Shotgun Label-Free Proteomic Analysis for Identification of a Potential Diagnostic Biomarker for Pancreatic Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Preparation of Protein Samples for Proteomics and Western Blot
2.4. Proteomic Analysis
2.5. Semiquantification of Identified Proteins
2.6. Bioinformatics
2.7. Western Blot
2.8. ELISA
2.9. Patients and Serum Specimens
2.10. Statistical Analysis
3. Results
3.1. Semiquantitative Comparison of Differentially Expressed Proteins in Normal Pancreatic Ductal Cells and PC Cells
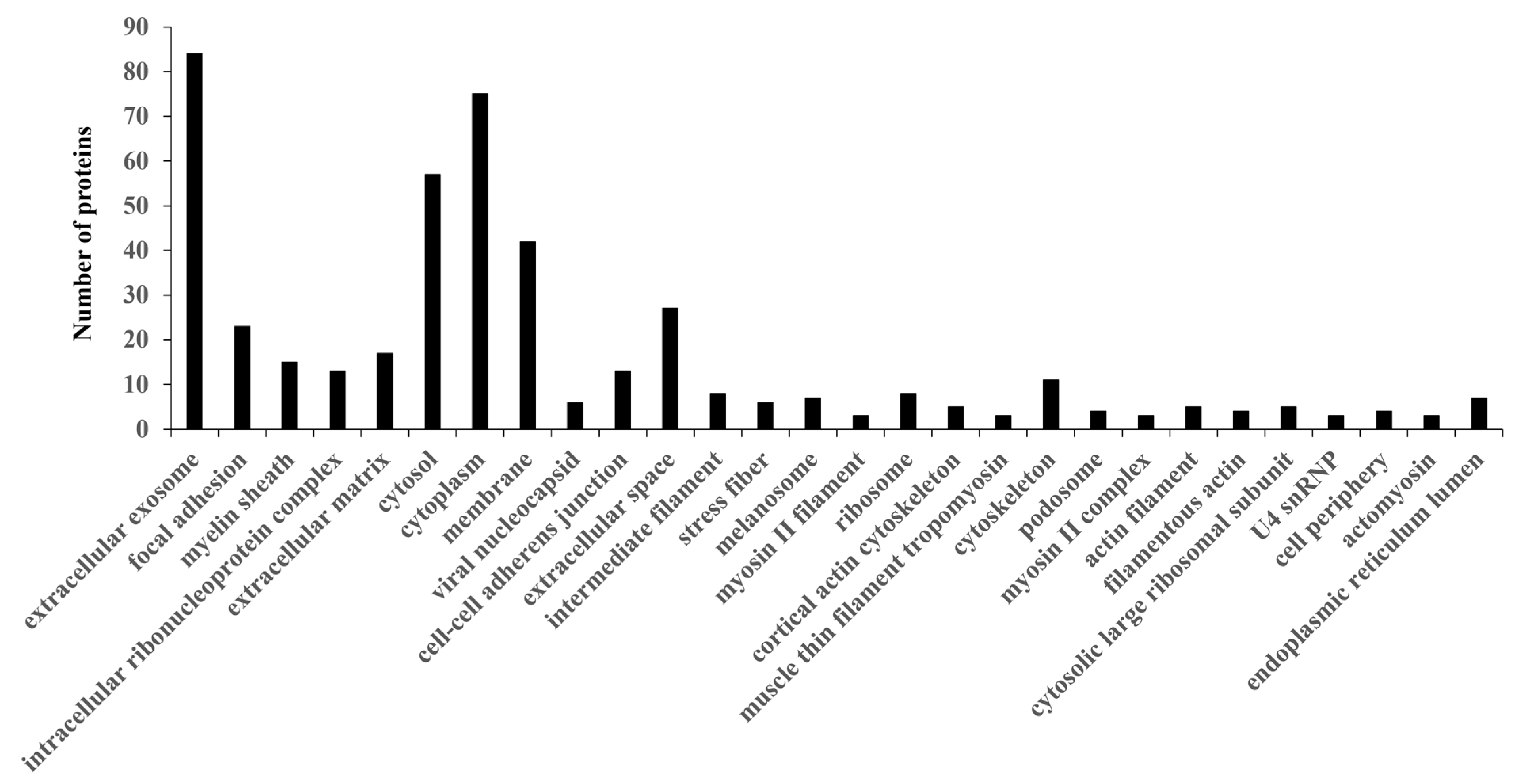
3.2. Functional Annotation of PC-Related Proteins
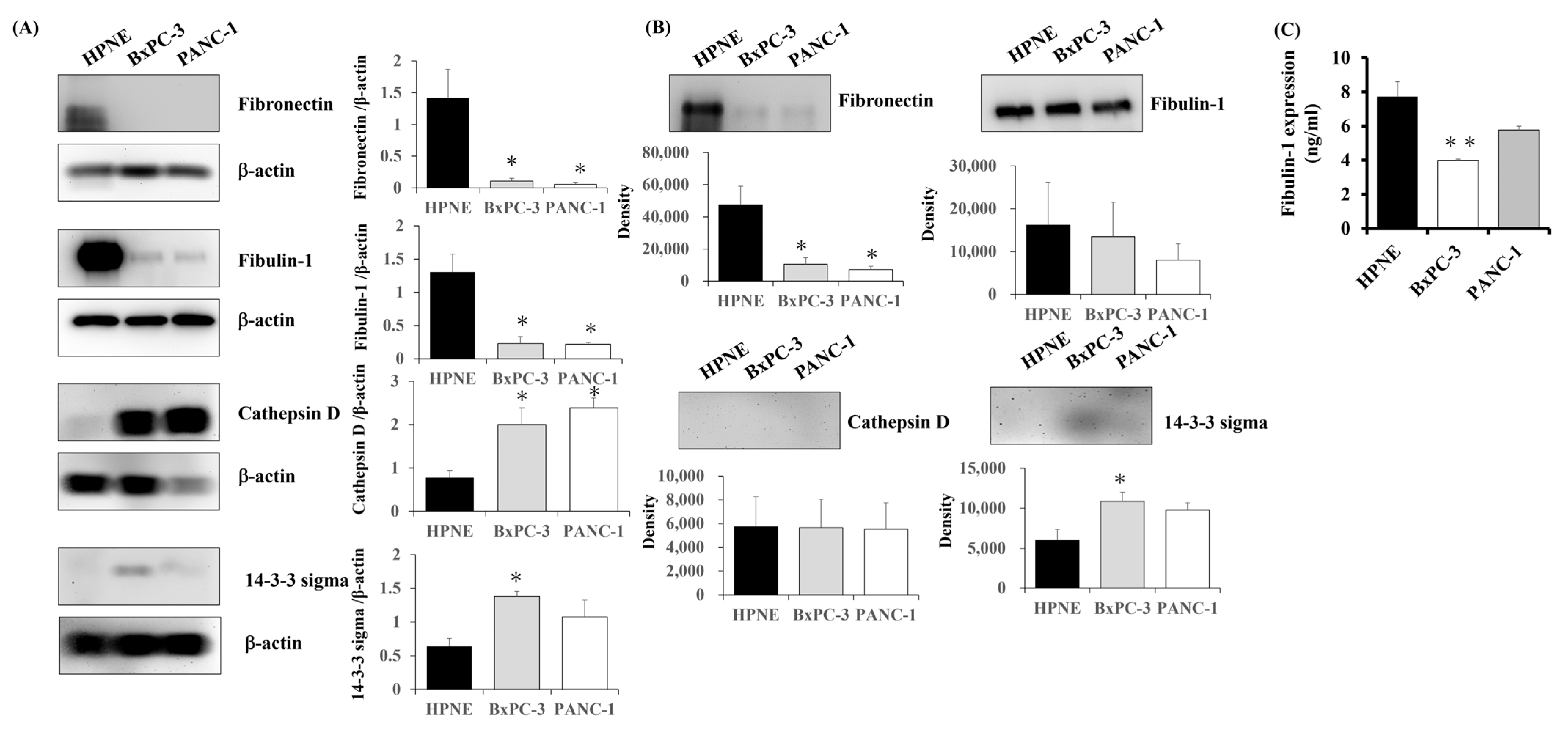
3.3. Expression of Candidate Proteins in PC Cells
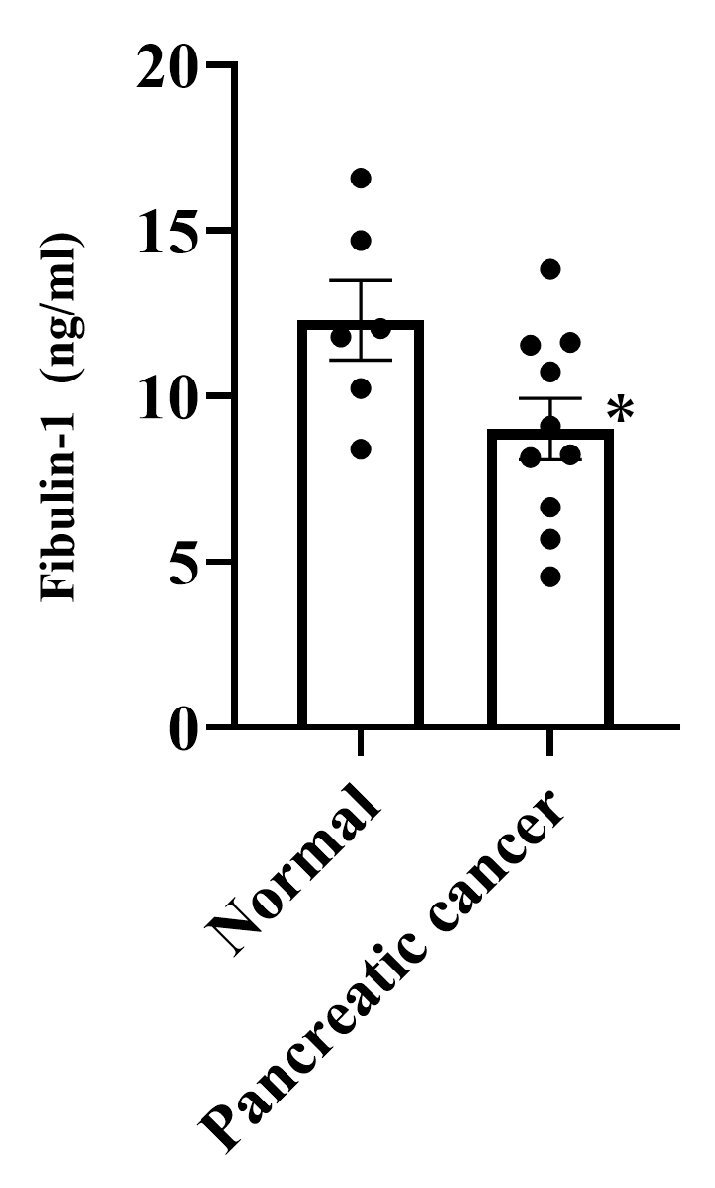
3.4. Expression of Fibulin-1 in PC Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| PC | Pancreatic cancer |
| CA | Carbohydrate antigen |
| LC | Liquid chromatography |
| MS | Mass spectrometry |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Tolue Ghasaban, F.; Ghanei, M.; Mahmoudian, R.A.; Taghehchian, N.; Abbaszadegan, M.R.; Moghbeli, M. MicroRNAs as the critical regulators of epithelial mesenchymal transition in pancreatic tumor cells. Heliyon 2024, 10, e30599. [Google Scholar] [CrossRef]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Gupta, T.; Murtaza, M. Advancing targeted therapies in pancreatic cancer: Leveraging molecular abberrations for therapeutic success. Prog. Biophys. Mol. Biol. 2025, 196, 19–32. [Google Scholar] [CrossRef]
- Wood, L.D.; Canto, M.I.; Jaffee, E.M.; Simeone, D.M. Pancreatic Cancer: Pathogenesis, Screening, Diagnosis, and Treatment. Gastroenterology 2022, 163, 386–402.e1. [Google Scholar] [CrossRef]
- Sugiura, T.; Uesaka, K.; Kanemoto, H.; Mizuno, T.; Sasaki, K.; Furukawa, H.; Matsunaga, K.; Maeda, A. Serum CA19-9 is a significant predictor among preoperative parameters for early recurrence after resection of pancreatic adenocarcinoma. J. Gastrointest. Surg. 2012, 16, 977–985. [Google Scholar] [CrossRef]
