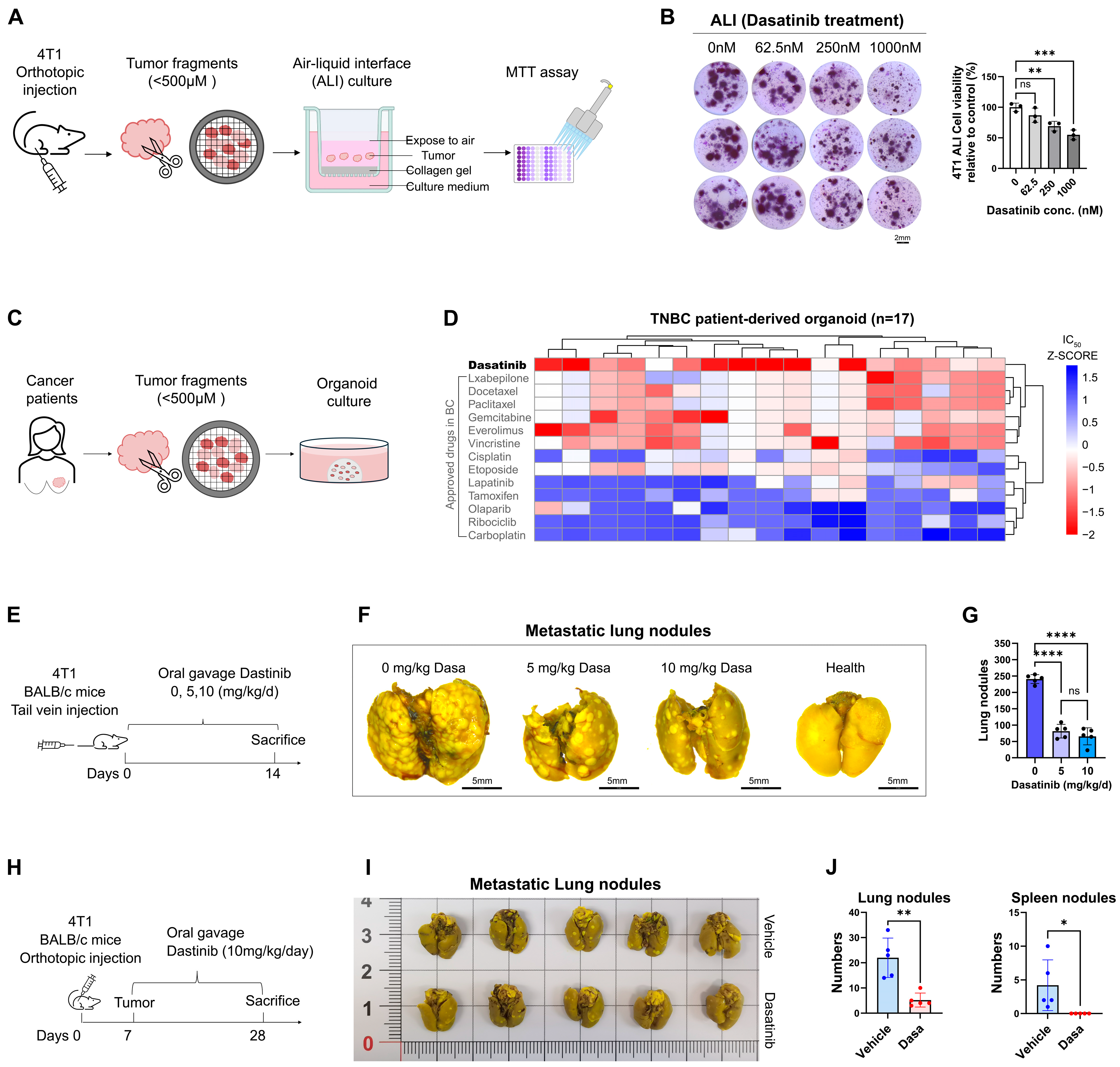
2.10. Animal Experiments
2.11. Three-Dimensional Ex Vivo Air–Liquid Interface (ALI) Culture
2.12. Public Data Collection and Processing
2.13. Sample Preparation and RNA-Seq Process
2.14. RNA-Seq Data Preprocessing and Analysis
2.15. Statistical Analysis
3. Results
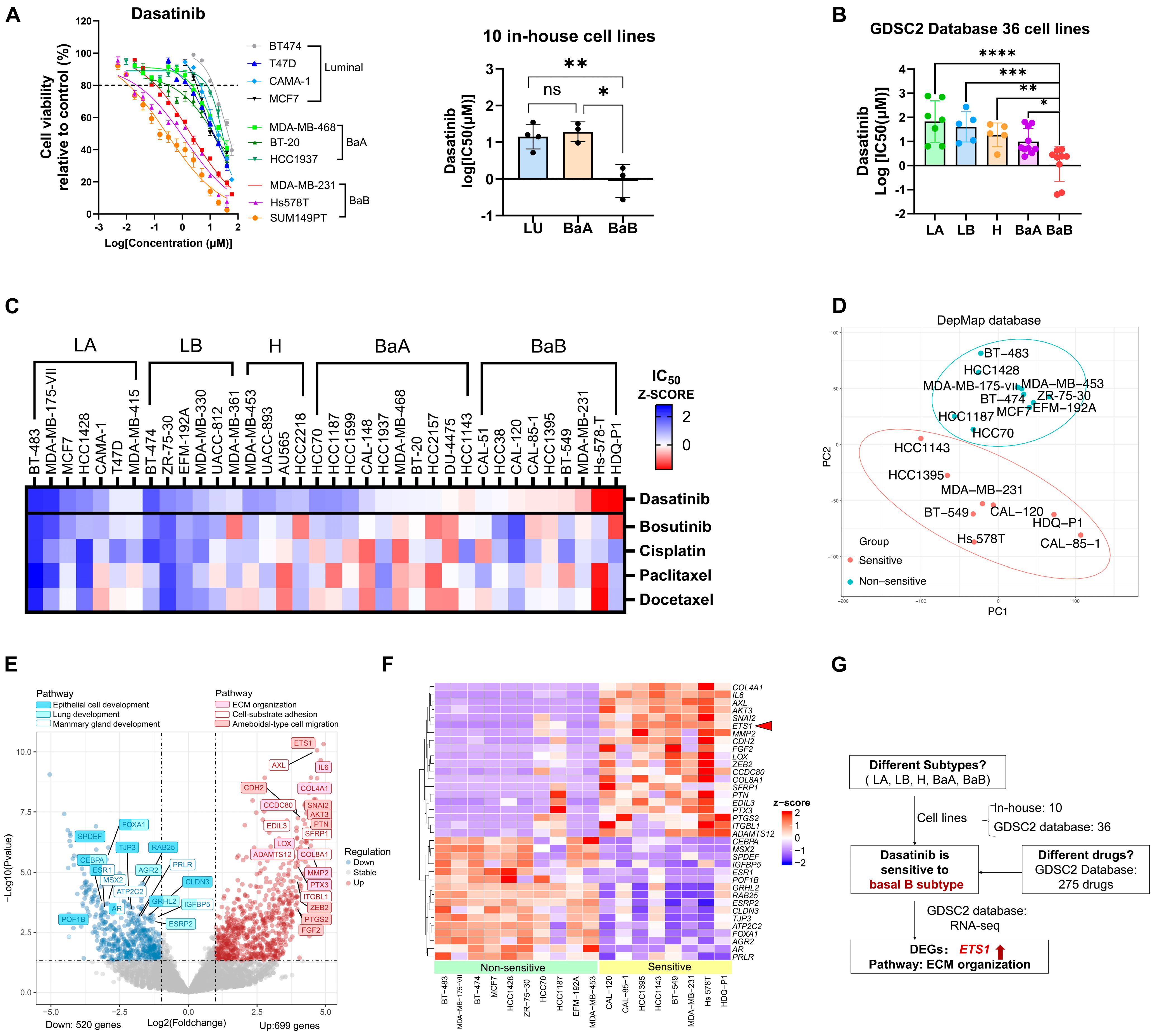
3.1. Large-Scale Drug Screening Identifies Dasatinib as an Inhibitor for Breast Cancer Cell Migration
3.2. Dasatinib Hinders Breast Cancer Cell Survival and Metastasis
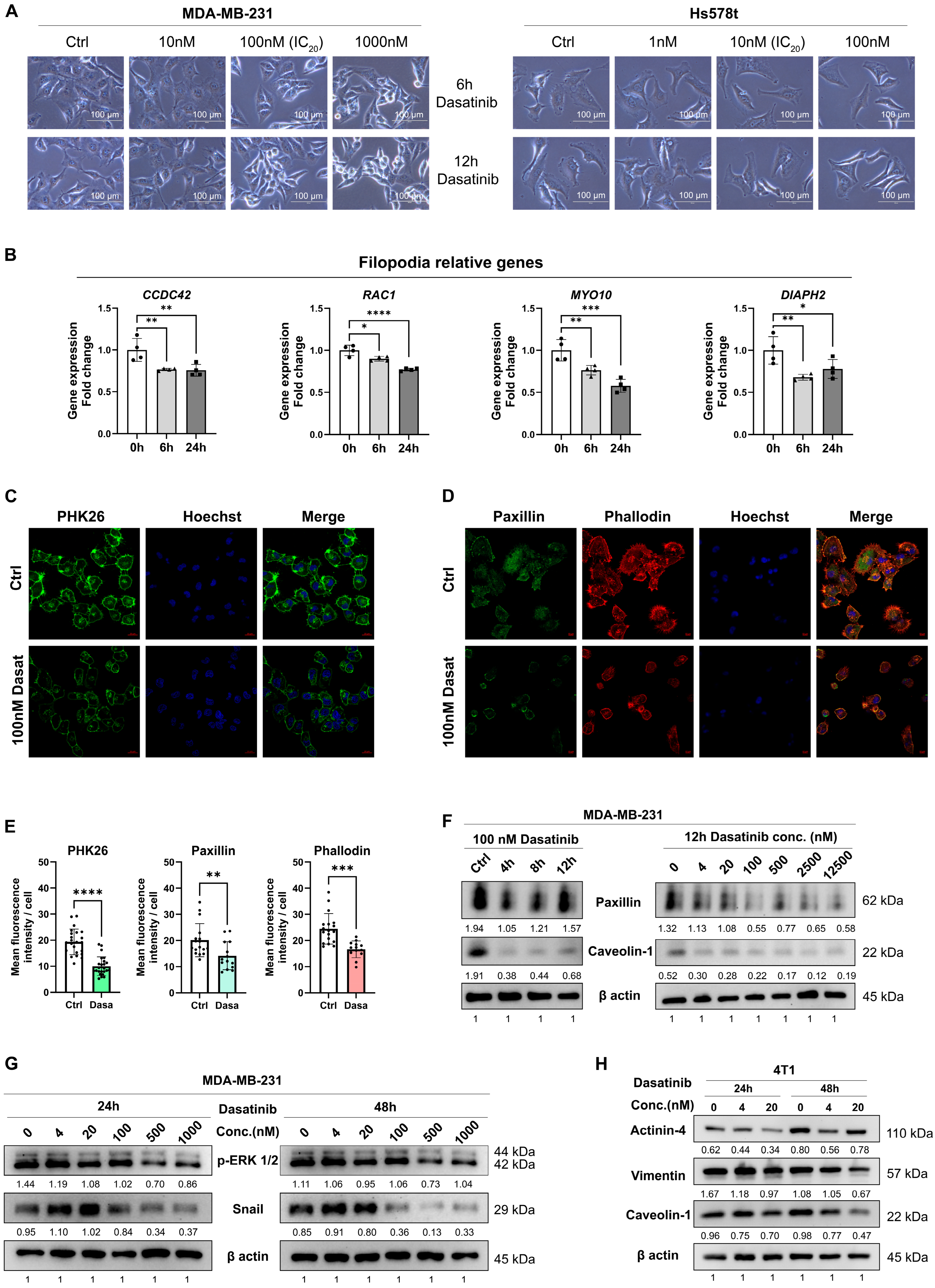
3.3. Dasatinib Prevents Metastasis by Disrupting the Actin Cytoskeleton
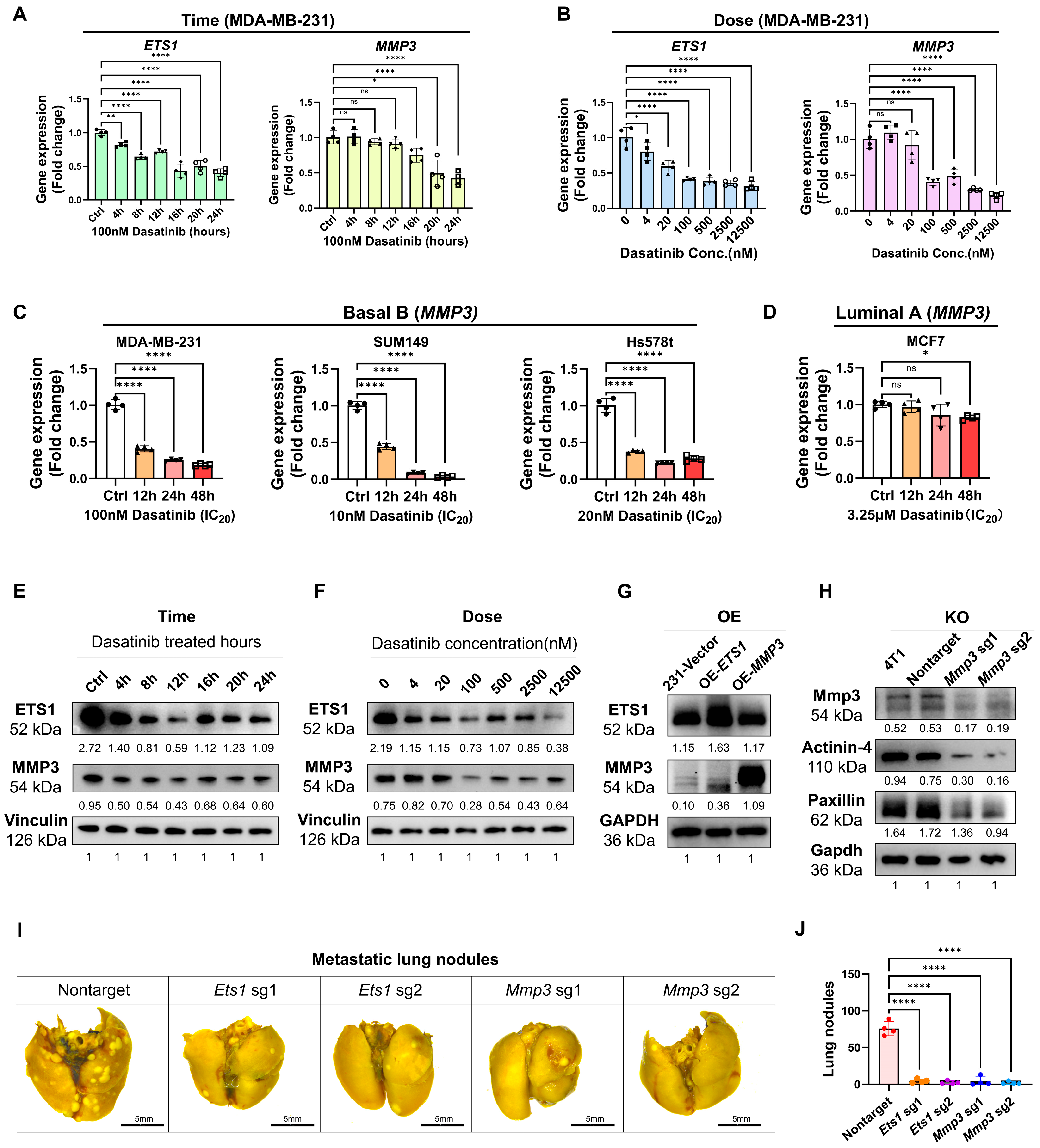
3.4. Dasatinib Exhibits High Sensitivity in the Basal B Breast Cancer with High ETS1 Expression
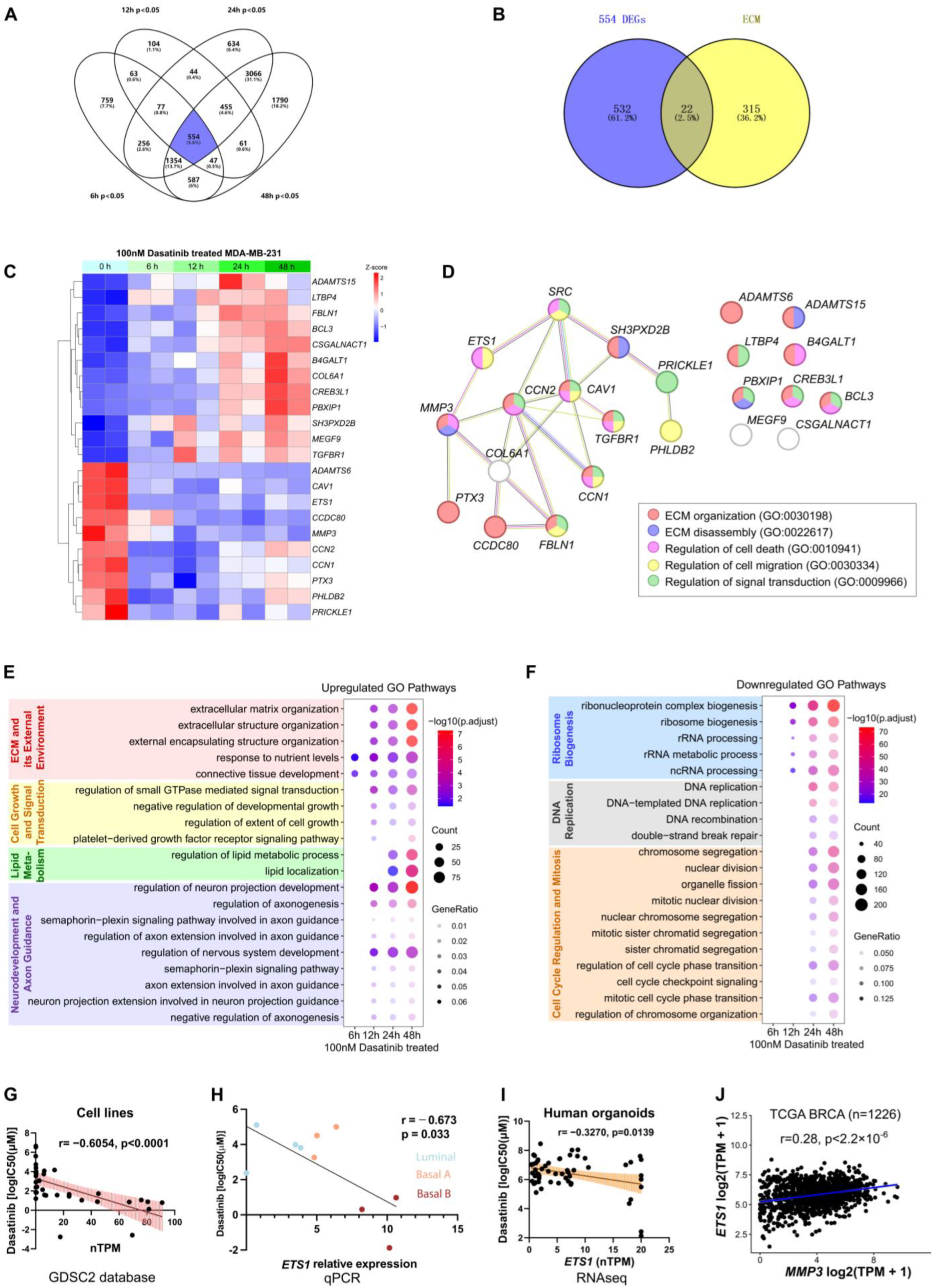
3.5. Dasatinib Modulates ECM Organization and Suppresses ETS1 and MMP3 Expression
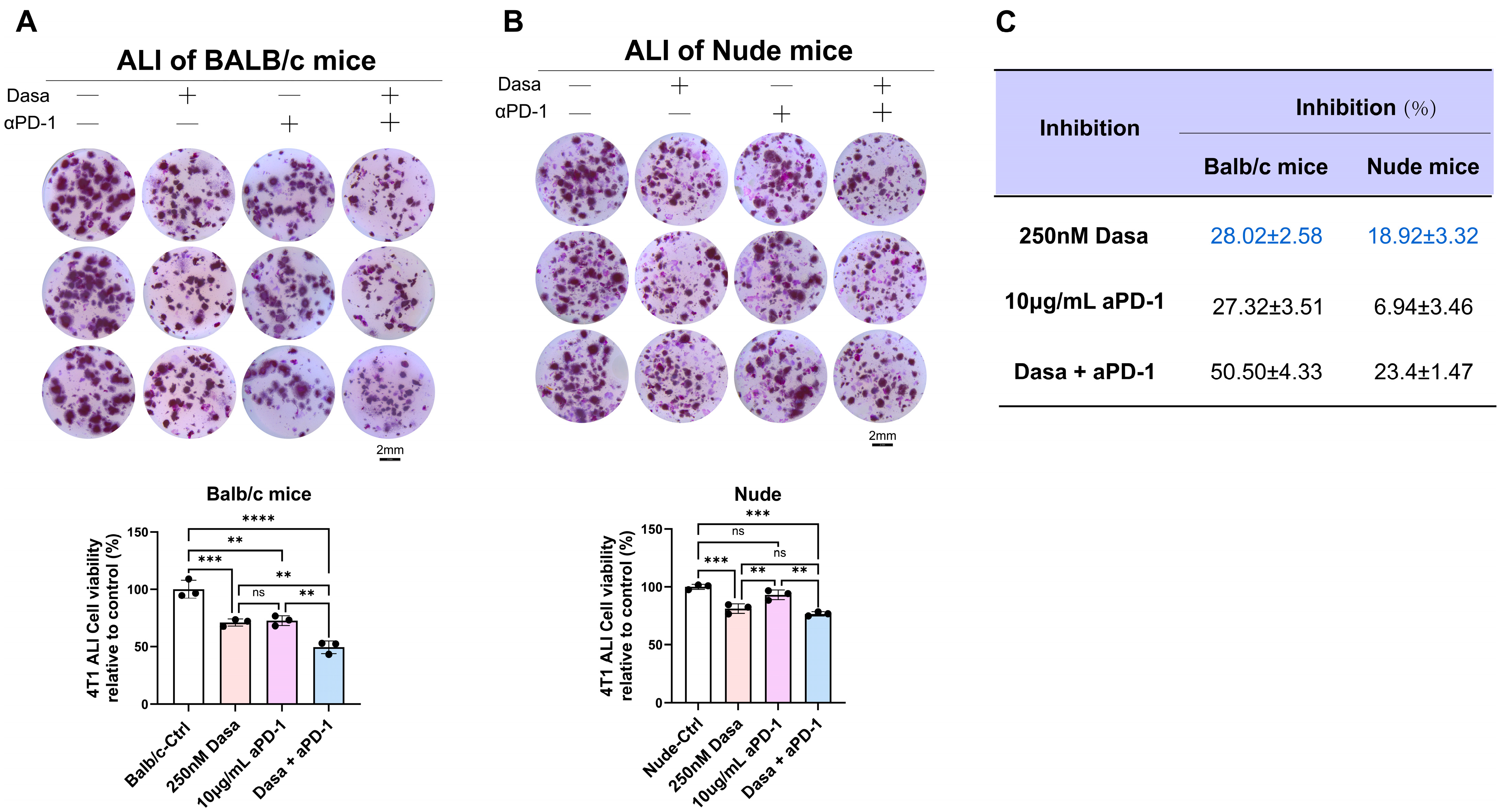
3.6. Dasatinib Potentiates Anti-Tumor Effects via the Immune Microenvironment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ALI | air–liquid interface |
| ECM | extracellular matrix |
| EMT | epithelial–mesenchymal transition |
| ETS1 | ETS proto-oncogene 1 |
| TME | Tumor microenvironment |
| TNBC | triple-negative breast cancer |
| PD-1 | anti-programmed cell death protein-1 |
| PDO | patient-derived organoid |
| MMP3 | matrix metalloproteinase-3 |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Nolan, E.; Lindeman, G.J.; Visvader, J.E. Deciphering breast cancer: From biology to the clinic. Cell 2023, 186, 1708–1728. [Google Scholar] [CrossRef] [PubMed]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Prat, A.; Adamo, B.; Cheang, M.C.; Anders, C.K.; Carey, L.A.; Perou, C.M. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013, 18, 123–133. [Google Scholar] [CrossRef]
- Dai, X.; Cheng, H.; Bai, Z.; Li, J. Breast Cancer Cell Line Classification and Its Relevance with Breast Tumor Subtyping. J. Cancer 2017, 8, 3131–3141. [Google Scholar] [CrossRef]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef]
- Li, Y.; Liu, F.; Cai, Q.; Deng, L.; Ouyang, Q.; Zhang, X.H.; Zheng, J. Invasion and metastasis in cancer: Molecular insights and therapeutic targets. Signal Transduct. Target. Ther. 2025, 10, 57. [Google Scholar] [CrossRef]
- Huang, J.; Luo, S.; Shen, J.; Lee, M.; Chen, R.; Ma, S.; Sun, L.Q.; Li, J.J. Cellular polarity pilots breast cancer progression and immunosuppression. Oncogene 2025, 44, 783–793. [Google Scholar] [CrossRef]
- Lamouille, S.; Xu, J.; Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014, 15, 178–196. [Google Scholar] [CrossRef]
