Nanopore Sequencing Reveals Novel Alternative Splice Variants of EZH2 in Pediatric Medulloblastoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. RNA Purification
2.3. cDNA Synthesis
2.4. RT–PCR
2.5. PCR Product Purification
2.6. Library Preparation and Nanopore Sequencing
2.7. Bioinformatic Analysis of Long-Read Sequencing Data
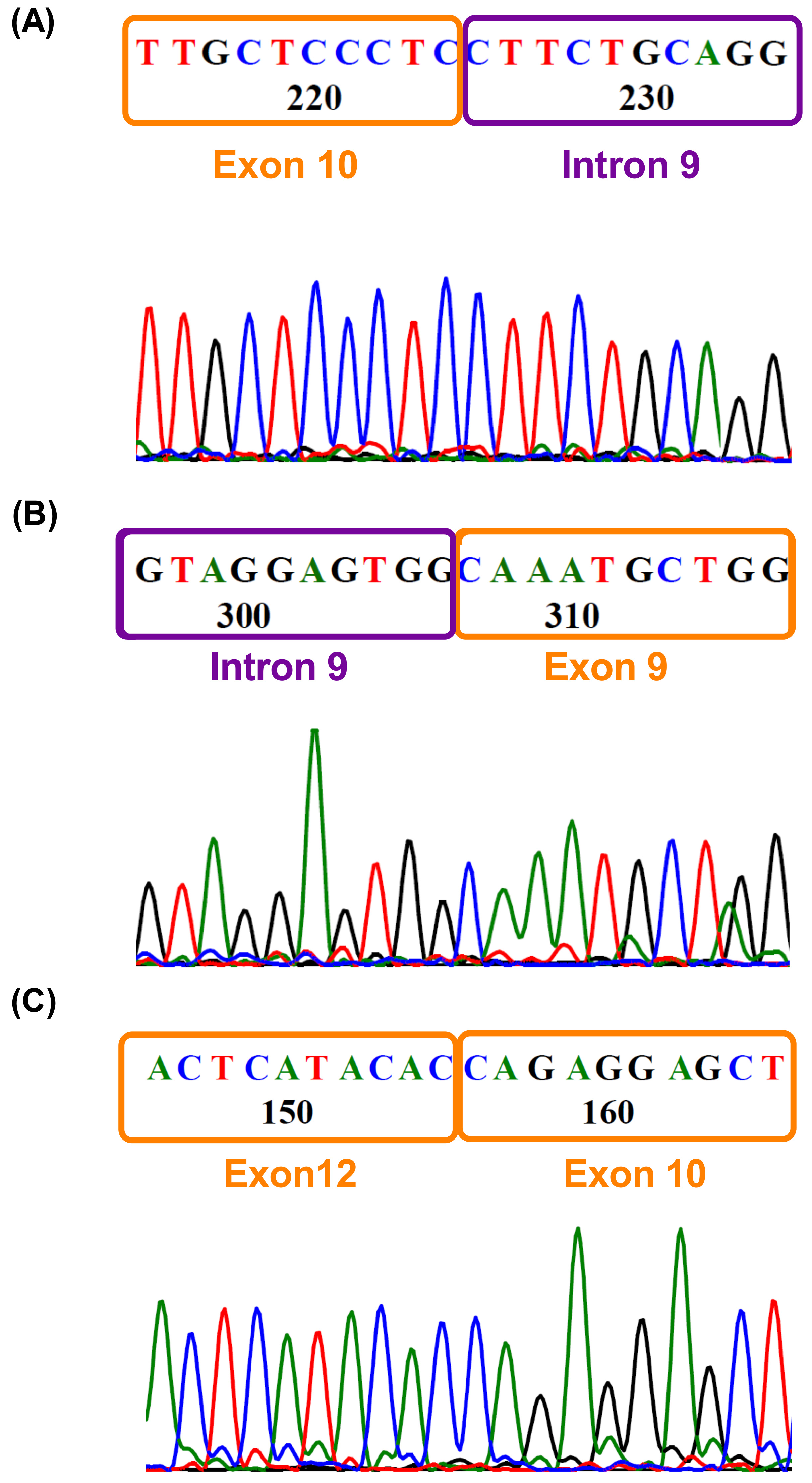
2.8. Sanger Sequencing
2.9. RNAseq Data Mining and Bioinformatic Analysis
2.10. Digital PCR
3. Results
3.1. Patient Cohort
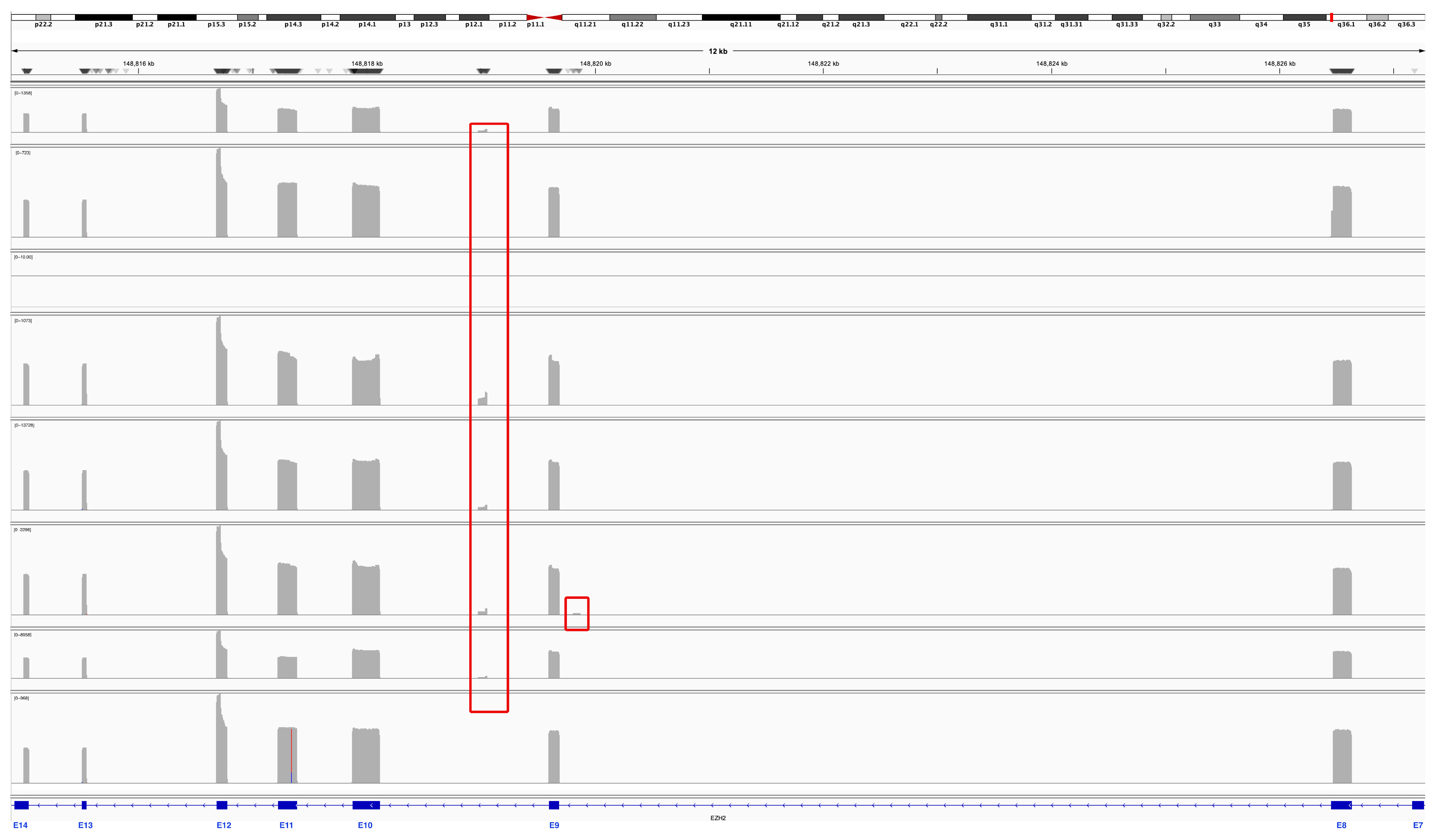
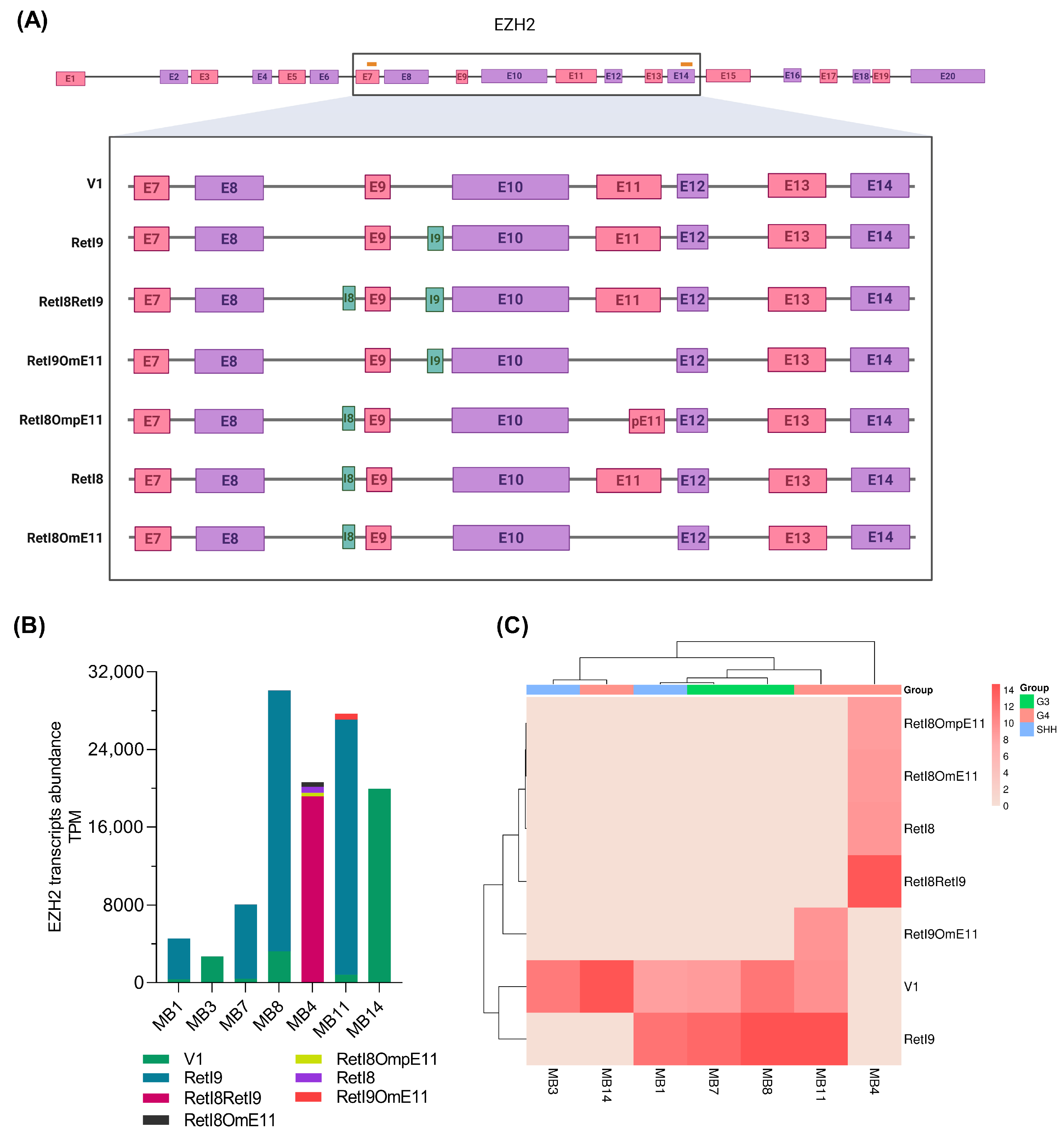
3.2. Nanopore Sequencing Reveals New Junction Patterns in EZH2 Transcripts Expressed in Childhood Medulloblastoma
3.3. Expression Profile of Alternative EZH2 Transcripts
3.4. Validation of New EZH2 Transcripts
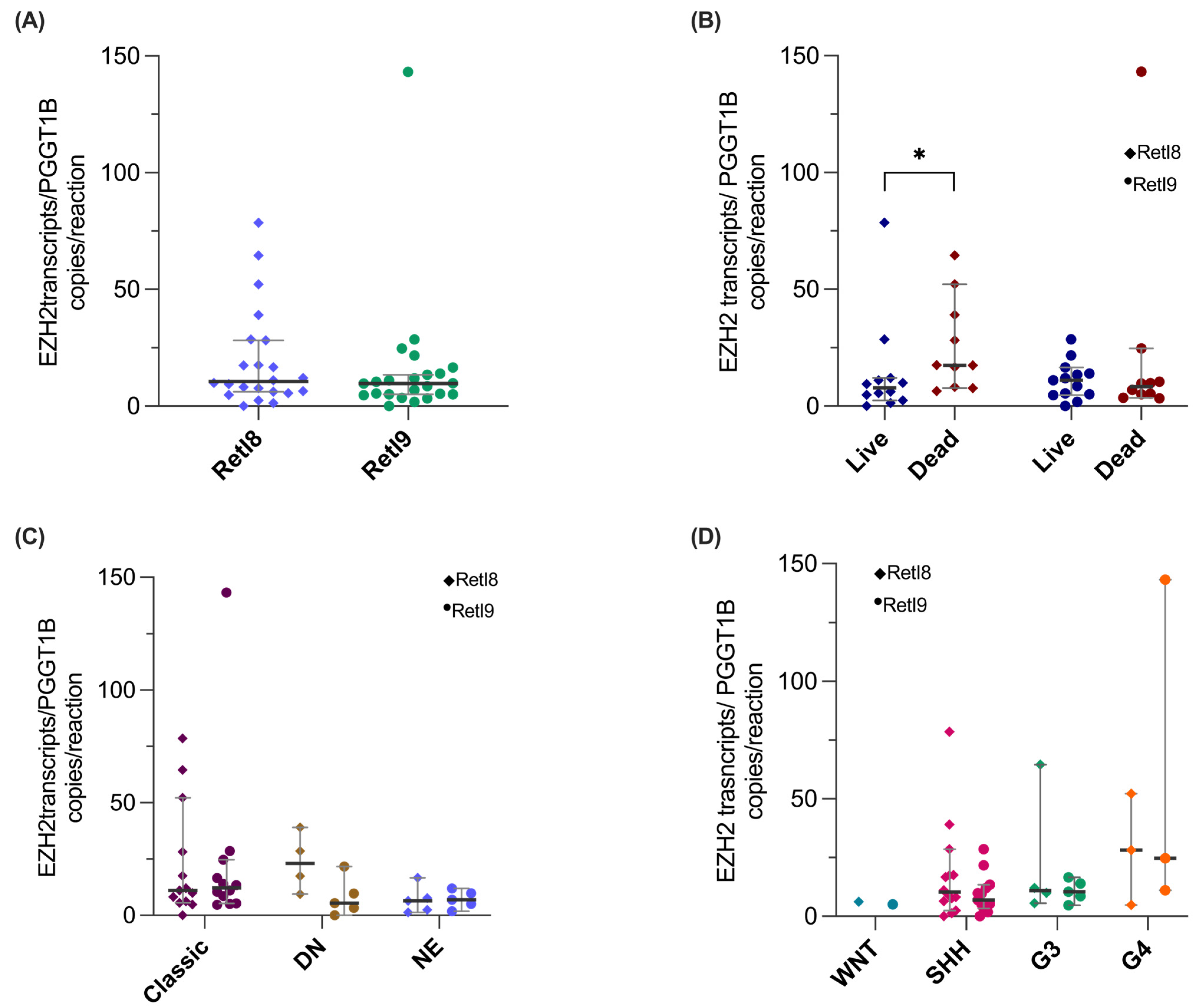
3.5. Quantification of the Novel EZH2 Transcripts RetI8 and RetI9 in Patients with Medulloblastoma
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Girardi, F.; Di Carlo, V.; Stiller, C.; Gatta, G.; Woods, R.R.; Visser, O.; Lacour, B.; Tucker, T.C.; Coleman, M.P.; Allemani, C. Global survival trends for brain tumors, by histology: Analysis of individual records for 67,776 children diagnosed in 61 countries during 2000–2014 (CONCORD-3). Neuro-Oncology 2023, 25, 593–606. [Google Scholar] [CrossRef]
- Rutkowski, S.; Bode, U.; Deinlein, F.; Ottensmeier, H.; Warmuth-Metz, M.; Soerensen, N.; Graf, N.; Emser, A.; Pietsch, T.; Wolff, J.E.; et al. Treatment of Early Childhood Medulloblastoma by Postoperative Chemotherapy Alone. N. Engl. J. Med. 2005, 352, 978–986. [Google Scholar] [CrossRef]
- Amayiri, N.; Swaidan, M.; Ibrahimi, A.; Hirmas, N.; Musharbash, A.; Bouffet, E.; Al-Hussaini, M.; Ramaswamy, V. Molecular Subgroup Is the Strongest Predictor of Medulloblastoma Outcome in a Resource-Limited Country. JCO Glob. Oncol 2021, 7, 1442–1453. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231. [Google Scholar] [CrossRef]
- Triscott, J.; Yip, S.; Johnston, D.; Michaud, J.; Rassekh, S.R.; Hukin, J.; Dunn, S.; Dunham, C. Histologic Correlates of Molecular Group 4 Pediatric Medulloblastoma: A Retrospective Canadian Review. Pediatr. Dev. Pathol. 2021, 24, 309–317. [Google Scholar] [CrossRef]
- Babaoglu, B.; Hanalioglu, S.; Varan, A.; Oguz, K.K.; Bilginer, B.; Dolgun, A.; Soylemezoglu, F. Molecular subgrouping based on immunohistochemistry in medulloblastoma: A single-center experience. Turk. Neurosurg. 2024, 34, 999–1008. [Google Scholar] [CrossRef]
- Harry, J.L.; Shezi, N.B.; Mwazha, A. Molecular classification of medulloblastoma using immunohistochemistry: A single centre study. Ann. Diagn. Pathol. 2025, 76, 152463. [Google Scholar] [CrossRef]