- Kondo, N.; Murakami, Y.; Uemura, K.; Nakagawa, N.; Takahashi, S.; Ohge, H.; Sueda, T. Comparison of the prognostic impact of pre- and post-operative CA19-9, SPan-1, and DUPAN-II levels in patients with pancreatic carcinoma. Pancreatology 2017, 17, 95–102. [Google Scholar] [CrossRef]
- Scara, S.; Bottoni, P.; Scatena, R. CA 19-9: Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Liu, C.; Guo, M.; Long, J.; Liu, Z.; Xiao, Z.; Jin, K.; Cheng, H.; Lu, Y.; Ni, Q.; et al. CA19-9-Low&Lewis (+) pancreatic cancer: A unique subtype. Cancer Lett. 2017, 385, 46–50. [Google Scholar] [CrossRef] [PubMed]
- Staal, B.; Liu, Y.; Barnett, D.; Hsueh, P.; He, Z.; Gao, C.; Partyka, K.; Hurd, M.W.; Singhi, A.D.; Drake, R.R.; et al. The sTRA Plasma Biomarker: Blinded Validation of Improved Accuracy Over CA19-9 in Pancreatic Cancer Diagnosis. Clin. Cancer Res. 2019, 25, 2745–2754. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Tong, Y.; Zhong, A.; Wang, Y.; Lu, R.; Guo, L. Identification of Serum microRNA-25 as a novel biomarker for pancreatic cancer. Medicine 2020, 99, e23863. [Google Scholar] [CrossRef]
- Nurmi, A.M.; Mustonen, H.; Haglund, C.; Seppanen, H. Changes in CRP and CA19-9 during Preoperative Oncological Therapy Predict Postoperative Survival in Pancreatic Ductal Adenocarcinoma. Oncology 2021, 99, 686–698. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yin, H.; Liu, S.; Shoucair, S.; Ding, N.; Ji, Y.; Zhang, J.; Wang, D.; Kuang, T.; Xu, X.; et al. Prognostic value of tumor-associated N1/N2 neutrophil plasticity in patients following radical resection of pancreas ductal adenocarcinoma. J. Immunother. Cancer 2022, 10, e005798. [Google Scholar] [CrossRef]
- Zhou, T.; Man, Q.; Li, X.; Xie, Y.; Hou, X.; Wang, H.; Yan, J.; Wei, X.; Bai, W.; Liu, Z.; et al. Artificial intelligence-based comprehensive analysis of immune-stemness-tumor budding profile to predict survival of patients with pancreatic adenocarcinoma. Cancer Biol. Med. 2023, 20, 196–217. [Google Scholar] [CrossRef]