- Winkler, J.; Abisoye-Ogunniyan, A.; Metcalf, K.J.; Werb, Z. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat. Commun. 2020, 11, 5120. [Google Scholar] [CrossRef]
- Pang, X.; He, X.; Qiu, Z.; Zhang, H.; Xie, R.; Liu, Z.; Gu, Y.; Zhao, N.; Xiang, Q.; Cui, Y. Targeting integrin pathways: Mechanisms and advances in therapy. Signal Transduct. Target. Ther. 2023, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Eshaq, A.M.; Flanagan, T.W.; Hassan, S.Y.; Al Asheikh, S.A.; Al-Amoudi, W.A.; Santourlidis, S.; Hassan, S.L.; Alamodi, M.O.; Bendhack, M.L.; Alamodi, M.O.; et al. Non-Receptor Tyrosine Kinases: Their Structure and Mechanistic Role in Tumor Progression and Resistance. Cancers 2024, 16, 2754. [Google Scholar] [CrossRef] [PubMed]
- Ortiz, M.A.; Mikhailova, T.; Li, X.; Porter, B.A.; Bah, A.; Kotula, L. Src family kinases, adaptor proteins and the actin cytoskeleton in epithelial-to-mesenchymal transition. Cell Commun. Signal 2021, 19, 67. [Google Scholar] [CrossRef] [PubMed]
- Aleshin, A.; Finn, R.S. SRC: A century of science brought to the clinic. Neoplasia 2010, 12, 599–607. [Google Scholar] [CrossRef]
- Hosford, S.R.; Miller, T.W. Clinical potential of novel therapeutic targets in breast cancer: CDK4/6, Src, JAK/STAT, PARP, HDAC, and PI3K/AKT/mTOR pathways. Pharmgenomics Pers. Med. 2014, 7, 203–215. [Google Scholar] [CrossRef]
- Young, A.I.; Law, A.M.; Castillo, L.; Chong, S.; Cullen, H.D.; Koehler, M.; Herzog, S.; Brummer, T.; Lee, E.F.; Fairlie, W.D.; et al. MCL-1 inhibition provides a new way to suppress breast cancer metastasis and increase sensitivity to dasatinib. Breast Cancer Res. 2016, 18, 125. [Google Scholar] [CrossRef]
- Tian, J.; Raffa, F.A.; Dai, M.; Moamer, A.; Khadang, B.; Hachim, I.Y.; Bakdounes, K.; Ali, S.; Jean-Claude, B.; Lebrun, J.J. Dasatinib sensitises triple negative breast cancer cells to chemotherapy by targeting breast cancer stem cells. Br. J. Cancer 2018, 119, 1495–1507. [Google Scholar] [CrossRef]
- Morris, P.G.; Rota, S.; Cadoo, K.; Zamora, S.; Patil, S.; D’Andrea, G.; Gilewski, T.; Bromberg, J.; Dang, C.; Dickler, M.; et al. Phase II Study of Paclitaxel and Dasatinib in Metastatic Breast Cancer. Clin. Breast Cancer 2018, 18, 387–394. [Google Scholar] [CrossRef]