- Tauziède-Espariat, A.; Huybrechts, S.; Indersie, E.; Dufour, C.; Puget, S.; Chivet, A.; Roux, A.; Pagès, M.; Gareton, A.; Chrétien, F.; et al. Diagnostic Accuracy of a Reduced Immunohistochemical Panel in Medulloblastoma Molecular Subtyping, Correlated to DNA-methylation Analysis. Am. J. Surg. Pathol. 2021, 45, 558–566. [Google Scholar] [CrossRef] [PubMed]
- Williamson, D.; Schwalbe, E.C.; Hicks, D.; Aldinger, K.A.; Lindsey, J.C.; Crosier, S.; Richardson, S.; Goddard, J.; Hill, R.M.; Castle, J.; et al. Medulloblastoma group 3 and 4 tumors comprise a clinically and biologically significant expression continuum reflecting human cer-ebellar development. Cell Rep. 2022, 40, 111162. [Google Scholar] [CrossRef] [PubMed]
- Hovestadt, V.; Jones, D.T.W.; Picelli, S.; Wang, W.; Kool, M.; Northcott, P.A.; Sultan, M.; Stachurski, K.; Ryzhova, M.; Warnatz, H.-J.; et al. Decoding the regulatory landscape of medulloblastoma using DNA methylation sequencing. Nature 2014, 510, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Zhang, Y. functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr. Opin. Genet. Dev. 2004, 14, 155–164. [Google Scholar] [CrossRef]
- Xu, K.; Wu, Z.J.; Groner, A.C.; He, H.H.; Cai, C.; Lis, R.T.; Wu, X.; Stack, E.C.; Loda, M.; Liu, T.; et al. EZH2 Oncogenic Activity in Castration-Resistant Prostate Cancer Cells Is Polycomb-Independent. Science 2012, 338, 1465–1469. [Google Scholar] [CrossRef]
- Kim, E.; Kim, M.; Woo, D.-H.; Shin, Y.; Shin, J.; Chang, N.; Oh, Y.T.; Kim, H.; Rheey, J.; Nakano, I.; et al. Phosphorylation of EZH2 Activates STAT3 Signaling via STAT3 Methylation and Promotes Tumorigenicity of Glioblastoma Stem-like Cells. Cancer Cell 2013, 23, 839–852. [Google Scholar] [CrossRef] [PubMed]
- He, A.; Shen, X.; Ma, Q.; Cao, J.; von Gise, A.; Zhou, P.; Wang, G.; Marquez, V.E.; Orkin, S.H.; Pu, W.T. PRC2 directly methylates GATA4 and represses its transcriptional activity. Genes. Dev. 2012, 26, 37–42. [Google Scholar] [CrossRef]
- Wagener, N.; Macher-Goeppinger, S.; Pritsch, M.; Hüsing, J.; Hoppe-Seyler, K.; Schirmacher, P.; Pfitzenmaier, J.; Haferkamp, A.; Hoppe-Seyler, F.; Hohenfellner, M. Enhancer of zeste homolog 2 (EZH2) expression is an independent prognostic factor in renal cell carcinoma. BMC Cancer 2010, 10, 524. [Google Scholar] [CrossRef] [PubMed]
- Kunju, L.P.; Cookingham, C.; A Toy, K.; Chen, W.; Sabel, M.S.; Kleer, C.G. EZH2 and ALDH-1 mark breast epithelium at risk for breast cancer development. Mod. Pathol. 2011, 24, 786–793. [Google Scholar] [CrossRef]
