Evolutionary Insight into Fatal Human Coronaviruses (hCoVs) with a Focus on Circulating SARS-CoV-2 Variants Under Monitoring (VUMs)
Abstract
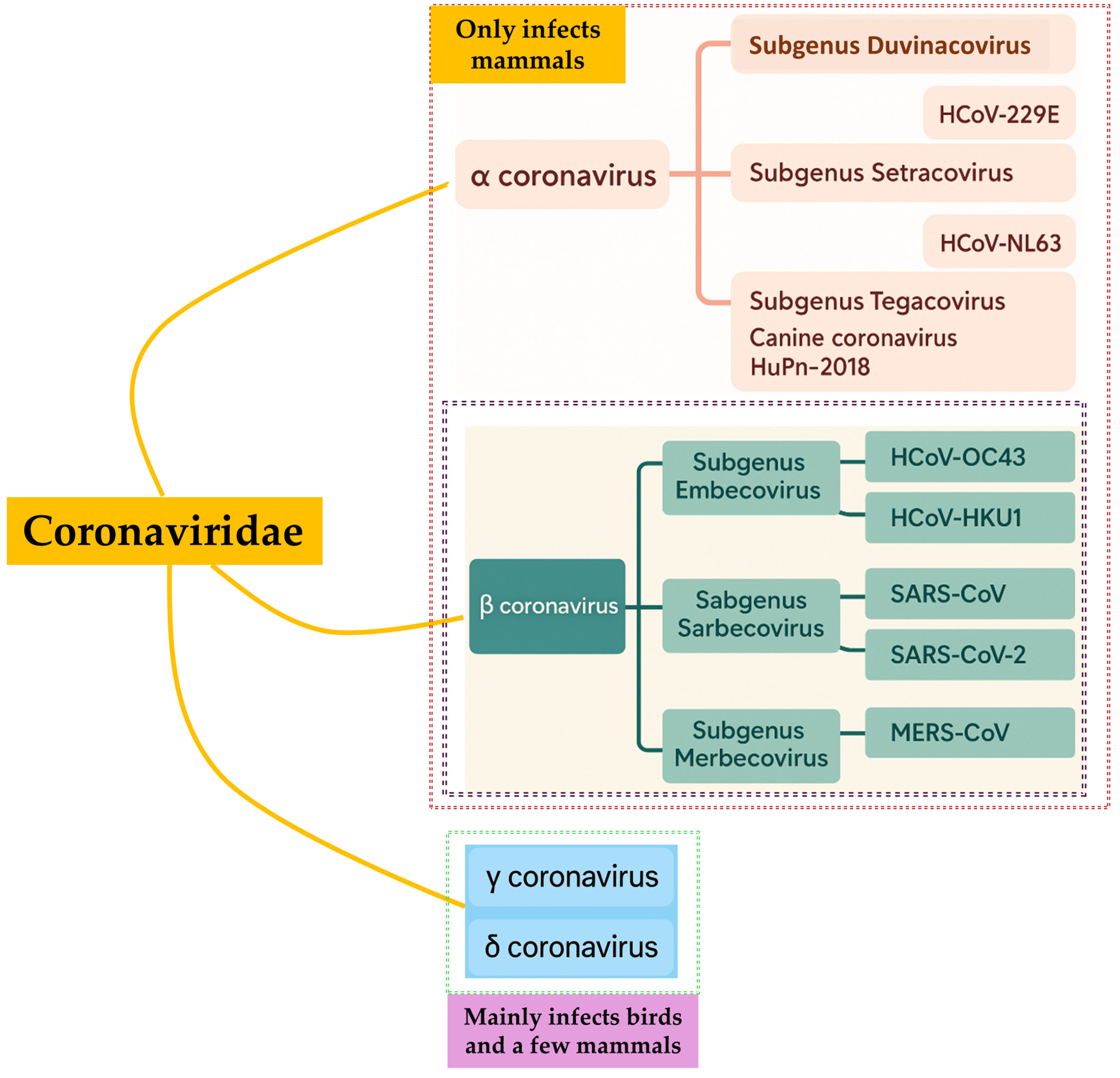
1. Introduction
2. SARS-CoV
2.1. Emergence as an Alarm to a Lurking Pandemic (SARS-CoV-2 Pandemic)
2.2. Genomic Organization
2.3. Evolution in SARS-CoV
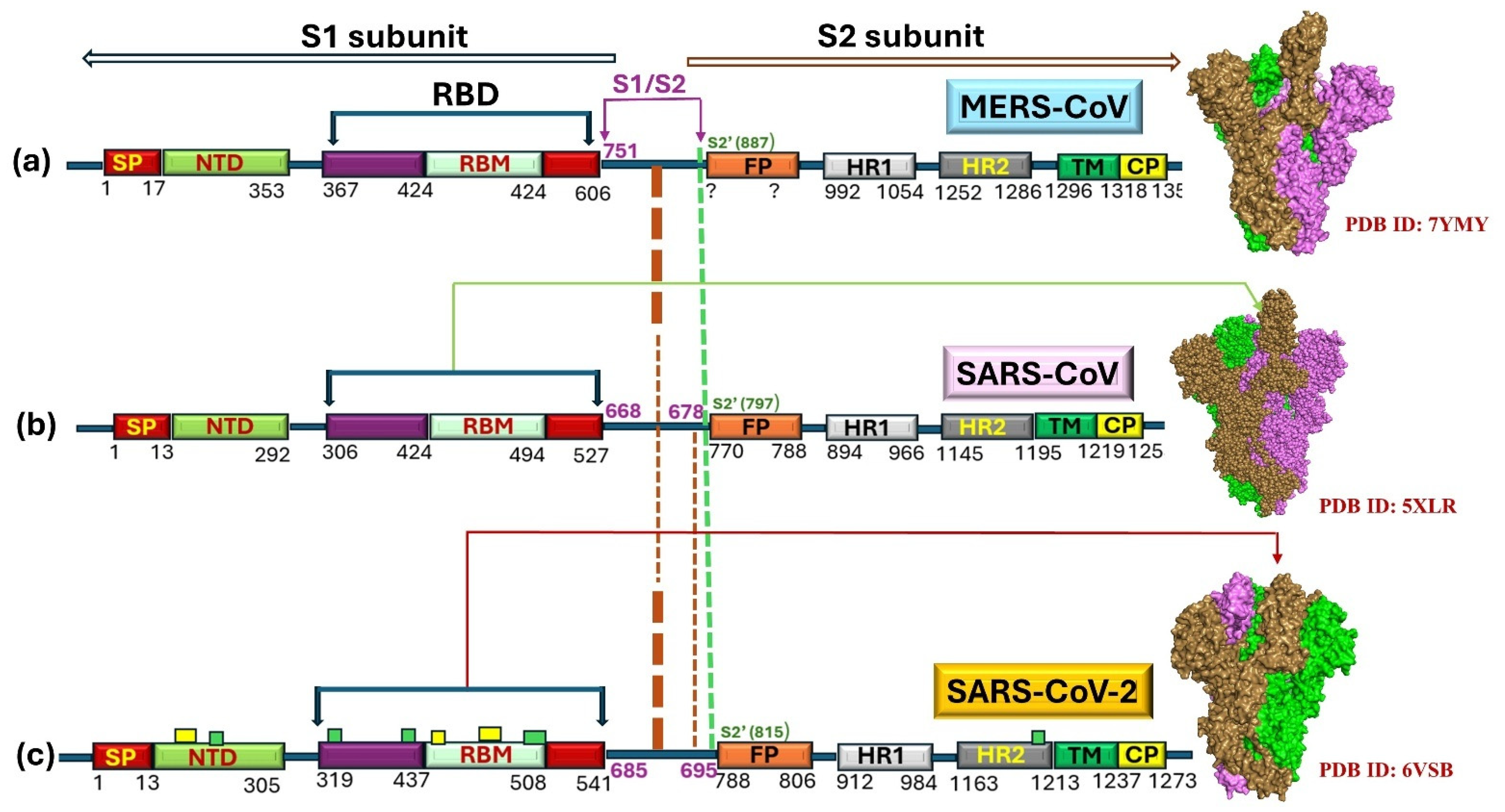
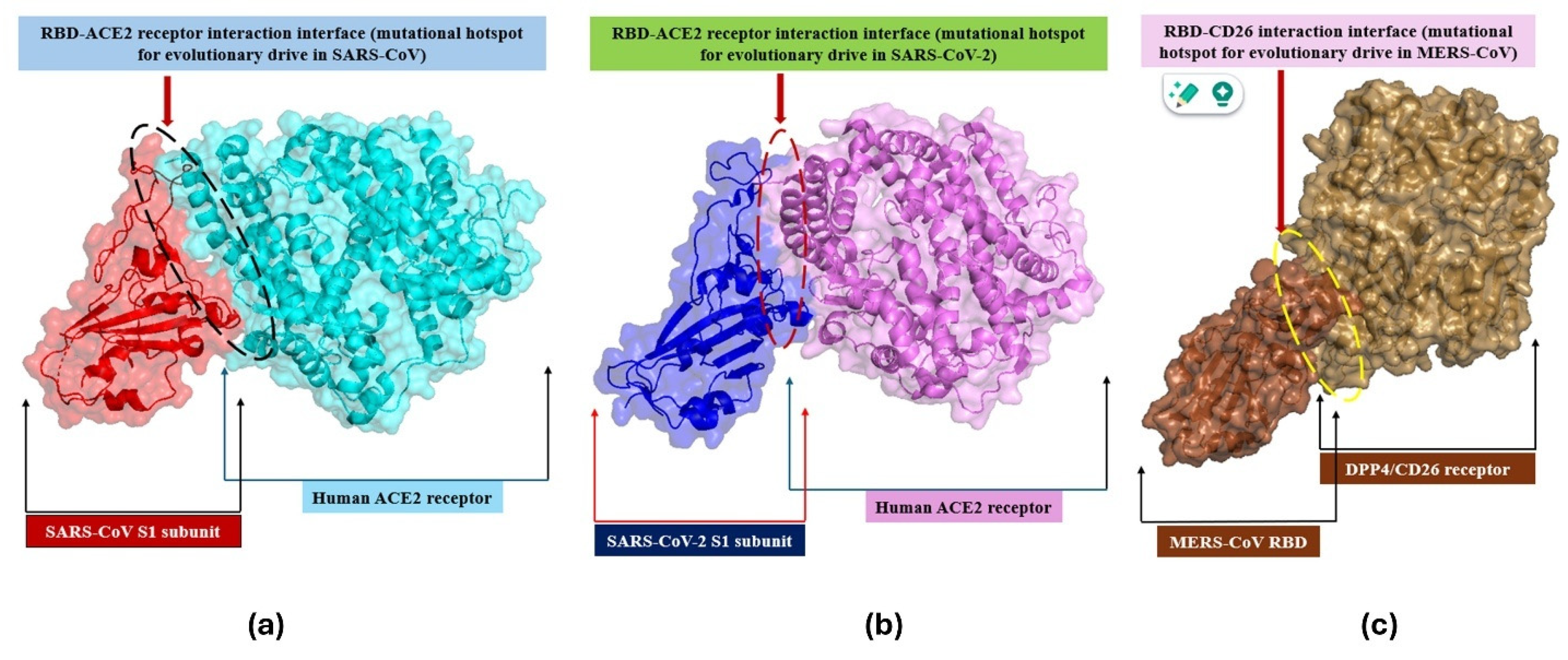
2.4. Evolution in Spike (S)-Protein
2.5. Natural Selection
2.6. Evolutionary Dynamics of SARS-CoV
3. MERS-CoV
3.1. Emergence
3.2. Diversity of MERS-CoV
3.3. Evolution of S-Protein
4. SARS-CoV-2’s Story of Emergence
4.1. Mutational Burden in Spike Protein Fuels Variant Generation and Alters Evolutionary Dynamics
4.2. Evolutionary Progression of SARS-CoV-2 Lineages
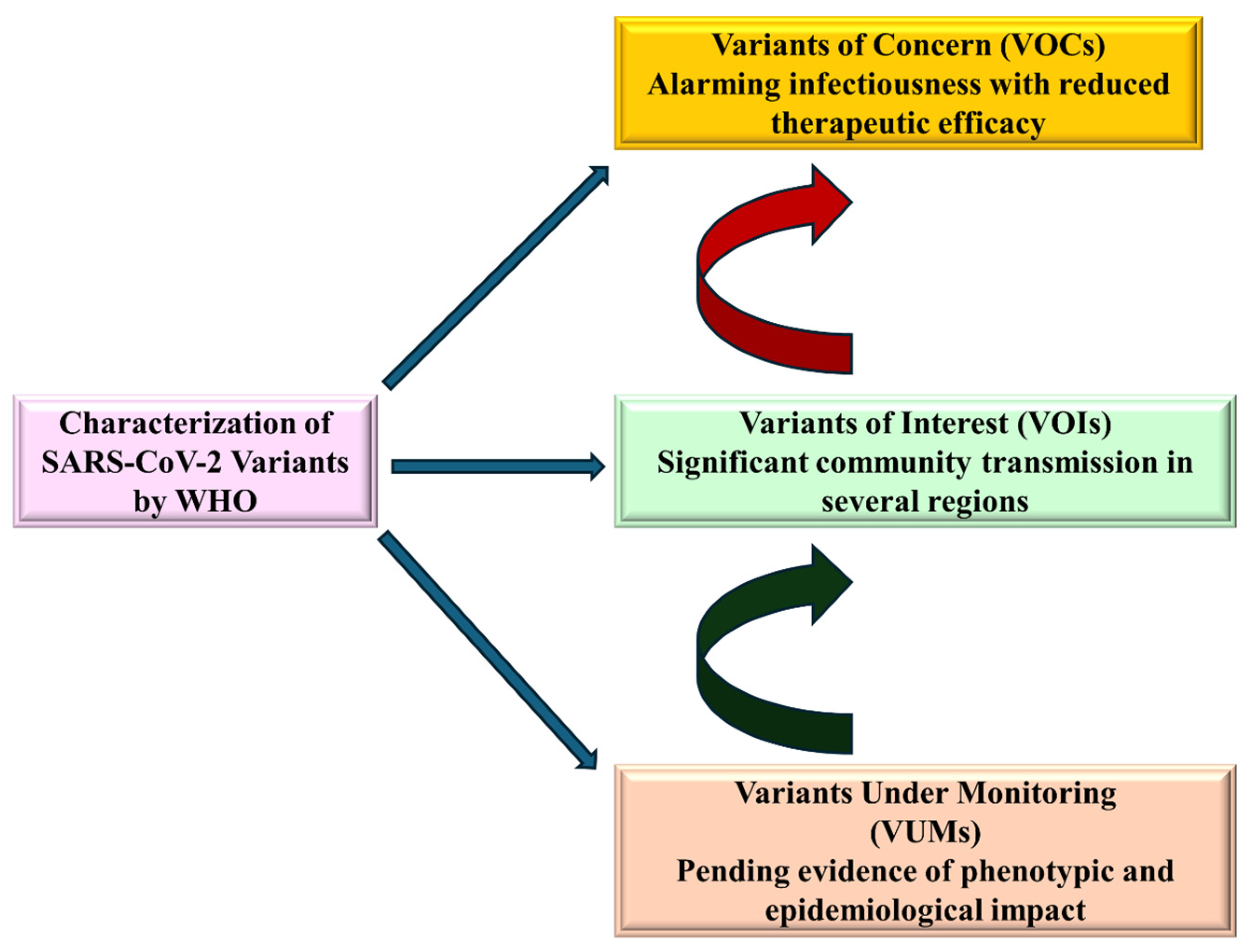
4.3. Circulating Variants of Monitoring (VUMs)
Critical Bioinformatics Analysis of Mutational Mapping and Structural Overlap of RBD of the Recently Evolved NB.1.8.1 Sub-Variant (VUM)
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, D.; Yi, S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shen, L.; Gu, X. Evolutionary Dynamics of MERS-CoV: Potential Recombination, Positive Selection and Transmission. Sci. Rep. 2016, 6, 25049. [Google Scholar] [CrossRef]
- Hussain, B.; Wu, C. Evolutionary and Phylogenetic Dynamics of SARS-CoV-2 Variants: A Genetic Comparative Study of Taiyuan and Wuhan Cities of China. Viruses 2024, 16, 907. [Google Scholar] [CrossRef]
- Alsolamy, S.; Arabi, Y.M. Infection with Middle East respiratory syndrome coronavirus. Can. J. Respir. Ther. 2015, 51, 102. [Google Scholar]
- Msemburi, W.; Karlinsky, A.; Knutson, V.; Aleshin-Guendel, S.; Chatterji, S.; Wakefield, J. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature 2023, 613, 130–137. [Google Scholar] [CrossRef]
- Russell, T.W.; Wu, J.T.; Clifford, S.; Edmunds, W.J.; Kucharski, A.J.; Jit, M. Effect of internationally imported cases on internal spread of COVID-19: A mathematical modelling study. Lancet Public Health 2021, 6, e12–e20. [Google Scholar] [CrossRef]
- Tang, G.; Liu, Z.; Chen, D. Human coronaviruses: Origin, host and receptor. J. Clin. Virol. 2022, 155, 105246. [Google Scholar] [CrossRef] [PubMed]
- Kistler, K.E.; Bedford, T. Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses OC43 and 229e. Elife 2021, 10, e64509. [Google Scholar] [CrossRef]
- Ye, R.Z.; Gong, C.; Cui, X.M.; Liu, J.Y.; Fan, H.; Xie, H.; Wang, Q.; Ren, Z.Y.; Zhang, Y.W.; Xia, L.Y.; et al. Continuous evolution and emerging lineage of seasonal human coronaviruses: A multicenter surveillance study. J. Med. Virol. 2023, 95, e28861. [Google Scholar] [CrossRef] [PubMed]
- Nickbakhsh, S.; Ho, A.; Marques, D.F.P.; McMenamin, J.; Gunson, R.N.; Murcia, P.R. Epidemiology of Seasonal Coronaviruses: Establishing the Context for the Emergence of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Jones, B.E.; Ying, J.; Stevens, V.; Haroldsen, C.; He, T.; Nevers, M.; Christensen, M.A.; Nelson, R.E.; Stoddard, G.J.; Sauer, B.C. Empirical anti-MRSA vs standard antibiotic therapy and risk of 30-day mortality in patients hospitalized for pneumonia. JAMA Intern. Med. 2020, 180, 552–560. [Google Scholar] [CrossRef]
- Habib, G.; Mahmood, K.; Gul, H.; Tariq, M.; Ain, Q.U.; Hayat, A.; Rehman, M.U. Pathophysiology of methicillin-resistant Staphylococcus aureus superinfection in COVID-19 patients. Pathophysiology 2022, 29, 405–413. [Google Scholar] [CrossRef]
- Drożdżal, S.; Rosik, J.; Lechowicz, K.; Machaj, F.; Szostak, B.; Przybyciński, J.; Lorzadeh, S.; Kotfis, K.; Ghavami, S.; Łos, M.J. An update on drugs with therapeutic potential for SARS-CoV-2 (COVID-19) treatment. Drug Resist. Updates 2021, 59, 100794. [Google Scholar] [CrossRef]
- Mubarak, A.; Alturaiki, W.; Hemida, M.G. Middle East respiratory syndrome coronavirus (MERS-CoV): Infection, immunological response, and vaccine development. J. Immunol. Res. 2019, 2019, 6491738. [Google Scholar] [CrossRef]
- Malik, J.A.; Ahmed, S.; Mir, A.; Shinde, M.; Bender, O.; Alshammari, F.; Ansari, M.; Anwar, S. The SARS-CoV-2 mutations versus vaccine effectiveness: New opportunities to new challenges. J. Infect. Public Health 2022, 15, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Poland, G.A.; Ovsyannikova, I.G.; Crooke, S.N.; Kennedy, R.B. SARS-CoV-2 vaccine development: Current status. Mayo Clin. Proc. 2020, 95, 2172–2188. [Google Scholar] [CrossRef]
- Finsterer, J. Neurological side effects of SARS-CoV-2 vaccinations. Acta Neurol. Scand. 2022, 145, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Menni, C.; Klaser, K.; May, A.; Polidori, L.; Capdevila, J.; Louca, P.; Sudre, C.H.; Nguyen, L.H.; Drew, D.A.; Merino, J. Vaccine side-effects and SARS-CoV-2 infection after vaccination in users of the COVID Symptom Study app in the UK: A prospective observational study. Lancet Infect. Dis. 2021, 21, 939–949. [Google Scholar] [CrossRef]
- Amanzio, M.; Mitsikostas, D.D.; Giovannelli, F.; Bartoli, M.; Cipriani, G.E.; Brown, W.A. Adverse events of active and placebo groups in SARS-CoV-2 vaccine randomized trials: A systematic review. Lancet Reg. Health Eur. 2022, 12, 100253. [Google Scholar] [CrossRef]
- Traore, A.; Charniga, K.; Grellet, S.; Terpant, G.; Da Cruz, H.; Lamy, A.; Thomas, N.; Gbaguidi, G.; Mercier, A.; Prudhomme, J. Monitoring SARS-CoV-2 variants with complementary surveillance systems: Risk evaluation of the Omicron JN. 1 variant in France, August 2023 to January 2024. Eurosurveillance 2025, 30, 2400293. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, A.; Mirzazadeh, A.; Tavakolpour, S. Genetics and genomics of SARS-CoV-2: A review of the literature with the special focus on genetic diversity and SARS-CoV-2 genome detection. Genomics 2021, 113, 1221–1232. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic variants of SARS-CoV-2—What do they mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef]
- Peiris, J.S.; Guan, Y.; Yuen, K. Severe acute respiratory syndrome. Nat. Med. 2004, 10, S88–S97. [Google Scholar] [CrossRef]
- Peiris, J.; Lai, S.; Poon, L.; Guan, Y.; Yam, L.; Lim, W.; Nicholls, J.; Yee, W.; Yan, W.; Cheung, M. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 2003, 361, 1319–1325. [Google Scholar] [CrossRef] [PubMed]