- Yugawa, K.; Maeda, T.; Nagata, S.; Sakai, A.; Taketani, K.; Yamaguchi, S.; Konishi, K.; Hashimoto, K. A novel combined carbohydrate antigen 19-9 and lymphocyte-to-monocyte ratio score can predict early recurrence of resectable pancreatic ductal adenocarcinoma. Surg. Today 2023, 53, 1199–1208. [Google Scholar] [CrossRef]
- Zhuang, J.; Wang, S.; Wang, Y.; Wu, Y.; Hu, R. Prognostic significance of preoperative lymphocytes, albumin, and neutrophils (LANR) index in resectable pancreatic ductal adenocarcinoma. BMC Cancer 2024, 24, 568. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Zhang, Z.X.; Chen, X.Y.; Xu, Y.L.; Yin, N.; Yang, J.; Zhu, D.M.; Li, D.C.; Zhou, J. A Panel of Three Biomarkers Identified by iTRAQ for the Early Diagnosis of Pancreatic Cancer. Proteomics. Clin. Appl. 2019, 13, e1800195. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.C.; Lu, Y.T.; Yeh, T.S.; Chan, Y.H.; Dash, S.; Yu, J.S. Identification of Fucosylated SERPINA1 as a Novel Plasma Marker for Pancreatic Cancer Using Lectin Affinity Capture Coupled with iTRAQ-Based Quantitative Glycoproteomics. Int. J. Mol. Sci. 2021, 22, 6079. [Google Scholar] [CrossRef]
- Yamamoto, T.; Nakanishi, S.; Mitamura, K.; Taga, A. Shotgun label-free proteomic analysis for identification of proteins in HaCaT human skin keratinocytes regulated by the administration of collagen from soft-shelled turtle. J. Biomed. Mater. Res. B Appl. Biomater. 2018, 106, 2403–2413. [Google Scholar] [CrossRef]
- Yamamoto, T.; Kudo, M.; Peng, W.X.; Takata, H.; Takakura, H.; Teduka, K.; Fujii, T.; Mitamura, K.; Taga, A.; Uchida, E.; et al. Identification of aldolase A as a potential diagnostic biomarker for colorectal cancer based on proteomic analysis using formalin-fixed paraffin-embedded tissue. Tumour Biol. 2016, 37, 13595–13606. [Google Scholar] [CrossRef]
- Nagai, N.; Yamamoto, T.; Mitamura, K.; Taga, A. Proteomic profile of the lens in a streptozotocin-induced diabetic rat model using shotgun proteomics. Biomed. Rep. 2017, 7, 445–450. [Google Scholar] [CrossRef]