- Varambally, S.; Dhanasekaran, S.M.; Zhou, M.; Barrette, T.R.; Kumar-Sinha, C.; Sanda, M.G.; Ghosh, D.; Pienta, K.J.; Sewalt, R.G.A.B.; Otte, A.P.; et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 2002, 419, 624–629. [Google Scholar] [CrossRef]
- Smits, M.; van Rijn, S.; Hulleman, E.; Biesmans, D.; van Vuurden, D.G.; Kool, M.; Haberler, C.; Aronica, E.; Vandertop, W.P.; Noske, D.P.; et al. EZH2-regulated DAB2IP is a medulloblastoma tumor suppressor and a positive marker for survival. Clin. Cancer Res. 2012, 18, 4048–4058. [Google Scholar] [CrossRef]
- Vo, B.H.T.; Li, C.; Morgan, M.A.; Theurillat, I.; Finkelstein, D.; Wright, S.; Hyle, J.; Smith, S.M.; Fan, Y.; Wang, Y.-D.; et al. Inactivation of Ezh2 Upregulates Gfi1 and Drives Aggressive Myc-Driven Group 3 Medulloblastoma. Cell Rep. 2017, 18, 2907–2917. [Google Scholar] [CrossRef] [PubMed]
- Miele, E.; Valente, S.; Alfano, V.; Silvano, M.; Mellini, P.; Borovika, D.; Marrocco, B.; Po, A.; Besharat, Z.M.; Catanzaro, G.; et al. The histone methyltransferase EZH2 as a druggable target in SHH medulloblastoma cancer stem cells. Oncotarget 2017, 8, 68557. [Google Scholar] [CrossRef]
- Nikoloski, G.; Langemeijer, S.M.C.; Kuiper, R.P.; Knops, R.; Massop, M.; Tönnissen, E.R.L.T.M.; van der Heijden, A.; Scheele, T.N.; Vandenberghe, P.; de Witte, T.; et al. Somatic mutations of the histone methyltransferase gene EZH2 in myelodysplastic syndromes. Nat. Genet. 2010, 42, 665–667. [Google Scholar] [CrossRef]
- Ernst, T.; Chase, A.J.; Score, J.; Hidalgo-Curtis, C.E.; Bryant, C.; Jones, A.V.; Waghorn, K.; Zoi, K.; Ross, F.M.; Reiter, A.; et al. Inactivating mutations of the his-tone methyltransferase gene EZH2 in myeloid disorders. Nat. Genet. 2010, 42, 722–726. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The mutational landscape of head and neck squamous cell carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Brunmeir, R.; Ying, L.; Yan, J.; Hee, Y.T.; Lin, B.; Kaur, H.; Leong, Q.Z.; Teo, W.W.; Choong, G.; Jen, W.-Y.; et al. EZH2 modulates mRNA splicing and exerts part of its oncogenic function through repression of splicing factors in CML: CHRONIC MYELOGENOUS LEUKE-MIA. Leukemia 2025, 39, 650–662. [Google Scholar] [CrossRef]
- Chen, K.; Xiao, H.; Zeng, J.; Yu, G.; Zhou, H.; Huang, C.; Yao, W.; Xiao, W.; Hu, J.; Guan, W.; et al. Alternative splicing of EZH2 pre-mRNA by SF3B3 contributes to the tumorigenic potential of renal cancer. Clin. Cancer Res. 2017, 23, 3428–3441. [Google Scholar] [CrossRef]