- Rota, P.A.; Oberste, M.S.; Monroe, S.S.; Nix, W.A.; Campagnoli, R.; Icenogle, J.P.; Penaranda, S.; Bankamp, B.; Maher, K.; Chen, M.-H. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 2003, 300, 1394–1399. [Google Scholar] [CrossRef] [PubMed]
- Marra, M.A.; Jones, S.J.; Astell, C.R.; Holt, R.A.; Brooks-Wilson, A.; Butterfield, Y.S.; Khattra, J.; Asano, J.K.; Barber, S.A.; Chan, S.Y. The genome sequence of the SARS-associated coronavirus. Science 2003, 300, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Zheng, B.; He, Y.; Liu, X.; Zhuang, Z.; Cheung, C.; Luo, S.; Li, P.H.; Zhang, L.; Guan, Y. Isolation and characterization of viruses related to the SARS coronavirus from animals in southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef]
- Lau, S.K.; Woo, P.C.; Li, K.S.; Huang, Y.; Tsoi, H.-W.; Wong, B.H.; Wong, S.S.; Leung, S.-Y.; Chan, K.-H.; Yuen, K.-Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc. Natl. Acad. Sci. USA 2005, 102, 14040–14045. [Google Scholar] [CrossRef]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef]
- Tang, X.; Zhang, J.; Zhang, S.; Wang, P.; Fan, X.; Li, L.; Li, G.; Dong, B.; Liu, W.; Cheung, C. Prevalence and genetic diversity of coronaviruses in bats from China. J. Virol. 2006, 80, 7481–7490. [Google Scholar] [CrossRef]
- Vijaykrishna, D.; Smith, G.J.; Zhang, J.X.; Peiris, J.; Chen, H.; Guan, Y. Evolutionary insights into the ecology of coronaviruses. J. Virol. 2007, 81, 4012–4020. [Google Scholar] [CrossRef] [PubMed]
- Gorbalenya, A.E.; Snijder, E.J.; Spaan, W.J. Severe acute respiratory syndrome coronavirus phylogeny: Toward consensus. J. Virol. 2004, 78, 7863–7866. [Google Scholar] [CrossRef]
- Yount, B.; Roberts, R.S.; Sims, A.C.; Deming, D.; Frieman, M.B.; Sparks, J.; Denison, M.R.; Davis, N.; Baric, R.S. Severe acute respiratory syndrome coronavirus group-specific open reading frames encode nonessential functions for replication in cell cultures and mice. J. Virol. 2005, 79, 14909–14922. [Google Scholar] [CrossRef]
- Lu, W.; Zheng, B.-J.; Xu, K.; Schwarz, W.; Du, L.; Wong, C.K.; Chen, J.; Duan, S.; Deubel, V.; Sun, B. Severe acute respiratory syndrome-associated coronavirus 3a protein forms an ion channel and modulates virus release. Proc. Natl. Acad. Sci. USA 2006, 103, 12540–12545. [Google Scholar] [CrossRef]
- Snijder, E.J.; Bredenbeek, P.J.; Dobbe, J.C.; Thiel, V.; Ziebuhr, J.; Poon, L.L.; Guan, Y.; Rozanov, M.; Spaan, W.J.; Gorbalenya, A.E. Unique and conserved features of genome and proteome of SARS-coronavirus, an early split-off from the coronavirus group 2 lineage. J. Mol. Biol. 2003, 331, 991–1004. [Google Scholar] [CrossRef]
- Yang, Z. Computational Molecular Evolution; OUP Oxford: Oxford, UK, 2006. [Google Scholar]
- Twiddy, S.S.; Farrar, J.J.; Chau, N.V.; Wills, B.; Gould, E.A.; Gritsun, T.; Lloyd, G.; Holmes, E.C. Phylogenetic relationships and differential selection pressures among genotypes of dengue-2 virus. Virology 2002, 298, 63–72. [Google Scholar] [CrossRef]
- Consortium, C.S.M.E. Molecular evolution of the SARS coronavirus during the course of the SARS epidemic in China. Science 2004, 303, 1666–1669. [Google Scholar] [CrossRef]
- Song, H.-D.; Tu, C.-C.; Zhang, G.-W.; Wang, S.-Y.; Zheng, K.; Lei, L.-C.; Chen, Q.-X.; Gao, Y.-W.; Zhou, H.-Q.; Xiang, H. Cross-host evolution of severe acute respiratory syndrome coronavirus in palm civet and human. Proc. Natl. Acad. Sci. USA 2005, 102, 2430–2435. [Google Scholar] [CrossRef]
- Zhang, C.-Y.; Wei, J.-F.; He, S.-H. Adaptive evolution of the spike gene of SARS coronavirus: Changes in positively selected sites in different epidemic groups. BMC Microbiol. 2006, 6, 88. [Google Scholar] [CrossRef] [PubMed]
- Sohpal, V.K. Comparative study: Nonsynonymous and synonymous substitution of SARS-CoV-2, SARS-CoV, and MERS-CoV genome. Genom. Inform. 2021, 19, e15. [Google Scholar] [CrossRef] [PubMed]
- Roy, C.; Mandal, S.M.; Mondal, S.K.; Mukherjee, S.; Mapder, T.; Ghosh, W.; Chakraborty, R. Trends of mutation accumulation across global SARS-CoV-2 genomes: Implications for the evolution of the novel coronavirus. Genomics 2020, 112, 5331–5342. [Google Scholar] [CrossRef]
- Li, F.; Berardi, M.; Li, W.; Farzan, M.; Dormitzer, P.R.; Harrison, S.C. Conformational states of the severe acute respiratory syndrome coronavirus spike protein ectodomain. J. Virol. 2006, 80, 6794–6800. [Google Scholar] [CrossRef]
- Reinke, L.M.; Spiegel, M.; Plegge, T.; Hartleib, A.; Nehlmeier, I.; Gierer, S.; Hoffmann, M.; Hofmann-Winkler, H.; Winkler, M.; Pöhlmann, S. Different residues in the SARS-CoV spike protein determine cleavage and activation by the host cell protease TMPRSS2. PLoS ONE 2017, 12, e0179177. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Shi, Z.; Yu, M.; Ren, W.; Smith, C.; Epstein, J.H.; Wang, H.; Crameri, G.; Hu, Z.; Zhang, H. Bats are natural reservoirs of SARS-like coronaviruses. Science 2005, 310, 676–679. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-F.; Shi, Z.; Zhang, S.; Field, H.; Daszak, P.; Eaton, B.T. Review of bats and SARS. Emerg. Infect. Dis. 2006, 12, 1834. [Google Scholar] [CrossRef]
- Wrapp, D.; Wang, N.; Corbett, K.S.; Goldsmith, J.A.; Hsieh, C.-L.; Abiona, O.; Graham, B.S.; McLellan, J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 2020, 367, 1260–1263. [Google Scholar] [CrossRef] [PubMed]
- Hatmal, M.m.M.; Alshaer, W.; Al-Hatamleh, M.A.; Hatmal, M.; Smadi, O.; Taha, M.O.; Oweida, A.J.; Boer, J.C.; Mohamud, R.; Plebanski, M. Comprehensive structural and molecular comparison of spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their interactions with ACE2. Cells 2020, 9, 2638. [Google Scholar] [CrossRef]
- Guruprasad, L. Evolutionary relationships and sequence-structure determinants in human SARS coronavirus-2 spike proteins for host receptor recognition. Proteins Struct. Funct. Bioinform. 2020, 88, 1387–1393. [Google Scholar] [CrossRef]
- Yang, Z. The power of phylogenetic comparison in revealing protein function. Proc. Natl. Acad. Sci. USA 2005, 102, 3179–3180. [Google Scholar] [CrossRef]
- Tolou, H.; Couissinier-Paris, P.; Durand, J.-P.; Mercier, V.; de Pina, J.-J.; De Micco, P.; Billoir, F.; Charrel, R.; De Lamballerie, X. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J. Gen. Virol. 2001, 82, 1283–1290. [Google Scholar] [CrossRef]
- Wu, K.; Peng, G.; Wilken, M.; Geraghty, R.J.; Li, F. Mechanisms of host receptor adaptation by severe acute respiratory syndrome coronavirus. J. Biol. Chem. 2012, 287, 8904–8911. [Google Scholar] [CrossRef]