- Wang, H.; Guo, S.; Kim, S.J.; Shao, F.; Ho, J.W.K.; Wong, K.U.; Miao, Z.; Hao, D.; Zhao, M.; Xu, J.; et al. Cisplatin prevents breast cancer metastasis through blocking early EMT and retards cancer growth together with paclitaxel. Theranostics 2021, 11, 2442–2459. [Google Scholar] [CrossRef]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e1916. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Soares, J.; Greninger, P.; Edelman, E.J.; Lightfoot, H.; Forbes, S.; Bindal, N.; Beare, D.; Smith, J.A.; Thompson, I.R.; et al. Genomics of Drug Sensitivity in Cancer (GDSC): A resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013, 41, D955–D961. [Google Scholar] [CrossRef]
- Ghandi, M.; Huang, F.W.; Jane-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R., 3rd; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H.; et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Kaller, M. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Loibl, S.; Poortmans, P.; Morrow, M.; Denkert, C.; Curigliano, G. Breast cancer. Lancet 2021, 397, 1750–1769, Erratum in: Lancet 2021, 397, 1710. https://doi.org/10.1016/S0140-6736(21)00838-2. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Zhang, X.; Ding, R.; Yang, L.; Lyu, X.; Zeng, J.; Lei, J.H.; Wang, L.; Bi, J.; Shao, N.; et al. Patient-Derived Organoids Can Guide Personalized-Therapies for Patients with Advanced Breast Cancer. Adv. Sci. 2021, 8, e2101176. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Xu, X.; Bi, Y.; Xu, J.; Qin, C.; Han, M. Systemic inflammation promotes lung metastasis via E-selectin upregulation in mouse breast cancer model. Cancer Biol. Ther. 2014, 15, 789–796. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Li, W.; Chen, C.S. Breast cancer animal models and applications. Zool. Res. 2020, 41, 477–494. [Google Scholar] [CrossRef] [PubMed]
- Anastassiadis, T.; Deacon, S.W.; Devarajan, K.; Ma, H.; Peterson, J.R. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat. Biotechnol. 2011, 29, 1039–1045. [Google Scholar] [CrossRef]
- Herschkowitz, J.I.; Simin, K.; Weigman, V.J.; Mikaelian, I.; Usary, J.; Hu, Z.; Rasmussen, K.E.; Jones, L.P.; Assefnia, S.; Chandrasekharan, S.; et al. Identification of conserved gene expression features between murine mammary carcinoma models and human breast tumors. Genome Biol. 2007, 8, R76. [Google Scholar] [CrossRef]
- Liang, J.; Yao, X.; Aouad, P.; Wang, B.E.; Crocker, L.; Chaudhuri, S.; Liang, Y.; Darmanis, S.; Giltnane, J.; Moore, H.M.; et al. ERalpha dysfunction caused by ESR1 mutations and therapeutic pressure promotes lineage plasticity in ER(+) breast cancer. Nat. Cancer 2025, 6, 357–371. [Google Scholar] [CrossRef]
- Wen, W.; Chen, Z.; Bao, J.; Long, Q.; Shu, X.O.; Zheng, W.; Guo, X. Genetic variations of DNA bindings of FOXA1 and co-factors in breast cancer susceptibility. Nat. Commun. 2021, 12, 5318. [Google Scholar] [CrossRef]