- Kawamura, T.; Nomura, M.; Tojo, H.; Fujii, K.; Hamasaki, H.; Mikami, S.; Bando, Y.; Kato, H.; Nishimura, T. Proteomic analysis of laser-microdissected paraffin-embedded tissues: (1) Stage-related protein candidates upon non-metastatic lung adenocarcinoma. J. Proteom. 2010, 73, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Old, W.M.; Meyer-Arendt, K.; Aveline-Wolf, L.; Pierce, K.G.; Mendoza, A.; Sevinsky, J.R.; Resing, K.A.; Ahn, N.G. Comparison of label-free methods for quantifying human proteins by shotgun proteomics. Mol. Cell. Proteom. 2005, 4, 1487–1502. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Huang da, W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Zhang, X.; Luo, Y.; Cen, Y.; Qiu, X.; Li, J.; Jie, M.; Yang, S.; Qin, S. MACC1 promotes pancreatic cancer metastasis by interacting with the EMT regulator SNAI1. Cell Death Dis. 2022, 13, 923. [Google Scholar] [CrossRef] [PubMed]
- Robin, F.; Angenard, G.; Cano, L.; Courtin-Tanguy, L.; Gaignard, E.; Khene, Z.E.; Bergeat, D.; Clement, B.; Boudjema, K.; Coulouarn, C.; et al. Molecular profiling of stroma highlights stratifin as a novel biomarker of poor prognosis in pancreatic ductal adenocarcinoma. Br. J. Cancer 2020, 123, 72–80. [Google Scholar] [CrossRef]
- Argraves, W.S.; Dickerson, K.; Burgess, W.H.; Ruoslahti, E. Fibulin, a Novel Protein That Interacts with the Fibronectin Receptor-Beta Subunit Cytoplasmic Domain. Cell 1989, 58, 623–629. [Google Scholar] [CrossRef]
- Liu, G.; Cooley, M.A.; Jarnicki, A.G.; Hsu, A.C.; Nair, P.M.; Haw, T.J.; Fricker, M.; Gellatly, S.L.; Kim, R.Y.; Inman, M.D.; et al. Fibulin-1 regulates the pathogenesis of tissue remodeling in respiratory diseases. JCI Insight 2016, 1, e86380. [Google Scholar] [CrossRef]
- Twal, W.O.; Czirok, A.; Hegedus, B.; Knaak, C.; Chintalapudi, M.R.; Okagawa, H.; Sugi, Y.; Argraves, W.S. Fibulin-1 suppression of fibronectin-regulated cell adhesion and motility. J. Cell Sci. 2001, 114, 4587–4598. [Google Scholar] [CrossRef]
- Hedayati, M.; Abooshahab, R.; Razavi, S.A.; Salehipour, P.; Ahmadikia, K.; Boroomand, S. Low level of plasma fibulin-1 in patients with thyroid lesions: A case-control study and literature review. Mol. Biol. Rep. 2020, 47, 8859–8866. [Google Scholar] [CrossRef]