- Wang, M.; Yan, M.; Xu, H.; Liang, W.; Kan, B.; Zheng, B.; Chen, H.; Zheng, H.; Xu, Y.; Zhang, E. SARS-CoV infection in a restaurant from palm civet. Emerg. Infect. Dis. 2005, 11, 1860. [Google Scholar] [CrossRef]
- Shi, Z.; Hu, Z. A review of studies on animal reservoirs of the SARS coronavirus. Virus Res. 2008, 133, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Gostin, L.O.; Lucey, D. Middle East respiratory syndrome: A global health challenge. JAMA 2015, 314, 771–772. [Google Scholar] [CrossRef] [PubMed]
- Zaki, A.M.; Van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820. [Google Scholar] [CrossRef]
- Bialek, S.R.; Allen, D.; Alvarado-Ramy, F.; Arthur, R.; Balajee, A.; Bell, D.; Best, S.; Blackmore, C.; Breakwell, L.; Cannons, A. First confirmed cases of Middle East respiratory syndrome coronavirus (MERS-CoV) infection in the United States, updated information on the epidemiology of MERS-CoV infection, and guidance for the public, clinicians, and public health authorities—May 2014. MMWR Morb. Mortal Wkly. Rep. 2014, 14, 1693–1699. [Google Scholar] [CrossRef]
- Azhar, E.I.; El-Kafrawy, S.A.; Farraj, S.A.; Hassan, A.M.; Al-Saeed, M.S.; Hashem, A.M.; Madani, T.A. Evidence for camel-to-human transmission of MERS coronavirus. N. Engl. J. Med. 2014, 370, 2499–2505. [Google Scholar] [CrossRef]
- Chu, D.K.; Poon, L.L.; Gomaa, M.M.; Shehata, M.M.; Perera, R.A.; Zeid, D.A.; El Rifay, A.S.; Siu, L.Y.; Guan, Y.; Webby, R.J. MERS coronaviruses in dromedary camels, Egypt. Emerg. Infect. Dis. 2014, 20, 1049. [Google Scholar] [CrossRef] [PubMed]
- Hussain, H.Y. Incidence and mortality rate of Middle East respiratory syndrome-corona virus (MERS-Cov), threatens and opportunities. J. Mycobac Dis. 2014, 4, 1000162. [Google Scholar] [CrossRef]
- Zhou, H.; Yang, J.; Zhou, C.; Chen, B.; Fang, H.; Chen, S.; Zhang, X.; Wang, L.; Zhang, L. A review of SARS-CoV2: Compared with SARS-CoV and MERS-CoV. Front. Med. 2021, 8, 628370. [Google Scholar] [CrossRef]
- Penttinen, P.; Kaasik-Aaslav, K.; Friaux, A.; Donachie, A.; Sudre, B.; Amato-Gauci, A.; Memish, Z.; Coulombier, D. Taking stock of the first 133 MERS coronavirus cases globally–Is the epidemic changing? Eurosurveillance 2013, 18, 20596. [Google Scholar] [CrossRef]
- Pereyaslov, D.; Rosin, P.; Palm, D.; Zeller, H.; Gross, D.; Brown, C.; Struelens, M. Laboratory capability and surveillance testing for Middle East respiratory syndrome coronavirus infection in the WHO European Region, June 2013. Eurosurveillance 2014, 19, 20923. [Google Scholar] [CrossRef]
- Hemida, M.; Perera, R.; Al Jassim, R.; Kayali, G.; Siu, L.; Wang, P.; Chu, K.; Perlman, S.; Ali, M.; Alnaeem, A. Seroepidemiology of Middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Eurosurveillance 2014, 19, 20828. [Google Scholar] [CrossRef]
- Lee, J.Y.; Kim, Y.-J.; Chung, E.H.; Kim, D.-W.; Jeong, I.; Kim, Y.; Yun, M.-R.; Kim, S.S.; Kim, G.; Joh, J.-S. The clinical and virological features of the first imported case causing MERS-CoV outbreak in South Korea, 2015. BMC Infect. Dis. 2017, 17, 498. [Google Scholar] [CrossRef]
- Mohd, H.A.; Al-Tawfiq, J.A.; Memish, Z.A. Middle East respiratory syndrome coronavirus (MERS-CoV) origin and animal reservoir. Virol. J. 2016, 13, 87. [Google Scholar] [CrossRef] [PubMed]
- Hemida, M.G.; Elmoslemany, A.; Al-Hizab, F.; Alnaeem, A.; Almathen, F.; Faye, B.; Chu, D.K.; Perera, R.A.; Peiris, M. Dromedary camels and the transmission of Middle East respiratory syndrome coronavirus (MERS-CoV). Transbound. Emerg. Dis. 2017, 64, 344–353. [Google Scholar] [CrossRef] [PubMed]
- Zumla, A.; Hui, D.S.; Perlman, S. Middle East respiratory syndrome. Lancet 2015, 386, 995–1007. [Google Scholar] [CrossRef]
- Du, L.; Tai, W.; Zhou, Y.; Jiang, S. Vaccines for the prevention against the threat of MERS-CoV. Expert. Rev. Vaccines 2016, 15, 1123–1134. [Google Scholar] [CrossRef] [PubMed]
- Baharoon, S.; Memish, Z.A. MERS-CoV as an emerging respiratory illness: A review of prevention methods. Travel. Med. Infect. Dis. 2019, 32, 101520. [Google Scholar] [CrossRef]
- Sabir, J.S.; Lam, T.T.-Y.; Ahmed, M.M.; Li, L.; Shen, Y.; EM Abo-Aba, S.; Qureshi, M.I.; Abu-Zeid, M.; Zhang, Y.; Khiyami, M.A. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 2016, 351, 81–84. [Google Scholar] [CrossRef]
- Widagdo, W.; Sooksawasdi Na Ayudhya, S.; Hundie, G.B.; Haagmans, B.L. Host determinants of MERS-CoV transmission and pathogenesis. Viruses 2019, 11, 280. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Yang, Y.; Zhou, Y.; Lu, L.; Li, F.; Jiang, S. MERS-CoV spike protein: A key target for antivirals. Expert. Opin. Ther. Targets 2017, 21, 131–143. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.I.; Kim, Y.-J.; Lemey, P.; Lee, I.; Park, S.; Bae, J.-Y.; Kim, D.; Kim, H.; Jang, S.-I.; Yang, J.-S. The recent ancestry of Middle East respiratory syndrome coronavirus in Korea has been shaped by recombination. Sci. Rep. 2016, 6, 18825. [Google Scholar] [CrossRef]
- Corman, V.M.; Ithete, N.L.; Richards, L.R.; Schoeman, M.C.; Preiser, W.; Drosten, C.; Drexler, J.F. Rooting the phylogenetic tree of middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J. Virol. 2014, 88, 11297–11303. [Google Scholar] [CrossRef]
- Sohrab, S.S.; Alsaqaf, F.; Hassan, A.M.; Tolah, A.M.; Bajrai, L.H.; Azhar, E.I. Genomic Diversity and Recombination Analysis of the Spike Protein Gene from Selected Human Coronaviruses. Biology 2024, 13, 282. [Google Scholar] [CrossRef]
- Forni, D.; Filippi, G.; Cagliani, R.; De Gioia, L.; Pozzoli, U.; Al-Daghri, N.; Clerici, M.; Sironi, M. The heptad repeat region is a major selection target in MERS-CoV and related coronaviruses. Sci. Rep. 2015, 5, 14480. [Google Scholar] [CrossRef]
- Cotten, M.; Watson, S.J.; Zumla, A.I.; Makhdoom, H.Q.; Palser, A.L.; Ong, S.H.; Al Rabeeah, A.A.; Alhakeem, R.F.; Assiri, A.; Al-Tawfiq, J.A. Spread, circulation, and evolution of the Middle East respiratory syndrome coronavirus. MBio 2014, 5, 10–1128. [Google Scholar] [CrossRef]
- Yamada, Y.; Liu, X.B.; Fang, S.G.; Tay, F.P.; Liu, D.X. Acquisition of cell–cell fusion activity by amino acid substitutions in spike protein determines the infectivity of a coronavirus in cultured cells. PLoS ONE 2009, 4, e6130. [Google Scholar] [CrossRef]