- Ye, T.; Li, J.; Feng, J.; Guo, J.; Wan, X.; Xie, D.; Liu, J. The subtype-specific molecular function of SPDEF in breast cancer and insights into prognostic significance. J. Cell Mol. Med. 2021, 25, 7307–7320. [Google Scholar] [CrossRef]
- Loh, C.Y.; Chai, J.Y.; Tang, T.F.; Wong, W.F.; Sethi, G.; Shanmugam, M.K.; Chong, P.P.; Looi, C.Y. The E-Cadherin and N-Cadherin Switch in Epithelial-to-Mesenchymal Transition: Signaling, Therapeutic Implications, and Challenges. Cells 2019, 8, 1118. [Google Scholar] [CrossRef]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. Emt: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef]
- Majumder, M.; Xin, X.; Liu, L.; Tutunea-Fatan, E.; Rodriguez-Torres, M.; Vincent, K.; Postovit, L.M.; Hess, D.; Lala, P.K. COX-2 Induces Breast Cancer Stem Cells via EP4/PI3K/AKT/NOTCH/WNT Axis. Stem Cells 2016, 34, 2290–2305. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, W.; Liu, S.; Chen, C. Targeting Breast Cancer Stem Cells. Int. J. Biol. Sci. 2023, 19, 552–570. [Google Scholar] [CrossRef]
- Bergholz, J.S.; Wang, Q.; Wang, Q.; Ramseier, M.; Prakadan, S.; Wang, W.; Fang, R.; Kabraji, S.; Zhou, Q.; Gray, G.K.; et al. PI3Kbeta controls immune evasion in PTEN-deficient breast tumours. Nature 2023, 617, 139–146. [Google Scholar] [CrossRef]
- Nobre, A.R.; Dalla, E.; Yang, J.; Huang, X.; Wullkopf, L.; Risson, E.; Razghandi, P.; Anton, M.L.; Zheng, W.; Seoane, J.A.; et al. ZFP281 drives a mesenchymal-like dormancy program in early disseminated breast cancer cells that prevents metastatic outgrowth in the lung. Nat. Cancer 2022, 3, 1165–1180. [Google Scholar] [CrossRef] [PubMed]
- Xia, H.; Chen, J.; Shi, M.; Gao, H.; Sekar, K.; Seshachalam, V.P.; Ooi, L.L.; Hui, K.M. EDIL3 is a novel regulator of epithelial-mesenchymal transition controlling early recurrence of hepatocellular carcinoma. J. Hepatol. 2015, 63, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Bacchetti, R.; Yuan, S.; Rainero, E. ADAMTS Proteases: Their Multifaceted Role in the Regulation of Cancer Metastasis. Dis. Res. 2024, 4, 40–52. [Google Scholar] [CrossRef]
- Li, Y.; Wu, T.; Peng, Z.; Tian, X.; Dai, Q.; Chen, M.; Zhu, J.; Xia, S.; Sun, A.; Yang, W.; et al. ETS1 is a prognostic biomarker of triple-negative breast cancer and promotes the triple-negative breast cancer progression through the YAP signaling. Am. J. Cancer Res. 2022, 12, 5074–5084. [Google Scholar] [PubMed]