- Wang, X.; Liu, W.; Zhan, C.; Zhang, Y.; Li, X.; Wang, Y.; Sheng, M.; Maqsood, M.; Shen, H.; Liang, A.; et al. Alternative splicing of EZH2 regulated by SNRPB me-diates hepatocellular carcinoma progression via BMP2 signaling pathway. iScience 2025, 28, 111626. [Google Scholar] [CrossRef]
- Rivera, O.D.; Mallory, M.J.; Quesnel-Vallières, M.; Chatrikhi, R.; Schultz, D.C.; Carroll, M.; Barash, Y.; Cherry, S.; Lynch, K.W. Alternative splicing redefines landscape of commonly mutated genes in acute myeloid leukemia. Proc. Natl. Acad. Sci. USA 2021, 118, e2014967118. [Google Scholar] [CrossRef] [PubMed]
- Wang, E.T.; Sandberg, R.; Luo, S.; Khrebtukova, I.; Zhang, L.; Mayr, C.; Kingsmore, S.F.; Schroth, G.P.; Burge, C.B. Alternative isoform regulation in hu-man tissue transcriptomes. Nature 2008, 456, 470–476. [Google Scholar] [CrossRef]
- Sveen, A.; Kilpinen, S.; Ruusulehto, A.; A Lothe, R.; I Skotheim, R. Aberrant RNA splicing in cancer; Expression changes and driver mutations of splicing factor genes. Oncogene 2016, 35, 2413–2427. [Google Scholar] [CrossRef] [PubMed]
- Villegas-Ruíz, V.; Olmos-Valdez, K.; Castro-López, K.A.; Saucedo-Tepanecatl, V.E.; Ramírez-Chiquito, J.C.; Pérez-López, E.I.; Medina-Vera, I.; Juárez-Méndez, S. Identification and Validation of Novel Reference Genes in Acute Lymphoblastic Leuke-mia for Droplet Digital PCR. Genes 2019, 10, 376. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2017, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.-C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved recon-struction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Madeira, F.; Madhusoodanan, N.; Lee, J.; Eusebi, A.; Niewielska, A.; Tivey, A.R.N.; Lopez, R.; Butcher, S. The EMBL-EBI Job Dispatch-er sequence analysis tools framework in 2024. Nucleic Acids Res. 2024, 52, W521–W525. [Google Scholar] [CrossRef]
- Tang, D.; Chen, M.; Huang, X.; Zhang, G.; Zeng, L.; Zhang, G.; Wu, S.; Wang, Y. SRplot: A free online platform for data visuali-zation and graphing. PLoS ONE 2023, 18, e0294236. [Google Scholar] [CrossRef]
- Jiménez-Ávila, C.E.; Villegas-Ruíz, V.; Zapata-Tarres, M.; Rubio-Portillo, A.E.; López, E.I.P.; Zenteno, J.C.; Juarez-Mendez, S. Cen-tromere-associated protein E expresses a novel mRNA isoform in acute lymphoblastic leukemia. Int. J. Mol. Epidemiol. Genet. 2018, 9, 43. [Google Scholar] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Hands-On: Genome-Wide Alternative Splicing Analysis Genome-Wide Alternative Splicing Analysis/Tran-Scriptomics n.d. Available online: https://training.galaxyproject.org/topics/transcriptomics/tutorials/differential-isoform-expression/tutorial.html#rna-seq-mapping-and-evaluation (accessed on 25 August 2025).