- McRoy, W.C.; Baric, R.S. Amino acid substitutions in the S2 subunit of mouse hepatitis virus variant V51 encode determinants of host range expansion. J. Virol. 2008, 82, 1414–1424. [Google Scholar] [CrossRef] [PubMed]
- Navas-Martin, S.; Hingley, S.T.; Weiss, S.R. Murine coronavirus evolution in vivo: Functional compensation of a detrimental amino acid substitution in the receptor binding domain of the spike glycoprotein. J. Virol. 2005, 79, 7629–7640. [Google Scholar] [CrossRef]
- Forni, D.; Cagliani, R.; Mozzi, A.; Pozzoli, U.; Al-Daghri, N.; Clerici, M.; Sironi, M. Extensive positive selection drives the evolution of nonstructural proteins in lineage C betacoronaviruses. J. Virol. 2016, 90, 3627–3639. [Google Scholar] [CrossRef] [PubMed]
- Báez-Santos, Y.M.; John, S.E.S.; Mesecar, A.D. The SARS-coronavirus papain-like protease: Structure, function and inhibition by designed antiviral compounds. Antivir. Res. 2015, 115, 21–38. [Google Scholar] [CrossRef]
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R. A novel coronavirus from patients with pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Gorbalenya, A.E.; Baker, S.C.; Baric, R.S.; de Groot, R.J.; Drosten, C.; Gulyaeva, A.A.; Haagmans, B.L.; Lauber, C.; Leontovich, A.M.; Neuman, B.W.; et al. The species Severe acute respiratory syndrome-related coronavirus: Classifying 2019-nCoV and naming it SARS-CoV-2. Nat. Microbiol. 2020, 5, 536–544. [Google Scholar] [CrossRef]
- Tai, D.Y. Pharmacologic treatment of SARS: Current knowledge and recommendations. Ann. Acad. Med. Singap. 2007, 36, 438. [Google Scholar] [CrossRef]
- Abduljalil, J.M.; Abduljalil, B.M. Epidemiology, genome, and clinical features of the pandemic SARS-CoV-2: A recent view. New Microbes New Infect. 2020, 35, 100672. [Google Scholar] [CrossRef]
- Petrosillo, N.; Viceconte, G.; Ergonul, O.; Ippolito, G.; Petersen, E. COVID-19, SARS and MERS: Are they closely related? Clin. Microbiol. Infect. 2020, 26, 729–734. [Google Scholar] [CrossRef]
- Chan, P.K.; Chan, M.C. Tracing the SARS-coronavirus. J. Thorac. Dis. 2013, 5, S118. [Google Scholar] [PubMed]
- Raj, V.S.; Osterhaus, A.D.; Fouchier, R.A.; Haagmans, B.L. MERS: Emergence of a novel human coronavirus. Curr. Opin. Virol. 2014, 5, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Meo, S.; Alhowikan, A.; Al-Khlaiwi, T.; Meo, I.; Halepoto, D.; Iqbal, M.; Usmani, A.; Hajjar, W.; Ahmed, N. Novel coronavirus 2019-nCoV: Prevalence, biological and clinical characteristics comparison with SARS-CoV and MERS-CoV. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2012–2019. [Google Scholar]
- Lupia, T.; Scabini, S.; Pinna, S.M.; Di Perri, G.; De Rosa, F.G.; Corcione, S. 2019 novel coronavirus (2019-nCoV) outbreak: A new challenge. J. Glob. Antimicrob. Resist. 2020, 21, 22–27. [Google Scholar] [CrossRef] [PubMed]
- Yuan, M.; Yin, W.; Tao, Z.; Tan, W.; Hu, Y. Association of radiologic findings with mortality of patients infected with 2019 novel coronavirus in Wuhan, China. PLoS ONE 2020, 15, e0230548. [Google Scholar] [CrossRef]
- Mahdy, M.A.; Younis, W.; Ewaida, Z. An overview of SARS-CoV-2 and animal infection. Front. Vet. Sci. 2020, 7, 596391. [Google Scholar] [CrossRef]
- Prince, T.; Smith, S.L.; Radford, A.D.; Solomon, T.; Hughes, G.L.; Patterson, E.I. SARS-CoV-2 infections in animals: Reservoirs for reverse zoonosis and models for study. Viruses 2021, 13, 494. [Google Scholar] [CrossRef]
- Zhao, J.; Cui, W.; Tian, B.P. The Potential Intermediate Hosts for SARS-CoV-2. Front. Microbiol. 2020, 11, 580137. [Google Scholar] [CrossRef]
- Sanjuán, R.; Domingo-Calap, P. Mechanisms of viral mutation. Cell. Mol. Life Sci. 2016, 73, 4433–4448. [Google Scholar] [CrossRef] [PubMed]
- Musso, N.; Maugeri, J.G.; Bongiorno, D.; Stracquadanio, S.; Bartoloni, G.; Stefani, S.; Di Stefano, E.D. SARS-CoV-2’s high rate of genetic mutation under immune selective pressure: From oropharyngeal B.1.1.7 to intrapulmonary B.1.533 in a vaccinated patient. Int. J. Infect. Dis. 2022, 118, 169–172. [Google Scholar] [CrossRef]
- Prabakaran, P.; Xiao, X.; Dimitrov, D.S. A model of the ACE2 structure and function as a SARS-CoV receptor. Biochem. Biophys. Res. Commun. 2004, 314, 235–241. [Google Scholar] [CrossRef]
- Scialo, F.; Daniele, A.; Amato, F.; Pastore, L.; Matera, M.G.; Cazzola, M.; Castaldo, G.; Bianco, A. ACE2: The major cell entry receptor for SARS-CoV-2. Lung 2020, 198, 867–877. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Z.; Yang, L.; Lian, X.; Xie, Y.; Li, S.; Xin, S.; Cao, P.; Lu, J. The MERS-CoV receptor DPP4 as a candidate binding target of the SARS-CoV-2 spike. Iscience 2020, 23, 101160. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wang, R.; Wang, M.; Wei, G.-W. Mutations strengthened SARS-CoV-2 infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef]
- Teng, S.; Sobitan, A.; Rhoades, R.; Liu, D.; Tang, Q. Systemic effects of missense mutations on SARS-CoV-2 spike glycoprotein stability and receptor-binding affinity. Brief. Bioinform. 2021, 22, 1239–1253. [Google Scholar] [CrossRef]
- Padhi, A.K.; Tripathi, T. Can SARS-CoV-2 accumulate mutations in the S-protein to increase pathogenicity? ACS Pharmacol. Transl. Sci. 2020, 3, 1023–1026. [Google Scholar] [CrossRef]
- Wang, S.; Xu, X.; Wei, C.; Li, S.; Zhao, J.; Zheng, Y.; Liu, X.; Zeng, X.; Yuan, W.; Peng, S. Molecular evolutionary characteristics of SARS-CoV-2 emerging in the United States. J. Med. Virol. 2022, 94, 310–317. [Google Scholar] [CrossRef]
- Wang, R.; Hozumi, Y.; Zheng, Y.-H.; Yin, C.; Wei, G.-W. Host immune response driving SARS-CoV-2 evolution. Viruses 2020, 12, 1095. [Google Scholar] [CrossRef] [PubMed]
- Akash, K.; Sharma, A.; Kumar, D.; Singh, S.K.; Gupta, G.; Chellappan, D.K.; Dua, K.; Nagraik, R. Molecular aspects of Omicron, vaccine development, and recombinant strain XE: A review. J. Med. Virol. 2022, 94, 4628. [Google Scholar]
- Mlcochova, P.; Kemp, S.A.; Dhar, M.S.; Papa, G.; Meng, B.; Ferreira, I.A.; Datir, R.; Collier, D.A.; Albecka, A.; Singh, S. SARS-CoV-2 B. 1.617. 2 Delta variant replication and immune evasion. Nature 2021, 599, 114–119. [Google Scholar] [CrossRef] [PubMed]
- Centres for Disease Control and Prevention. COVID-19 Data Tracker. Available online: https://www.cdc.gov/covid/php/variants/variants-and-genomic-surveillance.html (accessed on 12 June 2025).