- Yadav, M.; Sharma, A.; Patne, K.; Tabasum, S.; Suryavanshi, J.; Rawat, L.; Machaalani, M.; Eid, M.; Singh, R.P.; Choueiri, T.K.; et al. AXL signaling in cancer: From molecular insights to targeted therapies. Signal Transduct. Target. Ther. 2025, 10, 37. [Google Scholar] [CrossRef]
- Behrens, P.; Rothe, M.; Wellmann, A.; Krischler, J.; Wernert, N. The Ets-1 transcription factor is up-regulated together with MMP 1 and MMP 9 in the stroma of pre-invasive breast cancer. J. Pathol. 2001, 194, 43–50. [Google Scholar] [CrossRef]
- Dittmer, J. The biology of the Ets1 proto-oncogene. Mol. Cancer 2003, 2, 29. [Google Scholar] [CrossRef]
- Dittmer, J. The role of the transcription factor Ets1 in carcinoma. Semin. Cancer Biol. 2015, 35, 20–38. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.; Zhang, Q.; Huang, Y.; Song, J.; Tomaino, R.; Ehrenberger, T.; Lim, E.; Liu, W.; Bronson, R.T.; Bowden, M.; et al. Phosphorylation of ETS1 by Src family kinases prevents its recognition by the COP1 tumor suppressor. Cancer Cell 2014, 26, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Z.; Li, Y.; Zhang, S.; Wang, X.; Dou, H.; Yu, X.; Zhang, Z.; Yang, S.; Xiao, M. Extracellular matrix remodeling in tumor progression and immune escape: From mechanisms to treatments. Mol. Cancer 2023, 22, 48. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.L.B.; Gradishar, W.J. Triple-Negative Breast Cancer: Current Practice and Future Directions. J. Oncol. Pract. 2017, 13, 301–303. [Google Scholar] [CrossRef]
- Finn, R.S.; Bengala, C.; Ibrahim, N.; Roche, H.; Sparano, J.; Strauss, L.C.; Fairchild, J.; Sy, O.; Goldstein, L.J. Dasatinib as a single agent in triple-negative breast cancer: Results of an open-label phase 2 study. Clin. Cancer Res. 2011, 17, 6905–6913. [Google Scholar] [CrossRef]
- Kennedy, L.C.; Gadi, V. Dasatinib in breast cancer: Src-ing for response in all the wrong kinases. Ann. Transl. Med. 2018, 6, S60. [Google Scholar] [CrossRef]
- Puls, L.N.; Eadens, M.; Messersmith, W. Current status of SRC inhibitors in solid tumor malignancies. Oncologist 2011, 16, 566–578. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Finn, R.S.; Dering, J.; Ginther, C.; Wilson, C.A.; Glaspy, P.; Tchekmedyian, N.; Slamon, D.J. Dasatinib, an orally active small molecule inhibitor of both the src and abl kinases, selectively inhibits growth of basal-type/”triple-negative” breast cancer cell lines growing in vitro. Breast Cancer Res. Treat. 2007, 105, 319–326. [Google Scholar] [CrossRef]
- Yu, X.; Jin, J.; Zheng, Y.; Zhu, H.; Xu, H.; Ma, J.; Lan, Q.; Zhuang, Z.; Chen, C.C.; Li, M. GBP5 drives malignancy of glioblastoma via the Src/ERK1/2/MMP3 pathway. Cell Death Dis. 2021, 12, 203. [Google Scholar] [CrossRef]