- Jiang, T.; Zhang, Y.; Wang, J.; Du, J.; Raynald; Qiu, X.; Wang, Y.; Li, C. A retrospective study of progression-free and overall survival in pediatric medulloblastoma based on molecular subgroup classification: A single-institution expe-rience. Front. Neurol. 2017, 8, 198. [Google Scholar] [CrossRef]
- Kim, J.W.; Park, S.-H.; Choi, S.A.; Kim, S.-K.; Koh, E.J.; Won, J.-K.; Nam, S.M.; Phi, J.H. Molecular subgrouping of medulloblastoma in pediatric population using the NanoString assay and comparison with immunohistochemistry methods. BMC Cancer 2022, 22, 1221. [Google Scholar] [CrossRef]
- Lamba, N.; Groves, A.; Torre, M.; Yeo, K.K.; Iorgulescu, J.B. The epidemiology of primary and metastatic brain tu-mors in infancy through childhood. J. Neurooncol. 2022, 156, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Schwalbe, E.C.; Lindsey, J.C.; Nakjang, S.; Crosier, S.; Smith, A.J.; Hicks, D.; Rafiee, G.; Hill, R.M.; Iliasova, A.; Stone, T.; et al. Novel molecular subgroups for clin-ical classification and outcome prediction in childhood medulloblastoma: A cohort study. Lancet Oncol. 2017, 18, 958–971. [Google Scholar] [CrossRef] [PubMed]
- Kaur, K.; Jha, P.; Pathak, P.; Suri, V.; Sharma, M.C.; Garg, A.; Suri, A.; Sarkar, C. Approach to molecular subgrouping of medullo-blastomas: Comparison of NanoString nCounter assay versus combination of immunohistochemistry and fluorescence in-situ hybridization in resource constrained centres. J. Neurooncol. 2019, 143, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Rivera-Luna, R.; López, E.; Rivera-Marquez, H.; Rivera-Ortegón, F.; Altamirano-Alvarez, E.; Mercado, G.; Covarrubias, G.; Rueda-Franco, F.; Marhx-Bracho, A.; Gutiérrez, P. Survival of children under 3 years old with medulloblastoma: A study from the Mexican Cooperative Group for Childhood Malignancies (AMOHP). Child’s Nerv. Syst. 2002, 18, 38–42. [Google Scholar] [CrossRef]
- López-Aguilar, E.; Sepúlveda-Vildósola, A.C.; Rivera-Márquez, H.; Siordia-Reyes, G.; Betanzos-Cabrera, Y.; Cerecedo-Díaz, F.; del Angel, V.W.; Diegopérez-Ramírez, J. Clinical and Molecular Parameters for Risk Stratification in Mexican Children with Me-dulloblastoma. Arch. Med. Res. 2007, 38, 769–773. [Google Scholar] [CrossRef]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e6. [Google Scholar] [CrossRef]
- Northcott, P.A.; Korshunov, A.; Witt, H.; Hielscher, T.; Eberhart, C.G.; Mack, S.; Bouffet, E.; Clifford, S.C.; Hawkins, C.E.; French, P.; et al. Medulloblastoma Comprises Four Distinct Molecular Variants. J. Clin. Oncol. 2010, 29, 1408. [Google Scholar] [CrossRef]