- Thaweethai, T.; Selvaggi, C.A.; Ng, T.-C.; Cheng, D.; Cao, T.; Chibnik, L.B.; Shinnick, D.J.; Foulkes, A.S. Biomarker states and risk of death among individuals hospitalized with SARS-CoV-2 infection. BMC Infect. Dis. 2025, 25, 260. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Consortium, C.-G.U. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 entry into cells. Nat. Rev. Mol. Cell Biol. 2022, 23, 3–20. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e6. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, J.; Xiao, T.; Peng, H.; Sterling, S.M.; Walsh, R.M., Jr.; Rawson, S.; Rits-Volloch, S.; Chen, B. Distinct conformational states of SARS-CoV-2 spike protein. Science 2020, 369, 1586–1592. [Google Scholar] [CrossRef]
- Elrobaa, I.H.; New, K.J. COVID-19: Pulmonary and extra pulmonary manifestations. Front. Public Health 2021, 9, 711616. [Google Scholar] [CrossRef]
- Zabidi, N.Z.; Liew, H.L.; Farouk, I.A.; Puniyamurti, A.; Yip, A.J.W.; Wijesinghe, V.N.; Low, Z.Y.; Tang, J.W.; Chow, V.T.; Lal, S.K. Evolution of SARS-CoV-2 variants: Implications on immune escape, vaccination, therapeutic and diagnostic strategies. Viruses 2023, 15, 944. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Mannar, D.; Srivastava, S.S.; Berezuk, A.M.; Demers, J.-P.; Saville, J.W.; Leopold, K.; Li, W.; Dimitrov, D.S.; Tuttle, K.S. Cryo-electron microscopy structures of the N501Y SARS-CoV-2 spike protein in complex with ACE2 and 2 potent neutralizing antibodies. PLoS Biol. 2021, 19, e3001237. [Google Scholar] [CrossRef]
- Ahmed, W.S.; Philip, A.M.; Biswas, K.H. Decreased interfacial dynamics caused by the N501Y mutation in the SARS-CoV-2 S1 spike: ACE2 complex. Biophys. J. 2022, 121, 39a. [Google Scholar] [CrossRef]
- Ali, F.; Kasry, A.; Amin, M. The new SARS-CoV-2 strain shows a stronger binding affinity to ACE2 due to N501Y mutant. Med. Drug Discov. 2021, 10, 100086. [Google Scholar] [CrossRef]
- Cao, L.; Lou, J.; Chan, S.Y.; Zheng, H.; Liu, C.; Zhao, S.; Li, Q.; Mok, C.K.P.; Chan, R.W.Y.; Chong, M.K.C. Rapid evaluation of COVID-19 vaccine effectiveness against symptomatic infection with SARS-CoV-2 variants by analysis of genetic distance. Nat. Med. 2022, 28, 1715–1722. [Google Scholar] [CrossRef] [PubMed]
- Cox, M.; Peacock, T.P.; Harvey, W.T.; Hughes, J.; Wright, D.W.; Willett, B.J.; Thomson, E.; Gupta, R.K.; Peacock, S.J.; Robertson, D.L.; et al. SARS-CoV-2 variant evasion of monoclonal antibodies based on in vitro studies. Nat. Rev. Microbiol. 2023, 21, 112–124. [Google Scholar] [CrossRef]
- Chen, L.; He, Y.; Liu, H.; Shang, Y.; Guo, G. Potential immune evasion of the severe acute respiratory syndrome coronavirus 2 Omicron variants. Front. Immunol. 2024, 15, 1339660. [Google Scholar] [CrossRef]
- Focosi, D.; Maggi, F.; Franchini, M.; McConnell, S.; Casadevall, A. Analysis of Immune Escape Variants from Antibody-Based Therapeutics against COVID-19: A Systematic Review. Int. J. Mol. Sci. 2022, 23, 29. [Google Scholar] [CrossRef]
- Saha, A.; Ghosh Roy, S.; Dwivedi, R.; Tripathi, P.; Kumar, K.; Nambiar, S.M.; Pathak, R. Beyond the Pandemic Era: Recent Advances and Efficacy of SARS-CoV-2 Vaccines Against Emerging Variants of Concern. Vaccines 2025, 13, 424. [Google Scholar] [CrossRef]
- Tokhanbigli, S.; Salami Ghaleh, S.; Rahimian, K.; Mahmanzar, M.; Bayat, S.; Ahangarzadeh, S.; Moradi, B.; Mahmanzar, R.; Wang, Y.; Oliver, B.G.G.; et al. Intersecting SARS-CoV-2 spike mutations and global vaccine efficacy against COVID-19. Front. Immunol. 2025, 16, 1435873. [Google Scholar] [CrossRef]
- Carabelli, A.M.; Peacock, T.P.; Thorne, L.G.; Harvey, W.T.; Hughes, J.; de Silva, T.I.; Peacock, S.J.; Barclay, W.S.; de Silva, T.I.; Towers, G.J.; et al. SARS-CoV-2 variant biology: Immune escape, transmission and fitness. Nat. Rev. Microbiol. 2023, 21, 162–177. [Google Scholar] [CrossRef] [PubMed]
- Roederer, A.L.; Cao, Y.; St. Denis, K.; Sheehan, M.L.; Li, C.J.; Lam, E.C.; Gregory, D.J.; Poznansky, M.C.; Iafrate, A.J.; Canaday, D.H.; et al. Ongoing evolution of SARS-CoV-2 drives escape from mRNA vaccine-induced humoral immunity. Cell Rep. Med. 2024, 5, 101850. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Yu, Y.; Xu, Y.; Jian, F.; Song, W.; Yisimayi, A.; Wang, P.; Wang, J.; Liu, J.; Yu, L. Fast evolution of SARS-CoV-2 BA.2.86 to JN.1 under heavy immune pressure. Lancet Infect. Dis. 2024, 24, e70–e72. [Google Scholar] [CrossRef] [PubMed]
- Planas, D.; Staropoli, I.; Michel, V.; Lemoine, F.; Donati, F.; Prot, M.; Porrot, F.; Guivel-Benhassine, F.; Jeyarajah, B.; Brisebarre, A.; et al. Distinct evolution of SARS-CoV-2 Omicron XBB and BA.2.86/JN.1 lineages combining increased fitness and antibody evasion. Nat. Commun. 2024, 15, 2254. [Google Scholar] [CrossRef]
- Kaku, Y.; Okumura, K.; Padilla-Blanco, M.; Kosugi, Y.; Uriu, K.; Hinay, A.A.; Chen, L.; Plianchaisuk, A.; Kobiyama, K.; Ishii, K.J. Virological characteristics of the SARS-CoV-2 JN. 1 variant. Lancet Infect. Dis. 2024, 24, e82. [Google Scholar] [CrossRef]
- Kosugi, Y.; Plianchaisuk, A.; Putri, O.; Uriu, K.; Kaku, Y.; Hinay, A.A.; Chen, L.; Kuramochi, J.; Sadamasu, K.; Yoshimura, K. Characteristics of the SARS-CoV-2 omicron HK. 3 variant harbouring the FLip substitution. Lancet Microbe 2024, 5, e313. [Google Scholar] [CrossRef]
- Yameny, A.A. The COVID-19 JN.1 variant diagnosed in Egypt. J. Med. Life Sci. 2023, 5, 318–321. [Google Scholar] [CrossRef]
- Stringhini, S.; Wisniak, A.; Piumatti, G.; Azman, A.S.; Lauer, S.A.; Baysson, H.; De Ridder, D.; Petrovic, D.; Schrempft, S.; Marcus, K. Seroprevalence of anti-SARS-CoV-2 IgG antibodies in Geneva, Switzerland (SEROCoV-POP): A population-based study. Lancet 2020, 396, 313–319. [Google Scholar] [CrossRef]
- Chan, K.K.; Tan, T.J.; Narayanan, K.K.; Procko, E. An engineered decoy receptor for SARS-CoV-2 broadly binds protein S sequence variants. Sci. Adv. 2021, 7, eabf1738. [Google Scholar] [CrossRef]
- Yadav, P.D.; Sarkale, P.; Razdan, A.; Gupta, N.; Nyayanit, D.A.; Sahay, R.R.; Potdar, V.; Patil, D.Y.; Baradkar, S.; Kumar, A. Isolation and characterization of SARS-CoV-2 Beta variant from UAE travelers. J. Infect. Public Health 2022, 15, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Fujino, T.; Nomoto, H.; Kutsuna, S.; Ujiie, M.; Suzuki, T.; Sato, R.; Fujimoto, T.; Kuroda, M.; Wakita, T.; Ohmagari, N. Novel SARS-CoV-2 variant in travelers from Brazil to Japan. Emerg. Infect. Dis. 2021, 27, 1243. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, J.; Plante, K.S.; Plante, J.A.; Xie, X.; Zhang, X.; Ku, Z.; An, Z.; Scharton, D.; Schindewolf, C. The N501Y spike substitution enhances SARS-CoV-2 infection and transmission. Nature 2022, 602, 294–299. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Chu, A.W.-H.; Zhang, R.R.; Chan, W.-M.; Ip, J.D.; Tsoi, H.-W.; Chen, L.-L.; Cai, J.-P.; Lung, D.C.; Tam, A.R. The impact of spike N501Y mutation on neutralizing activity and RBD binding of SARS-CoV-2 convalescent serum. EBioMedicine 2021, 71, 103544. [Google Scholar] [CrossRef]