- Watany, M.M.; Elmashad, N.M.; Badawi, R.; Hawash, N. Serum FBLN1 and STK31 as biomarkers of colorectal cancer and their ability to noninvasively differentiate colorectal cancer from benign polyps. Clin. Chim. Acta 2018, 483, 151–155. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.Y.; Jin, H.; Liu, X.; Siu, J.M.T.; Wong, Y.P.; Ng, E.K.O.; Yu, J.; Leung, W.K.; Sung, J.J.Y.; Chan, F.K.L. Fibulin 1 is downregulated through promoter hypermethylation in gastric cancer. Br. J. Cancer 2008, 99, 2083–2087. [Google Scholar] [CrossRef]
- Wlazlinski, A.; Engers, R.; Hoffmann, M.J.; Hader, C.; Jung, V.; Muller, M.; Schulz, W.A. Downregulation of several fibulin genes in prostate cancer. Prostate 2007, 67, 1770–1780. [Google Scholar] [CrossRef] [PubMed]
- Hayashido, Y.; Lucas, A.; Rougeot, C.; Godyna, S.; Argraves, W.S.; Rochefort, H. Estradiol and fibulin-1 inhibit motility of human ovarian- and breast-cancer cells induced by fibronectin. International journal of cancer. J. Int. Cancer 1998, 75, 654–658. [Google Scholar] [CrossRef]
- Kanda, M.; Nomoto, S.; Okamura, Y.; Hayashi, M.; Hishida, M.; Fujii, T.; Nishikawa, Y.; Sugimoto, H.; Takeda, S.; Nakao, A. Promoter hypermethylation of fibulin 1 gene is associated with tumor progression in hepatocellular carcinoma. Mol. Carcinog. 2011, 50, 571–579. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, J.; Li, H.; Guan, W.; Xia, D.; Yu, G.; Xiao, H.; Lang, B.; Ma, X.; Liu, J.; et al. Fibulin-1 is down-regulated through promoter hypermethylation and suppresses renal cell carcinoma progression. J. Urol. 2013, 190, 291–301. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, J.; Li, H.; Xia, D.; Yu, G.; Yao, W.; Yang, Y.; Xiao, H.; Lang, B.; Ma, X.; et al. Fibulin-1 is epigenetically down-regulated and related with bladder cancer recurrence. BMC Cancer 2014, 14, 677. [Google Scholar] [CrossRef]