- Kool, M.; Korshunov, A.; Remke, M.; Jones, D.T.; Schlanstein, M.; Northcott, P.A.; Cho, Y.-J.; Koster, J.; Schouten-van Meeteren, A.; Van Vuurden, D.; et al. Molecular subgroups of medulloblastoma: An international meta-analysis of transcriptome, genetic aberrations, and clinical data of WNT, SHH, Group 3, and Group 4 medulloblastomas. Acta Neuropathol. 2012, 123, 473–484. [Google Scholar] [CrossRef]
- Northcott, P.A.; Shih, D.J.H.; Peacock, J.; Garzia, L.; Sorana Morrissy, A.; Zichner, T.; Stütz, A.M.; Korshunov, A.; Reimand, J.; Schumacher, S.E.; et al. Subgroup-specific struc-tural variation across 1,000 medulloblastoma genomes. Nature 2012, 487, 49–56. [Google Scholar] [CrossRef]
- Alimova, I.; Venkataraman, S.; Harris, P.; Marquez, V.E.; Northcott, P.A.; Dubuc, A.; Taylor, M.D.; Foreman, N.K.; Vibhakar, R. Targeting the enhancer of zeste homologue 2 in medulloblastoma. Int. J. Cancer 2012, 131, 1800–1809. [Google Scholar] [CrossRef]
- Qi, L.; Lindsay, H.; Kogiso, M.; Du, Y.; Braun, F.K.; Zhang, H.; Guo, L.; Zhao, S.; Injac, S.G.; Baxter, P.A.; et al. Evaluation of an EZH2 inhibitor in pa-tient-derived orthotopic xenograft models of pediatric brain tumors alone and in combination with chemo- and radiation therapies. Lab. Investig. 2022, 102, 185–193. [Google Scholar] [CrossRef]
- Shishido, K.; Purvis, I.J.; Velpula, K.K.; Venkataraman, S.; Vibhakar, R.; Asuthkar, S. Targeting B7-H3 through EZH2 inhibition in MYC-positive Group 3 medulloblastoma. Oncol. Rep. 2023, 49, 119. [Google Scholar] [CrossRef]
- Wu, H.; Zeng, H.; Dong, A.; Li, F.; He, H.; Senisterra, G.; Seitova, A.; Duan, S.; Brown, P.J.; Vedadi, M.; et al. Structure of the catalytic domain of EZH2 reveals conformational plasticity in cofactor and substrate binding sites and explains oncogenic mutations. PLoS ONE 2013, 8, e83737. [Google Scholar] [CrossRef]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent mutation of his-tone-modifying genes in non-Hodgkin lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef] [PubMed]
- Ntziachristos, P.; Tsirigos, A.; Van Vlierberghe, P.; Nedjic, J.; Trimarchi, T.; Flaherty, M.S.; Ferres-Marco, D.; Da Ros, V.G.; Tang, Z.; Siegle, J.; et al. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 2012, 18, 296–301. [Google Scholar] [CrossRef] [PubMed]
- Li, L.Y. EZH2: Novel therapeutic target for human cancer. Biomedicine 2014, 4, 1. [Google Scholar] [CrossRef]
- Tan, J.Z.; Yan, Y.; Wang, X.X.; Jiang, Y.; Xu, H.E. EZH2: Biology, disease, and structure-based drug discovery. Acta Pharmacol. Sin. 2014, 35, 161–174. [Google Scholar] [CrossRef]