- Lubinski, B.; Fernandes, M.H.; Frazier, L.; Tang, T.; Daniel, S.; Diel, D.G.; Jaimes, J.A.; Whittaker, G.R. Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. Iscience 2022, 25, 103589. [Google Scholar] [CrossRef]
- Tian, D.; Sun, Y.; Xu, H.; Ye, Q. The emergence and epidemic characteristics of the highly mutated SARS-CoV-2 Omicron variant. J. Med. Virol. 2022, 94, 2376–2383. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.L.; DeFalco, L.; Anderson, D.E.; Zhang, Y.; Aw, J.G.A.; Lim, S.Y.; Lim, X.N.; Tan, K.Y.; Zhang, T.; Chawla, T.; et al. Comprehensive mapping of SARS-CoV-2 interactions in vivo reveals functional virus-host interactions. Nat. Commun. 2021, 12, 5113. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Kuroda, M.; Armbrust, T.; Theiler, J.; Balaram, A.; Moreno, G.K.; Accola, M.A.; Iwatsuki-Horimoto, K.; Valdez, R.; Stoneman, E. Characterization of the SARS-CoV-2 B. 1.621 (Mu) variant. Sci. Transl. Med. 2022, 14, eabm4908. [Google Scholar] [CrossRef]
- Wibmer, C.K.; Ayres, F.; Hermanus, T.; Madzivhandila, M.; Kgagudi, P.; Oosthuysen, B.; Lambson, B.E.; de Oliveira, T.; Vermeulen, M.; van der Berg, K.; et al. SARS-CoV-2 501Y.V2 escapes neutralization by South African COVID-19 donor plasma. Nat. Med. 2021, 27, 622–625. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, J.; Jian, F.; Xiao, T.; Song, W.; Yisimayi, A.; Huang, W.; Li, Q.; Wang, P.; An, R. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature 2022, 602, 657–663. [Google Scholar] [CrossRef]
- Wang, P.; Nair, M.S.; Liu, L.; Iketani, S.; Luo, Y.; Guo, Y.; Wang, M.; Yu, J.; Zhang, B.; Kwong, P.D.; et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature 2021, 593, 130–135. [Google Scholar] [CrossRef]
- Expression of Concern: Long non coding RNA FAM3D-AS1 inhibits development of colorectal cancer through NF-kB signaling pathway. Biosci. Rep. 2021, 41, BSR-20190724_EOC. [CrossRef]
- Di Giacomo, S.; Mercatelli, D.; Rakhimov, A.; Giorgi, F.M. Preliminary report on severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Spike mutation T478K. J. Med. Virol. 2021, 93, 5638–5643. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Karuppanan, K.; Subramaniam, G. Omicron (BA.1) and sub-variants (BA.1.1, BA.2, and BA.3) of SARS-CoV-2 spike infectivity and pathogenicity: A comparative sequence and structural-based computational assessment. J. Med. Virol. 2022, 94, 4780–4791. [Google Scholar] [CrossRef]
- Saito, A.; Irie, T.; Suzuki, R.; Maemura, T.; Nasser, H.; Uriu, K.; Kosugi, Y.; Shirakawa, K.; Sadamasu, K.; Kimura, I. Enhanced fusogenicity and pathogenicity of SARS-CoV-2 Delta P681R mutation. Nature 2022, 602, 300–306. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, T.; Fang, Y.; Liu, J.; Ye, Q.; Ding, L. SARS-CoV-2 spike L452R mutation increases Omicron variant fusogenicity and infectivity as well as host glycolysis. Signal Transduct. Target. Ther. 2022, 7, 76. [Google Scholar] [CrossRef] [PubMed]
- Tchesnokova, V.; Kulasekara, H.; Larson, L.; Bowers, V.; Rechkina, E.; Kisiela, D.; Sledneva, Y.; Choudhury, D.; Maslova, I.; Deng, K. Acquisition of the L452R mutation in the ACE2-binding interface of Spike protein triggers recent massive expansion of SARS-Cov-2 variants. J. Clin. Microbiol. 2021, 59, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.E.; Dávila-Barclay, A.; Salvatierra, G.; González, L.; Cuicapuza, D.; Solís, L.; Marcos-Carbajal, P.; Huancachoque, J.; Maturrano, L.; Tsukayama, P. The emergence of SARS-CoV-2 variant lambda (C.37) in South America. Microbiol. Spectr. 2021, 9, e0078921. [Google Scholar] [CrossRef]
- Baj, A.; Novazzi, F.; Ferrante, F.D.; Genoni, A.; Cassani, G.; Prestia, M.; Colombo, A.; Capuano, R.; Zago, C.; Pasciuta, R. Introduction of SARS-COV-2 C. 37 (WHO VOI lambda) from Peru to Italy. J. Med. Virol. 2021, 93, 6460. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef]
- Alkhatib, M.; Svicher, V.; Salpini, R.; Ambrosio, F.A.; Bellocchi, M.C.; Carioti, L.; Piermatteo, L.; Scutari, R.; Costa, G.; Artese, A. SARS-CoV-2 variants and their relevant mutational profiles: Update summer 2021. Microbiol. Spectr. 2021, 9, e0109621. [Google Scholar] [CrossRef]
- De Gasparo, R.; Pedotti, M.; Simonelli, L.; Nickl, P.; Muecksch, F.; Cassaniti, I.; Percivalle, E.; Lorenzi, J.C.C.; Mazzola, F.; Magrì, D.; et al. Bispecific IgG neutralizes SARS-CoV-2 variants and prevents escape in mice. Nature 2021, 593, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Jian, F.; Wang, J.; Yu, Y.; Song, W.; Yisimayi, A.; Wang, J.; An, R.; Chen, X.; Zhang, N. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature 2023, 614, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Shaheen, N.; Mohamed, A.; Soliman, Y.; Abdelwahab, O.A.; Diab, R.A.; Desouki, M.T.; Rababah, A.A.; Khaity, A.; Hefnawy, M.T.; Swed, S. Could the new BA.2.75 sub-variant lead to another COVID-19 wave in the world?—Correspondence. Int. J. Surg. 2022, 105, 106861. [Google Scholar] [CrossRef]
- Kaku, Y.; Uriu, K.; Kosugi, Y.; Okumura, K.; Yamasoba, D.; Uwamino, Y.; Kuramochi, J.; Sadamasu, K.; Yoshimura, K.; Asakura, H. Virological characteristics of the SARS-CoV-2 KP.2 variant. Lancet Infect. Dis. 2024, 24, e416. [Google Scholar] [CrossRef]
- Kumar, P.; Jayan, J.; Sharma, R.K.; Gaidhane, A.M.; Zahiruddin, Q.S.; Rustagi, S.; Satapathy, P. The emerging challenge of FLiRT variants: KP. 1.1 and KP. 2 in the global pandemic landscape. QJM Int. J. Med. 2024, 117, 485–487. [Google Scholar] [CrossRef]
- Gillot, C.; David, C.; Dogné, J.-M.; Cabo, J.; Douxfils, J.; Favresse, J. Neutralizing antibodies against KP.2 and KP.3: Why the current vaccine needs an update. Clin. Chem. Lab. Med. (CCLM) 2025, 63, e82–e85. [Google Scholar] [CrossRef] [PubMed]
- Kaku, Y.; Yo, M.S.; Tolentino, J.E.; Uriu, K.; Okumura, K.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 KP.3, LB.1, and KP.2.3 variants. Lancet Infect. Dis. 2024, 24, e482–e483. [Google Scholar] [CrossRef]
- Chen, N.; Decker, K.E.; Schulz, S.R.; Kempf, A.; Nehlmeier, I.; Moldenhauer, A.-S.; Dopfer-Jablonka, A.; Behrens, G.M.; Stankov, M.V.; Manthey, L. Comparative Analysis of Host Cell Entry Efficiency and Neutralization Sensitivity of Emerging SARS-CoV-2 Lineages KP.2, KP.2.3, KP.3, and LB.1. Vaccines 2024, 12, 1236. [Google Scholar] [CrossRef]
- Hu, Y.; Zou, J.; Nguyen, M.D.; Chang, H.C.; Yeung, J.; Hao, H.; Shi, P.-Y.; Ren, P.; Xie, X. Comparative analysis of replication and immune evasion among SARS-CoV-2 subvariants BA. 2.86, JN. 1, KP. 2, and KP. 3. mBio 2025, 16, e0350324. [Google Scholar] [CrossRef] [PubMed]
- WHO. SARS-CoV-2 Variant Trackers. Currently Circulating Variants Under Monitoring (VUMs) (as of 23 May 2025). Available online: https://www.who.int/en/activities/tracking-SARS-CoV-2-variants/g#:~:text=%23Excludes%20JN.,as%20of%2014%20April%202025 (accessed on 12 June 2025).