- Liu, X.T.; Liu, T.T.; Wu, M.Y.; Chen, Q.X.; Zhuang, J.X.; Wang, Q. Identifying FBLN1 (Gene ID: 2192) as a Potential Melanoma Biomarker for Melanoma based on an Analysis of microRNA Expression Profiles in the GEO and TCGA Databases. Genet. Test. Mol. Biomark. 2021, 25, 68–78. [Google Scholar] [CrossRef]
- Moll, F.; Katsaros, D.; Lazennec, G.; Hellio, N.; Roger, P.; Giacalone, P.L.; Chalbos, D.; Maudelonde, T.; Rochefort, H.; Pujol, P. Estrogen induction and overexpression of fibulin-1C mRNA in ovarian cancer cells. Oncogene 2002, 21, 1097–1107. [Google Scholar] [CrossRef]
- Greene, L.M.; Twal, W.O.; Duffy, M.J.; McDermott, E.W.; Hill, A.D.; O’Higgins, N.J.; McCann, A.H.; Dervan, P.A.; Argraves, W.S.; Gallagher, W.M. Elevated expression and altered processing of fibulin-1 protein in human breast cancer. Br. J. Cancer 2003, 88, 871–878. [Google Scholar] [CrossRef]
- Bardin, A.; Moll, F.; Margueron, R.; Delfour, C.; Chu, M.L.; Maudelonde, T.; Cavailles, V.; Pujol, P. Transcriptional and posttranscriptional regulation of fibulin-1 by estrogens leads to differential induction of messenger ribonucleic acid variants in ovarian and breast cancer cells. Endocrinology 2005, 146, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Aksoy, A.; Artas, G.; Kuloglu, T.; Koc, M. Fibulin1 and mesothelin expressions in pancreas ductal adenocarcinoma. N. Clin. Istanb. 2023, 10, 314–321. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Zhang, J.; Guo, Q.; Wang, Q.; Zhao, H.; Xiao, F.; Han, M.; Cao, Y.; Ding, R.; Yang, A.; et al. Fibulin-1 deficiency alleviates liver fibrosis by inhibiting hepatic stellate cell activation via the p38 MAPK pathway. Cell. Mol. Life Sci. 2025, 82, 192. [Google Scholar] [CrossRef]
- Hessmann, E.; Buchholz, S.M.; Demir, I.E.; Singh, S.K.; Gress, T.M.; Ellenrieder, V.; Neesse, A. Microenvironmental Determinants of Pancreatic Cancer. Physiol. Rev. 2020, 100, 1707–1751. [Google Scholar] [CrossRef] [PubMed]
- Aspberg, A.; Adam, S.; Kostka, G.; Timpl, R.; Heinegård, D. Fibulin-1 is a ligand for the C-type lectin domains of aggrecan and versican. J. Biol. Chem. 1999, 274, 20444–20449. [Google Scholar] [CrossRef]