- Vandenbroucke, R.E.; Libert, C. Is there new hope for therapeutic matrix metalloproteinase inhibition? Nat. Rev. Drug Discov. 2014, 13, 904–927. [Google Scholar] [CrossRef]
- Jiang, J.; Wu, Q.; Rajasekaran, S.; Wu, R. MMP3 at the crossroads: Linking molecular pathways to disease diagnosis and therapy. Pharmacol. Res. 2025, 216, 107750. [Google Scholar] [CrossRef]
- Sternlicht, M.D.; Lochter, A.; Sympson, C.J.; Huey, B.; Rougier, J.P.; Gray, J.W.; Pinkel, D.; Bissell, M.J.; Werb, Z. The stromal proteinase MMP3/stromelysin-1 promotes mammary carcinogenesis. Cell 1999, 98, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Alaseem, A.; Alhazzani, K.; Dondapati, P.; Alobid, S.; Bishayee, A.; Rathinavelu, A. Matrix Metalloproteinases: A challenging paradigm of cancer management. Semin. Cancer Biol. 2019, 56, 100–115. [Google Scholar] [CrossRef] [PubMed]
- Kessenbrock, K.; Wang, C.Y.; Werb, Z. Matrix metalloproteinases in stem cell regulation and cancer. Matrix Biol. 2015, 44–46, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Fields, G.B. The Rebirth of Matrix Metalloproteinase Inhibitors: Moving Beyond the Dogma. Cells 2019, 8, 984. [Google Scholar] [CrossRef]
- Seehawer, M.; Li, Z.; Nishida, J.; Foidart, P.; Reiter, A.H.; Rojas-Jimenez, E.; Goyette, M.A.; Yan, P.; Raval, S.; Munoz Gomez, M.; et al. Loss of Kmt2c or Kmt2d drives brain metastasis via KDM6A-dependent upregulation of MMP3. Nat. Cell Biol. 2024, 26, 1165–1175. [Google Scholar] [CrossRef]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef]
- Linder, S.; Cervero, P.; Eddy, R.; Condeelis, J. Mechanisms and roles of podosomes and invadopodia. Nat. Rev. Mol. Cell Biol. 2023, 24, 86–106. [Google Scholar] [CrossRef]
- Lu, P.; Lu, Y. Born to Run? Diverse Modes of Epithelial Migration. Front. Cell Dev. Biol. 2021, 9, 704939. [Google Scholar] [CrossRef]
- SenGupta, S.; Parent, C.A.; Bear, J.E. The principles of directed cell migration. Nat. Rev. Mol. Cell Biol. 2021, 22, 529–547. [Google Scholar] [CrossRef]
- Crossley, R.M.; Johnson, S.; Tsingos, E.; Bell, Z.; Berardi, M.; Botticelli, M.; Braat, Q.J.S.; Metzcar, J.; Ruscone, M.; Yin, Y.; et al. Modeling the extracellular matrix in cell migration and morphogenesis: A guide for the curious biologist. Front. Cell Dev. Biol. 2024, 12, 1354132. [Google Scholar] [CrossRef] [PubMed]
- Mazaki, Y.; Hashimoto, S.; Sabe, H. Monocyte cells and cancer cells express novel paxillin isoforms with different binding properties to focal adhesion proteins. J. Biol. Chem. 1997, 272, 7437–7444. [Google Scholar] [CrossRef] [PubMed]