- Suthar, M.S.; Manning, K.E.; Ellis, M.L.; Jain, S.; Bechnak, K.; Vander Velden, J.; Laboune, F.; Henry, A.R.; Godbole, S.; Kim, S. The KP.2-adapted COVID-19 vaccine improves neutralising activity against the XEC variant. Lancet Infect. Dis. 2025, 25, e122–e123. [Google Scholar] [CrossRef]
- Branda, F.; Ciccozzi, M.; Scarpa, F. Features of the SARS-CoV-2 KP.3 variant mutations. Infect. Dis. 2024, 56, 894–896. [Google Scholar] [CrossRef]
- Branda, F.; Ciccozzi, M.; Scarpa, F. On the new SARS-CoV-2 variant KP. 3.1. 1: Focus on its genetic potential. Infect. Dis. 2024, 56, 903–906. [Google Scholar] [CrossRef]
- Li, P.; Faraone, J.N.; Hsu, C.C.; Chamblee, M.; Liu, Y.; Zheng, Y.-M.; Xu, Y.; Carlin, C.; Horowitz, J.C.; Mallampalli, R.K. Neutralization and spike stability of JN. 1-derived LB.1, KP.2.3, KP.3, and KP.3.1. 1 subvariants. mBio 2025, 16, e0046425. [Google Scholar] [CrossRef] [PubMed]
- Aljabali, A.A.; Lundstrom, K.; Hromić-Jahjefendić, A.; Abd El-Baky, N.; Nawn, D.; Hassan, S.S.; Rubio-Casillas, A.; Redwan, E.M.; Uversky, V.N. The XEC Variant: Genomic Evolution, Immune Evasion, and Public Health Implications. Viruses 2025, 17, 985. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Kaku, Y.; Okumura, K.; Uriu, K.; Zhu, Y.; Ito, J.; Sato, K. Virological characteristics of the SARS-CoV-2 LP.8.1 variant. Lancet Infect. Dis. 2025, 25, E193. [Google Scholar] [CrossRef]
- Uriu, K.; Okumura, K.; Uwamino, Y.; Chen, L.; Tolentino, J.E.; Asakura, H.; Nagashima, M.; Sadamasu, K.; Yoshimura, K.; Ito, J. Virological characteristics of the SARS-CoV-2 NB. 1.8. 1 variant. Lancet Infect. Dis. 2025, 25, e443. [Google Scholar] [CrossRef]
- Feng, Z.; Huang, J.; Baboo, S.; Diedrich, J.K.; Bangaru, S.; Paulson, J.C.; Yates, J.R., 3rd; Yuan, M.; Wilson, I.A.; Ward, A.B. Structural and Functional Insights into the Evolution of SARS-CoV-2 KP.3.1.1 Spike Protein. bioRxiv 2024. [Google Scholar] [CrossRef]
- Wang, Q.; Guo, Y.; Mellis, I.A.; Wu, M.; Mohri, H.; Gherasim, C.; Valdez, R.; Purpura, L.J.; Yin, M.T.; Gordon, A.; et al. Antibody evasiveness of SARS-CoV-2 subvariants KP.3.1.1 and XEC. Cell Rep. 2025, 44, 115543. [Google Scholar] [CrossRef] [PubMed]
- Lugano, D.; Kutima, B.; Kimani, M.; Sigilai, A.; Gitonga, J.; Karani, A.; Akech, D.; Karia, B.; Ziraba, A.K.; Maina, A. Evaluation of population immunity against SARS-CoV-2 variants, EG.5.1, FY.4, BA.2.86, JN.1, JN.1.4, and KP.3.1.1 using samples from two health demographic surveillance systems in Kenya. BMC Infect. Dis. 2024, 24, 1474. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, Y.; Jian, F.; Yang, S.; Song, W.; Wang, P.; Yu, L.; Shao, F.; Cao, Y. Enhanced immune evasion of SARS-CoV-2 variants KP. 3.1. 1 and XEC through N-terminal domain mutations. Lancet Infect. Dis. 2025, 25, e6–e7. [Google Scholar] [CrossRef] [PubMed]
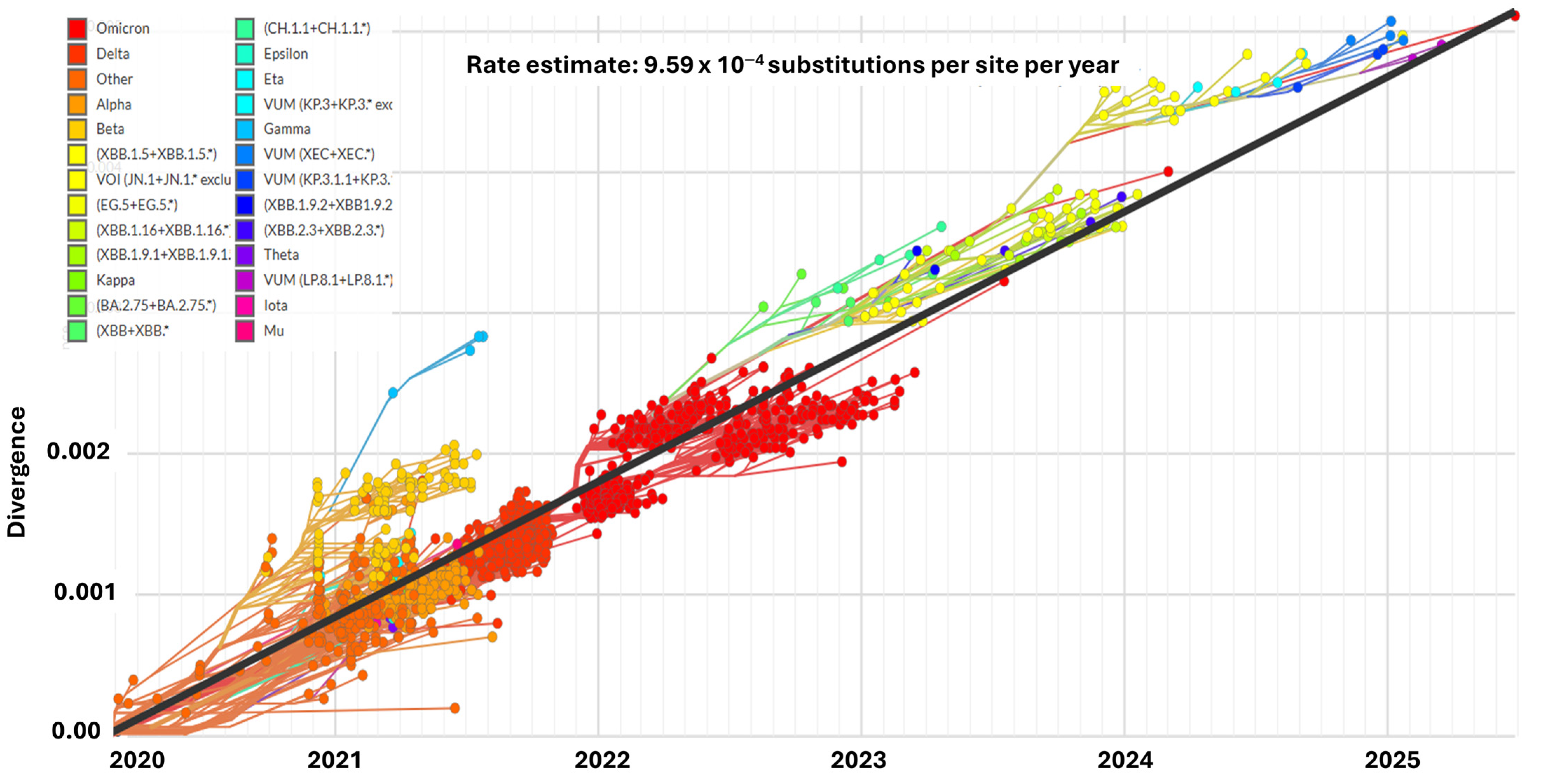
- Mutational Tracker. GISAID Initiative. Lineage Report. Available online: https://outbreak.info/situation-reports?xmin=2024-12-12&xmax=2025-06-12&pango=JN.1.18.1 (accessed on 12 June 2025).
- Nextstrain Clade. Genomic Epidemiology of SARS-CoV-2 with Clade-Focused Subsampling. Available online: https://nextstrain.org/ncov/gisaid/global/6m (accessed on 12 June 2025).
- Guo, C.; Yu, Y.; Liu, J.; Jian, F.; Yang, S.; Song, W.; Yu, L.; Shao, F.; Cao, Y. Antigenic and virological characteristics of SARS-CoV-2 variants BA. 3.2, XFG, and NB. 1.8. 1. Lancet Infect. Dis. 2025, 25, e374–e377. [Google Scholar] [CrossRef]
- Relan, P.; Motaze, N.V.; Kothari, K.; Askie, L.; Le Polain, O.; Van Kerkhove, M.D.; Diaz, J.; Tirupakuzhi Vijayaraghavan, B.K. Severity and outcomes of Omicron variant of SARS-CoV-2 compared to Delta variant and severity of Omicron sublineages: A systematic review and metanalysis. BMJ Glob. Health 2023, 8, e012328. [Google Scholar] [CrossRef]

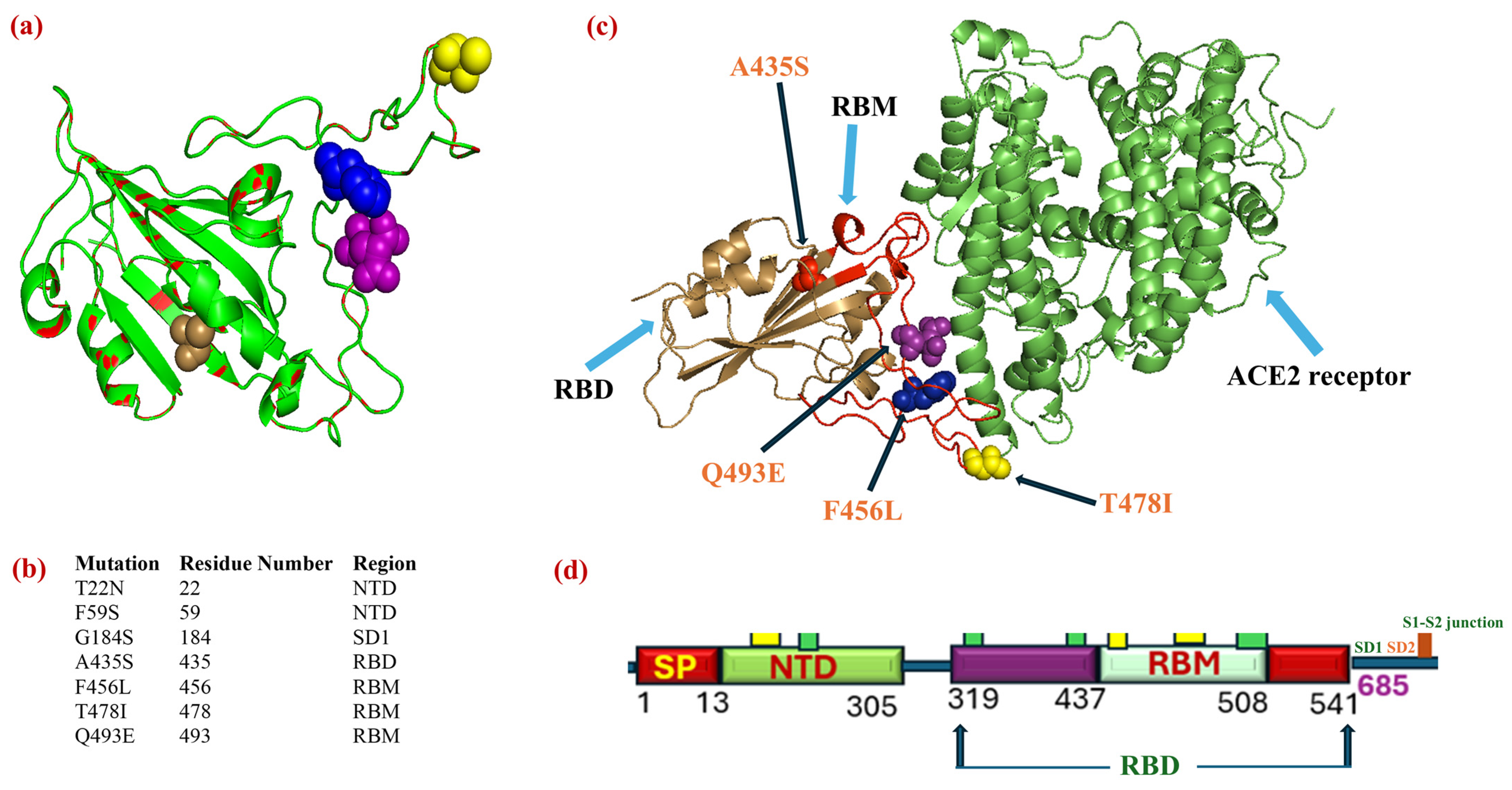
| VOCs or VOIs | Critical Mutations | Evolutionary and Clinical Implications | References |
|---|---|---|---|
| Alpha (α) (B.1.1.7 categorized as VOC). Gamma (γ) (P.1 categorized as VOC). Mu (VOI), Beta (β) (B.1.351/VOC). Omicron (VOC) | N501Y | Perform a major job during the first wave of VOCs. Higher transmissibility due to increased ACE2-receptor-RBD binding affinity. Vaccine protection was slightly diminished but remained effective, especially with a booster dose. | [134,135,136,137,138] |
| Alpha (α) (B.1.1.7 categorized as VOC), Omicron (B.1.1.529), Gamma (γ) grouped as VOC) | P681H | Increased transmissibility, infectivity, and severity. Enhancing protein cleavage by affecting furin-cleavage-site in the S-hot-spot, promoting virus-cell fusion and, therefore, viral entry into cells. | [136,139,140,141] |
| Alpha (α) (VOC), Gamma (γ) (P.1/VOC), Beta (β) (B.1.351/VOC), Mu (VOI) | E484K | Diminished binding affinity for neutralizing antibody. Remarkable immune escape and impact on vaccine protection. It impacts monoclonal therapy. Increases ACE2-receptor-RBD binding affinity. | [135,136,142,143] |
| Beta (β) also termed as B.1.351/VOC), Gamma (γ) categorized as VOC, Omicron (B.1.1.529), as well as Delta (δ) (VOC) | K417N | Enhanced ACE2-receptor-RBD binding affinity. Escape class 1 neutralizing antibody (nAb). | [135,136,143,144,145] |
| Gamma (γ) (P.1/VOC), Beta (β) (B.1.351/VOC), and Alpha (α)-sub-lineages. | L18F | Alter the configuration of the antigenic supersite in NTD. Evasion from NTD-specific nAbs. | [135,136,146,147] |
| Omicron (BA.2.75, XBB) and Delta (δ) (B.1.617.2/VOC) | T478K | It was part of a set of mutations triggering the spread of the Delta (δ) variant globally and persisted in Omicron variants/sub-variants, underscoring an evolutionary advantage. Aggrandize ACE2 binding and modify surface charge (electrostatic potential), possibly affecting antibody recognition and leading to evasion. | [147,148] |
| Delta (δ) (B.1.617.2/VOC) | P681R | cleavage of the S-protein into S1 and S2 by furin, leading to more efficient viral entry Facilitation of the furin-mediated spike protein cleavage, assisting viral fusion to the cell and potential entry. | [149] |
| Omicron (BA.4) as well as Delta (δ) (B.1.617.2 categorized as VOC) | L452R | Aggrandizes transmissibility and infectivity by stabilizing the ACE2-RBD interface. It was one of the clusters of mutations that fostered Delta’s greater transmissibility. Diminished Ab neutralization and downgraded cell-mediated immune response, allowing rapid viral replication | [150,151,152] |
| Lambda (γ) (VOI) | L542Q | Limited data are available. Slight immune evasion. Slight strengthening of the ACE2-RBD binding. | [153] |
| Lambda (γ) (C.37/VOI) | F490S | Alters receptor-binding motif (RBM) conformation, possibly affecting ACE2-Ab interaction, leading to immune evasion with nAb. | [154] |