| No. | Protein Name | Spectral Counting | Fold Change (Rsc) | |||
|---|---|---|---|---|---|---|
| HPNE | BxPC-3 | PANC-1 | BxpC-3 | PANC-1 | ||
| 1 | Fibronectin | 109 | 1 | 0 | −5.814 | −6.532 |
| 2 | Plasminogen activator inhibitor 1 | 20 | 0 | 0 | −4.267 | −4.136 |
| 3 | Collagen alpha-1(I) chain | 10 | 0 | 0 | −3.347 | −3.216 |
| 4 | Fibulin-1 | 3 | 0 | 0 | −1.941 | −1.810 |
| 5 | Serpin I2 | 3 | 0 | 0 | −1.941 | −1.810 |
| 6 | Collagen alpha-3(VI) chain | 3 | 0 | 0 | −1.941 | −1.810 |
| 7 | Gelsolin | 9 | 2 | 2 | −1.833 | −1.702 |
| 8 | Collagen alpha-1(III) chain | 2 | 0 | 0 | −1.554 | −1.422 |
| 9 | Dermcidin | 12 | 4 | 3 | −1.512 | −1.686 |
| 10 | Calreticulin | 14 | 6 | 4 | −1.249 | −1.584 |
| 11 | High mobility group protein B1 | 2 | 7 | 6 | 1.170 | 1.115 |
| 12 | Cathepsin D | 0 | 3 | 8 | 1.592 | 2.846 |
| 13 | Glucose-6-phosphate isomerase | 2 | 18 | 9 | 2.395 | 1.615 |
| 14 | 14-3-3 protein sigma | 0 | 33 | 3 | 4.608 | 1.723 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, T.; Boku, S.; Mitamura, K.; Taga, A. Shotgun Label-Free Proteomic Analysis for Identification of a Potential Diagnostic Biomarker for Pancreatic Cancer. Biomedicines 2025, 13, 2631. https://doi.org/10.3390/biomedicines13112631
Yamamoto T, Boku S, Mitamura K, Taga A. Shotgun Label-Free Proteomic Analysis for Identification of a Potential Diagnostic Biomarker for Pancreatic Cancer. Biomedicines. 2025; 13(11):2631. https://doi.org/10.3390/biomedicines13112631
Chicago/Turabian StyleYamamoto, Tetsushi, Shogen Boku, Kuniko Mitamura, and Atsushi Taga. 2025. "Shotgun Label-Free Proteomic Analysis for Identification of a Potential Diagnostic Biomarker for Pancreatic Cancer" Biomedicines 13, no. 11: 2631. https://doi.org/10.3390/biomedicines13112631
APA StyleYamamoto, T., Boku, S., Mitamura, K., & Taga, A. (2025). Shotgun Label-Free Proteomic Analysis for Identification of a Potential Diagnostic Biomarker for Pancreatic Cancer. Biomedicines, 13(11), 2631. https://doi.org/10.3390/biomedicines13112631