- Hekim, C.; Ilander, M.; Yan, J.; Michaud, E.; Smykla, R.; Vaha-Koskela, M.; Savola, P.; Tahtinen, S.; Saikko, L.; Hemminki, A.; et al. Dasatinib Changes Immune Cell Profiles Concomitant with Reduced Tumor Growth in Several Murine Solid Tumor Models. Cancer Immunol. Res. 2017, 5, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Han, X. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J. Clin. Investig. 2015, 125, 3384–3391. [Google Scholar] [CrossRef]
- Koller, P.; Baran, N.; Harutyunyan, K.; Cavazos, A.; Mallampati, S.; Chin, R.L.; Jiang, Z.; Sun, X.; Lee, H.H.; Hsu, J.L.; et al. PD-1 blockade in combination with dasatinib potentiates induction of anti-acute lymphocytic leukemia immunity. J. Immunother. Cancer 2023, 11, e006619. [Google Scholar] [CrossRef]
- Kadota, H.; Yuge, R.; Shimizu, D.; Miyamoto, R.; Otani, R.; Hiyama, Y.; Takigawa, H.; Hayashi, R.; Urabe, Y.; Kitadai, Y.; et al. Anti-Programmed Cell Death-1 Antibody and Dasatinib Combination Therapy Exhibits Efficacy in Metastatic Colorectal Cancer Mouse Models. Cancers 2022, 14, 6146. [Google Scholar] [CrossRef]
- Tu, M.M.; Lee, F.Y.F.; Jones, R.T.; Kimball, A.K.; Saravia, E.; Graziano, R.F.; Coleman, B.; Menard, K.; Yan, J.; Michaud, E.; et al. Targeting DDR2 enhances tumor response to anti-PD-1 immunotherapy. Sci. Adv. 2019, 5, eaav2437. [Google Scholar] [CrossRef]
- Redin, E.; Garmendia, I.; Lozano, T.; Serrano, D.; Senent, Y.; Redrado, M.; Villalba, M.; De Andrea, C.E.; Exposito, F.; Ajona, D.; et al. SRC family kinase (SFK) inhibitor dasatinib improves the antitumor activity of anti-PD-1 in NSCLC models by inhibiting Treg cell conversion and proliferation. J. Immunother. Cancer 2021, 9, e001496. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, J.; Chen, X.; Li, Z.; Gu, L.; Pan, D.; Zheng, X.; Zhang, Q.; Chen, R.; Zhang, H.; et al. Modulating tumor-stromal crosstalk via a redox-responsive nanomedicine for combination tumor therapy. J. Control Release 2023, 356, 525–541. [Google Scholar] [CrossRef]
- Liu, Y.; Li, L.; Liu, J.; Yang, M.; Wang, H.; Chu, X.; Zhou, J.; Huo, M.; Yin, T. Biomineralization-inspired dasatinib nanodrug with sequential infiltration for effective solid tumor treatment. Biomaterials 2021, 267, 120481. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, Z.; Pan, D.; Li, Y.; Zhou, J.; Chen, H.; Li, Z.; Zhu, M.; Li, C.; Qin, L.; et al. Dendritic Polymer-Based Nanomedicines Remodel the Tumor Stroma: Improve Drug Penetration and Enhance Antitumor Immune Response. Adv. Mater. 2024, 36, e2401304. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, J.; Wang, Y.; Wu, Y.; Li, Y.; Wang, B.; Liu, G.; Gong, Q.; Luo, K.; Jing, J. Stimuli-responsive polymer-dasatinib prodrug to reprogram cancer-associated fibroblasts for boosted immunotherapy. J. Control Release 2025, 381, 113606. [Google Scholar] [CrossRef]