| Lambda (VOI), alpha (α) (VOC), Beta (β) (VOC), Omicron (VOC), Gamma (γ) (VOC), Delta (δ) and (VOC) | D614G | It appeared in the initial state of the pandemic and became the most dominant mutation worldwide during the pandemic. It shows enhanced viral infectivity and replication. It enhances the transmission. It was observed to be allied with high viral load/increased infectivity, while the vaccine remained effective. Increased the S-protein’s open conformation stability and thus strengthened ACE2-RBD interaction. It was one of the major substitutions of the initial phase of the evolution. | [135,136,155] |
| Omicron (BA.1.1, BQ.1, and XBB.1.5/VOC) as well as Mu (μ) (B.1.621)/VOI) | R346K | Diminished neutralization by mAb and/or polyclonal sera. | [142] |
| Mu (B.1.621/VOI) | ins146N | Changes the closed–open S1-subunit conformation, fostering strengthened ACE2-RBD binding. | [142] |
| Sub-lineages evolved from Omicron (B.1.1.529/VOC) | G339D | Modify RBD’s local conformation and impact nAb binding and moderate evasion. | [144] |
| Omicron (B.1.1.529 categorized as VOC) | S477N | Aggrandized viral transmissibility (infectivity) via strengthening the ACE2-RBD interaction. | [156,157] |
| Omicron (BA.1, BA.2/VOC) | N440K | Enhanced viral fitness stabilizes ACE2-RBD binding. Foster resistance to mAb Evasion from natural or vaccine-induced nAb. | [144] |
| Omicron (BQ.1, BQ.1.1 sub-lineage, XBB.1.5, and XBB-sub-lineage, as well as CH.1.1/VOC) | R346T | Strongly allied with evasion from Class 3 mAb. Diminished Ab neutralization. | [158] |
| Omicron (XBB sub-lineage, XBB.1-sub-lineage, and XBB.1.5/VOC) | F486S | Enhanced neutralization of Abs (Class 1 & 2 nAb). Contributes to the increased fusogenicity. | [158] |
| Omicron (BA.1-sub-lineage and BA.2.75, as well as XBB-sub-lineage/VOC) | G446S | Foster resistance against Abs (Class 3 mAb and moderate evasion from polyclonal sera). | [158,159] |
| Omicron (BA.1/VOC) | R493Q | Evasion of Class 1 Ab. Enhances interaction with ACE2 receptor, easing adhesion to the cells | [158,159] |
| Pango-Lineage | Clade Information (Next-Strain) | Mutational-Fingerprint | The First Sampling Date | Variant Designation Date. Or the Risk Assessment Date | References ([166]) |
|---|---|---|---|---|---|
| KP.2-sub-lineage also referred to as N.1.11.1.2 | 24C | Lineage-JN.1 + S:Q493E, S:F456L, S:V1104L, S:R346T | 11 February 2024 | 3 May 2024 | [160,167] |
| KP.3-sub-lineage or (JN.1.11.1.2) | 24C | Lineage-JN.1 plus substitutions S:V1104L, S:F456L, S:Q493E, | 11 February 2024 | 3 May 2024 | [168] |
| KP.3.1.1 -sub-lineage or (JN.1.11.1.3.1.1) | 24C | KP.3 + deletion S:S31- | 27 March 2024 | 19 July 2024 | [169] |
| LB.1 also referred to as JN.1.9.2.1 | 24A | Lineage-JN.1 + S:Q183H, S:R346T, S:F456L substitution, and one S:S31- deletion. | 26 February 2024 | 28 June 2024 | [170] |
| XEC | 24F | JN.1 + S:F456L, S:T22N, S:F59S, S:V1104L, S:Q493E | 26 June 2024 | 24 September 2024 | [171] |
| LP.8.1 | 24B | JN1 + S:V445R, S:S31-, S:F186L, S:R346T, S:F456L, S:Q493E, S:V1104L, S:R190S, S:K1086R | 1 July 2024 | 24 January 2025 | [172] |
| NB.1.8.1 | 25B | JN1 + S:T22N, S:G184S, S:F59S, S:A435S, S:T478I, S:F456L, S:Q493E | 22 January 2025 | 23 May 2025 | [173] |
| Genomic Region | SARS-CoV-2 Sub-Lineage (VUMs) as of May 2025 | |||||
|---|---|---|---|---|---|---|
| JN.1 | KP.3 | KP.3.1.1 | XEC | LP.8.1 | NB.1.8.1 | |
| N | G204R | G204R | G204R | G204P | G204R | G204R |
| Q229K | Q229K | Q229K | Q229K | Q229K | - | |
| ORF3 | - | - | - | - | - | R138H |
| - | - | - | - | P178L | - | |
| NSP1 | - | - | - | - | - | K47R |
| NSP2 | - | - | - | - | - | S122F |
| A31D | A31D | A31D | A31D | A31D | - | |
| A419T | - | |||||
| NSP3 | V238L | V238L | V238L | V238L | V238L | - |
| - | - | - | - | - | A233Y | |
| - | - | - | - | - | A655V | |
| K1155R | K1155R | K1155R | K1155R | K1155R | - | |
| - | - | - | - | A1179V | - | |
| - | - | - | - | - | P1261Q | |
| T1465I | T1465I | T1465I | T1465I | |||
| N1708S | N1708S | N1708S | N1708S | N1708S | ||
| - | - | - | - | - | I1891V | |
| A1892T | A1892T | A1892T | A1892T | A1892T | - | |
| NSP4 | - | - | - | - | - | L438F |
| NSP6 | V24F | V24F | V24F | V24F | V24F | - |
| R252K | R252K | R252K | R252K | R252K | - | |
| NSP9 | T35I | T35I | T35I | T35I | T35I | - |
| - | - | - | - | - | P57S | |
| - | - | - | - | - | P80L | |
| NSP10 | - | - | S33C | - | - | - |
| NSP12 | - | - | - | - | - | D284Y |
| - | - | - | - | - | G671S | |
| NSP13 | - | - | - | - | - | S36P |
| S | - | - | - | T22N | - | T22N |
| - | - | S31del | - | S31del | ||
| - | - | - | S59S | S59S | ||
| - | - | - | - | G184S | ||
| - | - | - | - | F186L | - | |
| - | - | - | - | R190S | - | |
| - | - | - | - | R346T | - | |
| - | - | - | - | - | A435S | |
| V455H | V455H | V455H | V455H | V455R | V455H | |
| F456L | F456L | F456L | F456L | F456L | ||
| T478K | T478K | T478K | T478K | T478K | T478I | |
| Q493E | Q493E | Q493E | Q493E | Q493E | ||
| - | - | - | - | K1086R | - | |
| - | V1104L | V1104L | V1104L | V1104L | - | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Izhari, M.A.; Alghamdi, F.; Alodeani, E.A.; Salem, A.A.; Almontasheri, A.H.A.; Dardari, D.M.M.; Hadadi, M.A.A.; Gosady, A.R.A.; Alghamdi, W.A.; Alzahrani, B.A.; et al. Evolutionary Insight into Fatal Human Coronaviruses (hCoVs) with a Focus on Circulating SARS-CoV-2 Variants Under Monitoring (VUMs). Biomedicines 2025, 13, 2450. https://doi.org/10.3390/biomedicines13102450
Izhari MA, Alghamdi F, Alodeani EA, Salem AA, Almontasheri AHA, Dardari DMM, Hadadi MAA, Gosady ARA, Alghamdi WA, Alzahrani BA, et al. Evolutionary Insight into Fatal Human Coronaviruses (hCoVs) with a Focus on Circulating SARS-CoV-2 Variants Under Monitoring (VUMs). Biomedicines. 2025; 13(10):2450. https://doi.org/10.3390/biomedicines13102450
Chicago/Turabian StyleIzhari, Mohammad Asrar, Fahad Alghamdi, Essa Ajmi Alodeani, Ahmad A. Salem, Ahamad H. A. Almontasheri, Daifallah M. M. Dardari, Mansour A. A. Hadadi, Ahmed R. A. Gosady, Wael A. Alghamdi, Bakheet A. Alzahrani, and et al. 2025. "Evolutionary Insight into Fatal Human Coronaviruses (hCoVs) with a Focus on Circulating SARS-CoV-2 Variants Under Monitoring (VUMs)" Biomedicines 13, no. 10: 2450. https://doi.org/10.3390/biomedicines13102450
APA StyleIzhari, M. A., Alghamdi, F., Alodeani, E. A., Salem, A. A., Almontasheri, A. H. A., Dardari, D. M. M., Hadadi, M. A. A., Gosady, A. R. A., Alghamdi, W. A., Alzahrani, B. A., & Alzahrani, B. M. A. (2025). Evolutionary Insight into Fatal Human Coronaviruses (hCoVs) with a Focus on Circulating SARS-CoV-2 Variants Under Monitoring (VUMs). Biomedicines, 13(10), 2450. https://doi.org/10.3390/biomedicines13102450