- Gibson, W.T.; Hood, R.L.; Zhan, S.H.; Bulman, D.E.; Fejes, A.P.; Moore, R.; Mungall, A.J.; Eydoux, P.; Babul-Hirji, R.; An, J.; et al. Mutations in EZH2 cause weaver syn-drome. Am. J. Hum. Genet. 2012, 90, 110–118. [Google Scholar] [CrossRef]
- Kim, E.; Ilagan, J.O.; Liang, Y.; Daubner, G.M.; Lee, S.C.W.; Ramakrishnan, A.; Li, Y.; Chung, Y.R.; Micol, J.-B.; Murphy, M.E.; et al. Mutations Contribute to Myelodysplasia by Mutant-Specific Effects on Exon Recognition. Cancer Cell 2015, 27, 617–630. [Google Scholar] [CrossRef]
- Li, B.; Chng, W.J. EZH2 abnormalities in lymphoid malignancies: Underlying mechanisms and therapeutic im-plications. J. Hematol. Oncol. 2019, 12, 118. [Google Scholar] [CrossRef] [PubMed]
- Boyer, L.A.; Latek, R.R.; Peterson, C.L. The SANT domain: A unique histone-tail-binding module? Nat. Rev. Mol. Cell Biol. 2004, 5, 158–163. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, Y.; Malcovati, L.; Gallì, A.; Sato-Otsubo, A.; Kataoka, K.; Sato, Y.; Watatani, Y.; Suzuki, H.; Yoshizato, T.; Yoshida, K.; et al. Aberrant splicing and defective mRNA production induced by somatic spliceosome mutations in myelodysplasia. Nat. Commun. 2018, 9, 3649. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, D.; Zhang, Z.; Kaluz, S.; Yu, B.; Devi, N.S.; Olson, J.J.; Van Meir, E.G. EZH2 targeting in medulloblastoma reduces tumor growth through epigenetic reactivation of the BAI1/p53 tumor suppressor pathway. Oncogene 2019, 39, 1041. [Google Scholar] [CrossRef]
- Kurmasheva, R.T.; Sammons, M.; Favours, E.; Wu, J.; Kurmashev, D.; Cosmopoulos, K.; Keilhack, H.; Klaus, C.R.; Houghton, P.J.; Smith, M.A. Initial testing (stage 1) of tazemetostat (EPZ-6438), a novel EZH2 inhibitor, by the Pediatric Preclinical Testing Program. Pediatr. Blood Cancer 2017, 64, e26218. [Google Scholar] [CrossRef]
- Poulikakos, P.I.; Persaud, Y.; Janakiraman, M.; Kong, X.; Ng, C.; Moriceau, G.; Shi, H.; Atefi, M.; Titz, B.; Gabay, M.T.; et al. RAF inhibitor resistance is medi-ated by dimerization of aberrantly spliced BRAF(V600E). Nature 2011, 480, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.D.; Ceniccola, K.; Hwang, S.; Andrawis, R.; Horvath, A.; Freedman, J.A.; Olender, J.; Knapp, S.; Ching, T.; Garmire, L.; et al. Alternative splicing promotes tumour aggressiveness and drug resistance in African American prostate cancer. Nat. Commun. 2017, 8, 15921. [Google Scholar] [CrossRef]
- Sotillo, E.; Barrett, D.M.; Black, K.L.; Bagashev, A.; Oldridge, D.; Wu, G.; Sussman, R.; LaNauze, C.; Ruella, M.; Gazzara, M.R.; et al. Convergence of acquired mutations and alternative splicing of CD19 enables resistance to CART-19 immunotherapy. Cancer Discov. 2015, 5, 1282–1295. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.D.; Lee, N.H. Aberrant RNA Splicing in Cancer and Drug Resistance. Cancers 2018, 10, 458. [Google Scholar] [CrossRef] [PubMed]