| Gene | Name | Direction | Sequence |
|---|---|---|---|
| Human ETS1 | HGLibA_15640 | sg1 | AGAGTCGGCTTGAGATCGA |
| HGLibA_15641 | sg2 | TGGAAACCACAGTTCATTCG | |
| Human MMP3 | HGLibA_29533 | sg1 | GCATGGGCCAAAACATTTCC |
| HGLibA_29534 | sg2 | GTTCTGAAGTGACCAACATC | |
| Mouse Ets1 | MGLibA_16711 | sg1 | CAGAAACCCACGTCCGGGAC |
| MGLibA_16712 | sg2 | CTTACTGATGAAGTAATCCG | |
| Mouse Mmp3 | MGLibA_31582 | sg1 | ACTTTGACGATGATGAACGA |
| MGLibA_31583 | sg2 | AATAGGTACCAACCTATTCC |
| Primers | Direction | Sequence |
|---|---|---|
| Human 18S | Forward | AGTCCCTGCCCTTTGTACACA |
| Reverse | CGATCCGAGGGCCTCACTA | |
| Human MMP3 | Forward | CAGGCTTTCCCAAGCAAATAG |
| Reverse | CTCCAACTGTGAAGATCCAGTAA | |
| Human ETS1 | Forward | GATAGTTGTGATCGCCTCACC |
| Reverse | GTCCTCTGAGTCGAAGCTGTC | |
| Mouse Actb | Forward | GGCTGTATTCCCCTCCATCG |
| Reverse | CCAGTTGGTAACAATGCCATGT | |
| Mouse Mmp3 | Forward | ACATGGAGACTTTGTCCCTTTTG |
| Reverse | TTGGCTGAGTGGTAGAGTCCC | |
| Mouse Ets1 | Forward | GACCTCTTTATCCACTCTGGGA |
| Reverse | GTGGTCCTTTCTCCTACGATCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, X.; Sun, H.; Yu, F.; Feng, Y.; Guo, S.; Lei, J.H.; Miao, K.; Ip, K.-U.; Li, L.; Li, H.; et al. Dasatinib Inhibits Basal B Breast Cancer Through ETS1-Mediated Extracellular Matrix Remodeling. Biomedicines 2025, 13, 2888. https://doi.org/10.3390/biomedicines13122888
Guo X, Sun H, Yu F, Feng Y, Guo S, Lei JH, Miao K, Ip K-U, Li L, Li H, et al. Dasatinib Inhibits Basal B Breast Cancer Through ETS1-Mediated Extracellular Matrix Remodeling. Biomedicines. 2025; 13(12):2888. https://doi.org/10.3390/biomedicines13122888
Chicago/Turabian StyleGuo, Xinyu, Heng Sun, Feng Yu, Yangyang Feng, Sen Guo, Josh Haipeng Lei, Kai Miao, Ka-U Ip, Ling Li, Hanghang Li, and et al. 2025. "Dasatinib Inhibits Basal B Breast Cancer Through ETS1-Mediated Extracellular Matrix Remodeling" Biomedicines 13, no. 12: 2888. https://doi.org/10.3390/biomedicines13122888
APA StyleGuo, X., Sun, H., Yu, F., Feng, Y., Guo, S., Lei, J. H., Miao, K., Ip, K.-U., Li, L., Li, H., Liao, X., Xu, X., Zhou, R., & Deng, C.-X. (2025). Dasatinib Inhibits Basal B Breast Cancer Through ETS1-Mediated Extracellular Matrix Remodeling. Biomedicines, 13(12), 2888. https://doi.org/10.3390/biomedicines13122888