| Gene | Sequence (5′->3′) | Tm (°C) | Product Size (bp) |
|---|---|---|---|
| EZH2_RetI8 | Fw CTAGAGCTGTTTCTGTGTTCT Rv CAGTAAGAGCCTGAAGGAAAG | 60 60 | 84 |
| EZH2_RetI9 | Fw CCAGCATTTGCCACTCCTACCT Rv TTTGCTCCCTCCTTCTGCAGGT | 60 60 | 103 |
| PGGT1B | Fw CTGTGGTTTCCGAGGCTCTT Rv GCATGAGAGGCCAGTGTAGG | 60 60 | 108 |
| Medulloblastoma Patients | N = 23 (100%) |
|---|---|
| Gender | |
| Male | 16 (69.6%) |
| Female | 7 (30.4%) |
| Age | |
| Age at diagnosis (median, range) | 6 (0.5–15) years |
| Most common age at diagnosis | 1 year (13%) |
| ≤3 years | 8 (34.8%) |
| >3 years | 15 (65.2%) |
| Molecular group | |
| WNT | 1 (4.4%) |
| SHH | 14 (60.9%) |
| G3 | 5 (21.7%) |
| G4 | 3 (13.0%) |
| Histological group | |
| Classic | 13 (56.6%) |
| Nodular Desmoplastic | 5 (21.7%) |
| Extensive nodularity | 5 (21.7%) |
| Survival | |
| Alive | 13 (56.5%) |
| Dead | 10 (43.5%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramírez-Chiquito, J.C.; Rosete-Ambriz, S.A.; Olguín-García, A.C.; Eguía-Aguilar, M.d.P.; Niembro-Zuñiga, A.M.; Marhx-Bracho, A.; Perezpeña-Diazconti, M.; Juárez-Méndez, S. Nanopore Sequencing Reveals Novel Alternative Splice Variants of EZH2 in Pediatric Medulloblastoma. Biomedicines 2025, 13, 2461. https://doi.org/10.3390/biomedicines13102461
Ramírez-Chiquito JC, Rosete-Ambriz SA, Olguín-García AC, Eguía-Aguilar MdP, Niembro-Zuñiga AM, Marhx-Bracho A, Perezpeña-Diazconti M, Juárez-Méndez S. Nanopore Sequencing Reveals Novel Alternative Splice Variants of EZH2 in Pediatric Medulloblastoma. Biomedicines. 2025; 13(10):2461. https://doi.org/10.3390/biomedicines13102461
Chicago/Turabian StyleRamírez-Chiquito, Josselen Carina, Sergio Antony Rosete-Ambriz, Ana Consuelo Olguín-García, María del Pilar Eguía-Aguilar, Ana Maria Niembro-Zuñiga, Alfonso Marhx-Bracho, Mario Perezpeña-Diazconti, and Sergio Juárez-Méndez. 2025. "Nanopore Sequencing Reveals Novel Alternative Splice Variants of EZH2 in Pediatric Medulloblastoma" Biomedicines 13, no. 10: 2461. https://doi.org/10.3390/biomedicines13102461
APA StyleRamírez-Chiquito, J. C., Rosete-Ambriz, S. A., Olguín-García, A. C., Eguía-Aguilar, M. d. P., Niembro-Zuñiga, A. M., Marhx-Bracho, A., Perezpeña-Diazconti, M., & Juárez-Méndez, S. (2025). Nanopore Sequencing Reveals Novel Alternative Splice Variants of EZH2 in Pediatric Medulloblastoma. Biomedicines, 13(10), 2461. https://doi.org/10.3390/biomedicines13102461





